

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
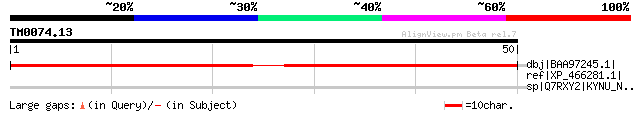
Query= TM0074.13
(50 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA97245.1| unnamed protein product [Arabidopsis thaliana] g... 62 5e-09
ref|XP_466281.1| putative pRGR1 [Oryza sativa (japonica cultivar... 35 0.71
sp|Q7RXY2|KYNU_NEUCR Putative kynureninase (L-kynurenine hydrola... 31 7.8
>dbj|BAA97245.1| unnamed protein product [Arabidopsis thaliana]
gi|26450165|dbj|BAC42201.1| unknown protein
[Arabidopsis thaliana] gi|28827714|gb|AAO50701.1|
unknown protein [Arabidopsis thaliana]
gi|15237787|ref|NP_197746.1| expressed protein
[Arabidopsis thaliana]
Length = 175
Score = 61.6 bits (148), Expect = 5e-09
Identities = 28/50 (56%), Positives = 41/50 (82%), Gaps = 3/50 (6%)
Query: 1 MEKMSKAFEKVKIMVGMEVEDEEQQAAALDNDNSFAFMDEFNRNCTLSTK 50
M+KM++A EK+K++VGMEVEDE+Q A D ++S +FM++ NRNC L+TK
Sbjct: 1 MDKMNQAIEKMKMLVGMEVEDEQQ---AADEESSLSFMEDLNRNCALTTK 47
>ref|XP_466281.1| putative pRGR1 [Oryza sativa (japonica cultivar-group)]
gi|46390354|dbj|BAD15819.1| putative pRGR1 [Oryza
sativa (japonica cultivar-group)]
Length = 171
Score = 34.7 bits (78), Expect = 0.71
Identities = 17/49 (34%), Positives = 27/49 (54%), Gaps = 7/49 (14%)
Query: 1 MEKMSKAFEKVKIMVGMEVEDEEQQAAALDNDNSFAFMDEFNRNCTLST 49
M+ M A E+ +++VGMEV++E +F D+ RNC L+T
Sbjct: 1 MDTMRGALERARMLVGMEVDEESA-------PEEQSFFDDVTRNCALTT 42
>sp|Q7RXY2|KYNU_NEUCR Putative kynureninase (L-kynurenine hydrolase)
gi|32403872|ref|XP_322549.1| hypothetical protein
[Neurospora crassa] gi|28917859|gb|EAA27546.1|
hypothetical protein [Neurospora crassa]
Length = 468
Score = 31.2 bits (69), Expect = 7.8
Identities = 15/25 (60%), Positives = 20/25 (80%), Gaps = 1/25 (4%)
Query: 19 VEDEEQQAAALDNDNSFAFM-DEFN 42
V+D +QA ALDN++S AF+ DEFN
Sbjct: 6 VQDARKQAEALDNEDSIAFVRDEFN 30
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.125 0.327
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,104,536
Number of Sequences: 2540612
Number of extensions: 1944184
Number of successful extensions: 3644
Number of sequences better than 10.0: 3
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 3641
Number of HSP's gapped (non-prelim): 3
length of query: 50
length of database: 863,360,394
effective HSP length: 26
effective length of query: 24
effective length of database: 797,304,482
effective search space: 19135307568
effective search space used: 19135307568
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0074.13


