

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0072a.1
(224 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
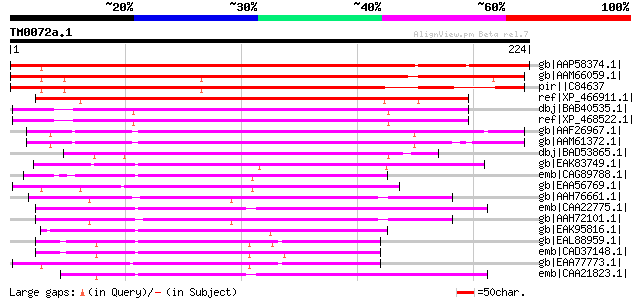
gb|AAP58374.1| RPA 32kDa [Pisum sativum] 347 1e-94
gb|AAM66059.1| putative replication protein A1 [Arabidopsis thal... 199 4e-50
pir||C84637 probable replication protein A1 [imported] - Arabido... 189 7e-47
ref|XP_466911.1| putative replication protein A2 [Oryza sativa (... 167 2e-40
dbj|BAB40535.1| replication protein A 30kDa [Oryza sativa (japon... 149 4e-35
ref|XP_468522.1| replication protein A2 [Oryza sativa (japonica ... 149 6e-35
gb|AAF26967.1| putative replication factor A [Arabidopsis thaliana] 143 4e-33
gb|AAM61372.1| putative replication factor A [Arabidopsis thalia... 142 7e-33
dbj|BAD53865.1| putative replication protein A2 [Oryza sativa (j... 116 5e-25
gb|EAK83749.1| hypothetical protein UM02579.1 [Ustilago maydis 5... 109 5e-23
emb|CAG89788.1| unnamed protein product [Debaryomyces hansenii C... 94 2e-18
gb|EAA56769.1| hypothetical protein MG07124.4 [Magnaporthe grise... 92 8e-18
gb|AAH76661.1| Replication protein A2, 32kDa [Xenopus tropicalis... 92 1e-17
emb|CAA22775.1| ssb2 [Schizosaccharomyces pombe] gi|1502415|gb|A... 92 1e-17
gb|AAH72101.1| MGC79017 protein [Xenopus laevis] 91 2e-17
gb|EAK95816.1| hypothetical protein CaO19.2267 [Candida albicans... 91 3e-17
gb|EAL88959.1| possible replication factor-a protein [Aspergillu... 90 5e-17
emb|CAD37148.1| possible replication factor-a protein [Aspergill... 89 7e-17
gb|EAA77773.1| hypothetical protein FG09724.1 [Gibberella zeae P... 89 9e-17
emb|CAA21823.1| ssb2 [Schizosaccharomyces pombe] 88 2e-16
>gb|AAP58374.1| RPA 32kDa [Pisum sativum]
Length = 273
Score = 347 bits (891), Expect = 1e-94
Identities = 171/226 (75%), Positives = 199/226 (87%), Gaps = 4/226 (1%)
Query: 1 MFSVTQFDSSSG--GGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSF 58
MFS +QFDSS+ GGGFTS+QLN+SSPAP K R+SQGLVPVTVKQI+EASQSGDEKS+F
Sbjct: 1 MFSSSQFDSSNAFSGGGFTSSQLNESSPAPTKSRDSQGLVPVTVKQISEASQSGDEKSNF 60
Query: 59 VINGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMY 118
INGV+LTNVTLVGMV+EK ERNTDVNFVLDDGTGRIKCRRW+N+ FD++E+EE+ N MY
Sbjct: 61 AINGVDLTNVTLVGMVYEKTERNTDVNFVLDDGTGRIKCRRWVNETFDSKEMEEVSNDMY 120
Query: 119 VRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASD 178
VRV G+LKSFQGV+Q+ AFSVRPV NFDEIPFHFIDCIH HL +K+K+EGT ST PP ++
Sbjct: 121 VRVYGNLKSFQGVKQLGAFSVRPVTNFDEIPFHFIDCIHSHLLAKVKLEGTPSTYPP-TN 179
Query: 179 SSLKTPVRNTSNGSQAPSSTPAYAQYGVDGLKDCDKLVIDYLQQHS 224
SS+ TPV++ NGSQAPS+ P Y QY DGL+DCDKLVIDYLQQHS
Sbjct: 180 SSINTPVKSALNGSQAPSN-PGYTQYSTDGLEDCDKLVIDYLQQHS 224
>gb|AAM66059.1| putative replication protein A1 [Arabidopsis thaliana]
gi|26450159|dbj|BAC42198.1| putative replication protein
A1 [Arabidopsis thaliana] gi|28973057|gb|AAO63853.1|
putative replication protein A1 [Arabidopsis thaliana]
gi|20197764|gb|AAD18120.2| putative replication protein
A1 [Arabidopsis thaliana] gi|18400560|ref|NP_565571.1|
replication protein, putative [Arabidopsis thaliana]
Length = 279
Score = 199 bits (507), Expect = 4e-50
Identities = 106/230 (46%), Positives = 151/230 (65%), Gaps = 12/230 (5%)
Query: 1 MFSVTQFDSSSG--GGGFTSTQLN---DSSPAPQKGRESQGLVPVTVKQINEASQSGDEK 55
MFS +QF+ +SG GGGF S+Q + +SS + K R+ QGLVPVTVKQI E QS EK
Sbjct: 1 MFSSSQFEPNSGFSGGGFMSSQPSQAYESSSSTAKNRDFQGLVPVTVKQITECFQSSGEK 60
Query: 56 SSFVINGVELTNVTLVGMVFEKAERN-TDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIM 114
S VING+ LTNV+LVG+V +K E T+V F LDDGTGRI C+RW+++ FD +E+E +
Sbjct: 61 SGLVINGISLTNVSLVGLVCDKDESKVTEVRFTLDDGTGRIDCKRWVSETFDAREMESVR 120
Query: 115 NGMYVRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDP 174
+G YVR++GHLK+FQG Q+ FSVRP+++F+E+ FH+I+CIH + ++ +
Sbjct: 121 DGTYVRLSGHLKTFQGKTQLLVFSVRPIMDFNEVTFHYIECIHFYSQNSESQRQQVGD-- 178
Query: 175 PASDSSLKTPVRNTSNGSQAPSSTPAYAQYGVD--GLKDCDKLVIDYLQQ 222
S+ T + SN +QA P + D G K+ D +++DYL+Q
Sbjct: 179 --VTQSVNTTFQGGSNTNQATLLNPVVSSQNNDGNGRKNLDDMILDYLKQ 226
>pir||C84637 probable replication protein A1 [imported] - Arabidopsis thaliana
Length = 250
Score = 189 bits (479), Expect = 7e-47
Identities = 102/228 (44%), Positives = 145/228 (62%), Gaps = 37/228 (16%)
Query: 1 MFSVTQFDSSSG--GGGFTSTQLN---DSSPAPQKGRESQGLVPVTVKQINEASQSGDEK 55
MFS +QF+ +SG GGGF S+Q + +SS + K R+ QGLVPVTVKQI E QS EK
Sbjct: 1 MFSSSQFEPNSGFSGGGFMSSQPSQAYESSSSTAKNRDFQGLVPVTVKQITECFQSSGEK 60
Query: 56 SSFVINGVELTNVTLVGMVFEKAERN-TDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIM 114
S VING+ LTNV+LVG+V +K E T+V F LDDGTGRI C+RW+++ FD +E+E +
Sbjct: 61 SGLVINGISLTNVSLVGLVCDKDESKVTEVRFTLDDGTGRIDCKRWVSETFDAREMESVR 120
Query: 115 NGMYVRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDP 174
+G YVR++GHLK+FQG Q+ FSVRP+++F+E+ FH+I+CIH + ++
Sbjct: 121 DGTYVRLSGHLKTFQGKTQLLVFSVRPIMDFNEVTFHYIECIHFYSQN------------ 168
Query: 175 PASDSSLKTPVRNTSNGSQAPSSTPAYAQYGVDGLKDCDKLVIDYLQQ 222
S+S + + N NG K+ D +++DYL+Q
Sbjct: 169 --SESQVVSSQNNDGNGR-----------------KNLDDMILDYLKQ 197
>ref|XP_466911.1| putative replication protein A2 [Oryza sativa (japonica
cultivar-group)] gi|49388178|dbj|BAD25304.1| putative
replication protein A2 [Oryza sativa (japonica
cultivar-group)]
Length = 298
Score = 167 bits (424), Expect = 2e-40
Identities = 88/203 (43%), Positives = 128/203 (62%), Gaps = 16/203 (7%)
Query: 12 GGGGFTSTQLNDSSPAPQ-------KGRESQGLVPVTVKQINEASQSGDEKSSFVINGVE 64
GGGGF +Q +++ K R +Q L+P+TVKQI +ASQ+ D+KS+F +NG+E
Sbjct: 26 GGGGFMPSQATNAAEGTSGGGGGFPKSRNAQALLPLTVKQIMDASQTNDDKSNFAVNGME 85
Query: 65 LTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGH 124
++ V LVG + K +R TDV+F LDDGTGR+ RW ND+ DT+E+ +I NG YV VNG
Sbjct: 86 VSTVRLVGRMLNKLDRVTDVSFTLDDGTGRVPVNRWENDSTDTKEMADIQNGDYVIVNGG 145
Query: 125 LKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHL---RSKLKMEGTISTDPP------ 175
LK FQG RQV A+SVR + NF+++ HF+ C+H HL R K ++ +T P
Sbjct: 146 LKGFQGKRQVVAYSVRRITNFNDVTHHFLHCVHVHLELTRPKSQVNANTATGTPNQTMPR 205
Query: 176 ASDSSLKTPVRNTSNGSQAPSST 198
S + ++P+ N ++ AP +T
Sbjct: 206 DSMAYNQSPLTNQASTFSAPQNT 228
>dbj|BAB40535.1| replication protein A 30kDa [Oryza sativa (japonica
cultivar-group)]
Length = 279
Score = 149 bits (377), Expect = 4e-35
Identities = 84/216 (38%), Positives = 118/216 (53%), Gaps = 27/216 (12%)
Query: 2 FSVTQFDSSSGGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSG---DEKSSF 58
FS +QF SS ++T P K R + +P+TVKQI+EA QSG ++ + F
Sbjct: 10 FSPSQFTSSQNAAADSTT--------PSKSRGASSTMPLTVKQISEAQQSGITGEKGAPF 61
Query: 59 VINGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMY 118
V++GVE NV LVG+V K ERNTDV+F++DDGTGR+ RW+ND D+ E + NGMY
Sbjct: 62 VVDGVETANVRLVGLVSGKTERNTDVSFMIDDGTGRLDFIRWVNDGADSAETAAVQNGMY 121
Query: 119 VRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRS---------------- 162
V V G LK Q ++ AF++RPV +++E+ HFI C+ HL +
Sbjct: 122 VSVIGSLKGLQERKRATAFAIRPVTDYNEVTLHFIQCVRMHLENTKSQIGSPAKTYSAMG 181
Query: 163 KLKMEGTISTDPPASDSSLKTPVRNTSNGSQAPSST 198
G P S S PV + +NGS+ +T
Sbjct: 182 SSSSNGFSEMTTPTSVKSNPAPVLSVTNGSKTDLNT 217
>ref|XP_468522.1| replication protein A2 [Oryza sativa (japonica cultivar-group)]
gi|9801268|emb|CAC03572.1| replication protein A2 [Oryza
sativa] gi|48716324|dbj|BAD22936.1| replication protein
A2 [Oryza sativa (japonica cultivar-group)]
Length = 279
Score = 149 bits (376), Expect = 6e-35
Identities = 84/216 (38%), Positives = 117/216 (53%), Gaps = 27/216 (12%)
Query: 2 FSVTQFDSSSGGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSG---DEKSSF 58
FS +QF SS ++T P K R + +P+TVKQI+EA QSG ++ + F
Sbjct: 10 FSPSQFTSSQNAAADSTT--------PSKSRGASSTMPLTVKQISEAQQSGITGEKGAPF 61
Query: 59 VINGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMY 118
V++GVE NV LVG+V K ERNTDV+F +DDGTGR+ RW+ND D+ E + NGMY
Sbjct: 62 VVDGVETANVRLVGLVSGKTERNTDVSFTIDDGTGRLDFIRWVNDGADSAETAAVQNGMY 121
Query: 119 VRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRS---------------- 162
V V G LK Q ++ AF++RPV +++E+ HFI C+ HL +
Sbjct: 122 VSVIGSLKGLQERKRATAFAIRPVTDYNEVTLHFIQCVRMHLENTKSQIGSPAKTYSAMG 181
Query: 163 KLKMEGTISTDPPASDSSLKTPVRNTSNGSQAPSST 198
G P S S PV + +NGS+ +T
Sbjct: 182 SSSSNGFSEMTTPTSVKSNPAPVLSVTNGSKTDLNT 217
>gb|AAF26967.1| putative replication factor A [Arabidopsis thaliana]
Length = 282
Score = 143 bits (360), Expect = 4e-33
Identities = 83/224 (37%), Positives = 133/224 (59%), Gaps = 13/224 (5%)
Query: 8 DSSSGGGGF----TSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGV 63
+++ GGGF +TQ ++SS + K R+ + L+P+T+KQ++ AS +G+ S+F I+GV
Sbjct: 9 NAAFAGGGFMPSQATTQAHESSSS-LKNRDVRTLLPLTLKQLSSASTTGE--SNFSIDGV 65
Query: 64 ELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNG 123
++ V +VG + R T V+FV+DDGTG + C RW + +T+E+E + GMYVR++G
Sbjct: 66 DIKTVVIVGRISRMENRITQVDFVVDDGTGWVDCVRWCHARQETEEMEAVKLGMYVRLHG 125
Query: 124 HLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTD-----PPASD 178
HLK FQG R V FSVRPV +F+EI HF +C++ H+ + G+I+ D P
Sbjct: 126 HLKIFQGKRSVNVFSVRPVTDFNEIVHHFTECMYVHMYNTKLRGGSITQDTATPRPQMPY 185
Query: 179 SSLKTPVRNTSNGSQAPSSTPAYAQYGVDGLKDCDKLVIDYLQQ 222
S++ TP + G + Q+ D + + V++YL Q
Sbjct: 186 STMPTPAKPYQTGPSNQNLFQFPNQFN-DSMHGVKQTVLNYLNQ 228
>gb|AAM61372.1| putative replication factor A [Arabidopsis thaliana]
gi|26450657|dbj|BAC42439.1| putative replication factor
A [Arabidopsis thaliana] gi|18396383|ref|NP_566188.1|
replication protein-related [Arabidopsis thaliana]
Length = 278
Score = 142 bits (358), Expect = 7e-33
Identities = 84/224 (37%), Positives = 134/224 (59%), Gaps = 17/224 (7%)
Query: 8 DSSSGGGGF----TSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGV 63
+++ GGGF +TQ ++SS + K R+ + L+P+T+KQ++ AS +G+ S+F I+GV
Sbjct: 9 NAAFAGGGFMPSQATTQAHESSSS-LKNRDVRTLLPLTLKQLSSASTTGE--SNFSIDGV 65
Query: 64 ELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNG 123
++ V +VG + R T V+FV+DDGTG + C RW + +T+E+E + GMYVR++G
Sbjct: 66 DIKTVVIVGRISRMENRITQVDFVVDDGTGWVDCVRWCHARQETEEMEAVKLGMYVRLHG 125
Query: 124 HLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTD-----PPASD 178
HLK FQG R V FSVRPV +F+EI HF +C++ H+ + G+I+ D P
Sbjct: 126 HLKIFQGKRSVNVFSVRPVTDFNEIVHHFTECMYVHMYNTKLRGGSITQDTATPRPQMPY 185
Query: 179 SSLKTPVRNTSNGSQAPSSTPAYAQYGVDGLKDCDKLVIDYLQQ 222
S++ TP + G PS+ + D + + V++YL Q
Sbjct: 186 STMPTPAKPYQTG---PSN--QFPNQFNDSMHGVKQTVLNYLNQ 224
>dbj|BAD53865.1| putative replication protein A2 [Oryza sativa (japonica
cultivar-group)]
Length = 438
Score = 116 bits (290), Expect = 5e-25
Identities = 68/170 (40%), Positives = 99/170 (58%), Gaps = 11/170 (6%)
Query: 24 SSPAPQKGRESQ--GLVPVTVKQINEA-----SQSGDEKSSFVINGVELTNVTLVGMVFE 76
++P+P K R+ + G VP TV I+ + + G F I+GVE TNV ++G V
Sbjct: 35 AAPSPSKPRDPRFSGCVPATVLHISRSFAAALAADGGGDPVFSIDGVETTNVRVLGRVVS 94
Query: 77 KAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLKSFQGVRQVAA 136
R+TDV F LDD TG+I RWI D DT++ I G+YV+V +L FQ +Q A
Sbjct: 95 VVSRDTDVCFTLDDSTGKIPLVRWITDQSDTRDTSYIQEGVYVKVQVNLMGFQAKKQGLA 154
Query: 137 FSVRPVVNFDEIPFHFIDCIHHHLRS-KLKMEGTISTDPPASDSSLKTPV 185
S+RP+ NF+E+ HFI+C+H HL S + KM+ + PP+ ++ T V
Sbjct: 155 RSIRPINNFNEVVLHFIECMHVHLESVQSKMQRQL---PPSVQTNEYTHV 201
>gb|EAK83749.1| hypothetical protein UM02579.1 [Ustilago maydis 521]
gi|49071810|ref|XP_400194.1| hypothetical protein
UM02579.1 [Ustilago maydis 521]
Length = 285
Score = 109 bits (273), Expect = 5e-23
Identities = 64/201 (31%), Positives = 113/201 (55%), Gaps = 8/201 (3%)
Query: 11 SGGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTNVTL 70
+ GGGF + + SP+ +K + L PVT++QI A Q + + F+++G EL +T
Sbjct: 23 ASGGGFLANGSQNDSPSGKKAGNNT-LRPVTIRQILNAEQPHPD-AEFILDGAELGQLTF 80
Query: 71 VGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFD-TQEVEEIMNGMYVRVNGHLKSFQ 129
V +V + T+V + ++DGTG+I+ R+W++ + D + + EI N +YVRV G LKSFQ
Sbjct: 81 VAVVRNISRNATNVAYSVEDGTGQIEVRQWLDSSSDDSSKASEIRNNVYVRVLGTLKSFQ 140
Query: 130 GVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLR-----SKLKMEGTISTDPPASDSSLKTP 184
R +++ +RPV++++E+ FH ++ +H HL+ + ++G ST L++
Sbjct: 141 NRRSISSGHMRPVIDYNEVMFHRLEAVHAHLQATRGPTASALKGATSTALFNQAGGLESN 200
Query: 185 VRNTSNGSQAPSSTPAYAQYG 205
N +GS+ Y G
Sbjct: 201 DMNAYSGSKKQDVNDLYKNLG 221
>emb|CAG89788.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50425571|ref|XP_461381.1| unnamed protein product
[Debaryomyces hansenii]
Length = 260
Score = 94.4 bits (233), Expect = 2e-18
Identities = 55/159 (34%), Positives = 89/159 (55%), Gaps = 8/159 (5%)
Query: 7 FDSSSGGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELT 66
F++ G GGF + N+ S QK + L PVT+KQI EA+Q + F ++ V L
Sbjct: 16 FNTDIGNGGFQNE--NNGS---QKQQLRSSLTPVTIKQITEATQPIPD-GEFQVHNVNLN 69
Query: 67 NVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWIND--AFDTQEVEEIMNGMYVRVNGH 124
V+ VG+V + + + ++DGTG I+ R+WI+D A D E E+ Y+ V G
Sbjct: 70 LVSFVGVVRNVQDITSAILITIEDGTGSIEVRKWIDDSNAMDENEQEKYEMNKYIYVTGA 129
Query: 125 LKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSK 163
LK F G + + ++RP+ + +EI +H + I HHL+++
Sbjct: 130 LKEFNGKKNLQHATMRPITDHNEILYHHLSAISHHLKAQ 168
>gb|EAA56769.1| hypothetical protein MG07124.4 [Magnaporthe grisea 70-15]
gi|39971617|ref|XP_367199.1| hypothetical protein
MG07124.4 [Magnaporthe grisea 70-15]
Length = 275
Score = 92.4 bits (228), Expect = 8e-18
Identities = 59/172 (34%), Positives = 91/172 (52%), Gaps = 6/172 (3%)
Query: 2 FSVTQFDSSSG--GGGFTSTQLNDSSPAPQ-KGRESQGLVPVTVKQINEASQSGDEKSSF 58
FS T +++ GGGF S P K + + L PVTVKQ+ + Q+ + F
Sbjct: 9 FSRTGYNAQGAEDGGGFMGGSQQGSQGGPGGKSYQDECLKPVTVKQLLDV-QAPYPDADF 67
Query: 59 VINGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWIND--AFDTQEVEEIMNG 116
+++G +T +TLVG V + T++ + +DDGTG I +RWI+ A D +
Sbjct: 68 LLDGRAITQITLVGQVRSINPQPTNITYRIDDGTGTIDVKRWIDPEKAEDADAASQHQPD 127
Query: 117 MYVRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEG 168
YVRV G LK+F R V A VRPV +F+E+ +H ++ + HL + + G
Sbjct: 128 SYVRVWGKLKAFNNRRHVGALFVRPVEDFNEVNYHMLEVAYVHLDAVRQSSG 179
>gb|AAH76661.1| Replication protein A2, 32kDa [Xenopus tropicalis]
gi|55742354|ref|NP_001006795.1| replication protein A2,
32kDa [Xenopus tropicalis]
Length = 275
Score = 92.0 bits (227), Expect = 1e-17
Identities = 61/189 (32%), Positives = 97/189 (51%), Gaps = 13/189 (6%)
Query: 9 SSSGGGGFTSTQLNDSSPAPQKGRE-----SQGLVPVTVKQINEASQSGDEKSSFVINGV 63
S GGGG+ + SPAP +G + SQ +VP TV Q+ A+Q+ + F I
Sbjct: 15 SGMGGGGYMQSPGGFGSPAPTQGEKKSRSRSQQIVPCTVSQLLSATQNDE---MFRIGEA 71
Query: 64 ELTNVTLVGMVFEKAERNTDVNFVLDDGTGR-IKCRRWINDAFDTQEVEEIMNGMYVRVN 122
EL+ VT+VG+V + T++ + +DD T + R+W++ + E + G YV+V
Sbjct: 72 ELSQVTIVGIVRHAEKAPTNILYKVDDMTAAPMDVRQWVDTDEASCENMVVPPGSYVKVA 131
Query: 123 GHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASDSSLK 182
GHL+SFQ + V AF + PV + +E H ++ +H H + M + S +L
Sbjct: 132 GHLRSFQNKKSVVAFKIAPVDDMNEFVSHMLEVVHAH----MTMNSQGAPSGGGSAVALN 187
Query: 183 TPVRNTSNG 191
TP R +G
Sbjct: 188 TPGRLGDSG 196
>emb|CAA22775.1| ssb2 [Schizosaccharomyces pombe] gi|1502415|gb|AAC49438.1|
single-stranded DNA binding protein p30 subunit
gi|7493273|pir||T41124 single-stranded DNA binding
protein 30K chain [validated] - fission yeast
(Schizosaccharomyces pombe)
gi|2498849|sp|Q92373|RFA2_SCHPO Replication factor-A
protein 2 (Single-stranded DNA-binding protein P30
subunit)
Length = 279
Score = 91.7 bits (226), Expect = 1e-17
Identities = 58/195 (29%), Positives = 93/195 (46%), Gaps = 5/195 (2%)
Query: 12 GGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTNVTLV 71
GG GF + ++ L PVT+KQI ASQ + + F I+GVE+ VT V
Sbjct: 25 GGAGFNEYDQSSQPSVDRQQGAGNKLRPVTIKQILNASQVHAD-AEFKIDGVEVGQVTFV 83
Query: 72 GMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLKSFQGV 131
G++ + T+ + ++DGTG I+ R W + + E+ YVRV G++K F G
Sbjct: 84 GVLRNIHAQTTNTTYQIEDGTGMIEVRHWEH----IDALSELATDTYVRVYGNIKIFSGK 139
Query: 132 RQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASDSSLKTPVRNTSNG 191
+A+ +R + + +E+ FHF++ I HL K + P +S N S+
Sbjct: 140 IYIASQYIRTIKDHNEVHFHFLEAIAVHLHFTQKANAVNGANAPGYGTSNALGYNNISSN 199
Query: 192 SQAPSSTPAYAQYGV 206
A S A+Y +
Sbjct: 200 GAANSLEQKLAEYSL 214
>gb|AAH72101.1| MGC79017 protein [Xenopus laevis]
Length = 276
Score = 91.3 bits (225), Expect = 2e-17
Identities = 60/186 (32%), Positives = 96/186 (51%), Gaps = 13/186 (6%)
Query: 12 GGGGFTSTQLNDSSPAPQKGRE-----SQGLVPVTVKQINEASQSGDEKSSFVINGVELT 66
GGGG+ + SPAP +G + SQ +VP TV Q+ A+Q+ + F I EL+
Sbjct: 19 GGGGYMQSPGGFGSPAPTQGEKKSRSRSQQIVPCTVSQLLSATQNDEV---FRIGEAELS 75
Query: 67 NVTLVGMVFEKAERNTDVNFVLDDGTGR-IKCRRWINDAFDTQEVEEIMNGMYVRVNGHL 125
VT+VG+V + T++ + +DD T + R+W++ + E + G YV+V GHL
Sbjct: 76 QVTIVGIVRHAEKAPTNILYKVDDMTAAPMDVRQWVDTDEASCENMVVPPGSYVKVAGHL 135
Query: 126 KSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASDSSLKTPV 185
+SFQ + V AF + PV + +E H ++ +H H + M + S +L TP
Sbjct: 136 RSFQNKKSVVAFKIAPVEDMNEFVSHMLEVVHAH----MAMNSQGAPSGGGSTVALSTPG 191
Query: 186 RNTSNG 191
R +G
Sbjct: 192 RVGDSG 197
>gb|EAK95816.1| hypothetical protein CaO19.2267 [Candida albicans SC5314]
gi|46436389|gb|EAK95752.1| hypothetical protein
CaO19.9807 [Candida albicans SC5314]
Length = 272
Score = 90.5 bits (223), Expect = 3e-17
Identities = 51/154 (33%), Positives = 82/154 (53%), Gaps = 6/154 (3%)
Query: 14 GGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTNVTLVGM 73
GGF +T+ SS + Q L PVT+KQIN+A+Q + F +N VEL ++ VG+
Sbjct: 20 GGF-NTEHAGSSQRQTTTQVRQSLTPVTIKQINDATQPVPD-GEFKVNNVELNMISFVGV 77
Query: 74 VFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVE----EIMNGMYVRVNGHLKSFQ 129
V N + ++DGTG I R+W+++ + E + M G YV V G LK F
Sbjct: 78 VRNVENTNASIAVTIEDGTGSIDVRKWVDETISSAEEDFEKYNEMKGKYVYVGGSLKQFN 137
Query: 130 GVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSK 163
+ V S+ + + ++I +H + I HHL+++
Sbjct: 138 NRKTVQNASISLITDSNQIVYHHLSAIEHHLKAQ 171
>gb|EAL88959.1| possible replication factor-a protein [Aspergillus fumigatus Af293]
Length = 289
Score = 89.7 bits (221), Expect = 5e-17
Identities = 58/161 (36%), Positives = 89/161 (55%), Gaps = 16/161 (9%)
Query: 12 GGGGFTSTQLNDSSPAPQKGRESQG-LVPVTVKQINEASQSGDEKSSFVINGVELTNVTL 70
GGGGF ++N SPA KG + L P+TVKQ +ASQ E ++F I+G + +V
Sbjct: 27 GGGGFMPGEMN--SPAGGKGDNNNATLRPITVKQALDASQPYPE-AAFQIDGADAASVCF 83
Query: 71 VGMVFEKAERNTDVNFVLDDGTGRIKCRRWIN-DAFDTQEVEE----------IMNGMYV 119
+G V + ++T+V + +DDGTG I+ ++WI+ DT E ++ +NG +
Sbjct: 84 IGQVRNISSQSTNVTYKIDDGTGEIEVKQWIDTQTADTMETDDGKAGTGKNQVELNG-FA 142
Query: 120 RVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHL 160
+V G +KSF R V A VRP N +E+ H ++ HL
Sbjct: 143 KVFGKIKSFGNKRYVGAHCVRPTTNLNEVHCHLLEAAAIHL 183
>emb|CAD37148.1| possible replication factor-a protein [Aspergillus fumigatus]
Length = 274
Score = 89.4 bits (220), Expect = 7e-17
Identities = 57/152 (37%), Positives = 86/152 (56%), Gaps = 6/152 (3%)
Query: 12 GGGGFTSTQLNDSSPAPQKGRESQG-LVPVTVKQINEASQSGDEKSSFVINGVELTNVTL 70
GGGGF ++N SPA KG + L P+TVKQ +ASQ E ++F I+G + +V
Sbjct: 20 GGGGFMPGEMN--SPAGGKGDNNNATLRPITVKQALDASQPYPE-AAFQIDGADAASVCF 76
Query: 71 VGMVFEKAERNTDVNFVLDDGTGRIKCRRWIN-DAFDTQEVEEIMNGM-YVRVNGHLKSF 128
+G V + ++T+V + +DDGTG I+ ++WI+ DT E ++ G +V G +KSF
Sbjct: 77 IGQVRNISSQSTNVTYKIDDGTGEIEVKQWIDTQTADTMETDDGKAGTGKNQVFGKIKSF 136
Query: 129 QGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHL 160
R V A VRP N +E+ H ++ HL
Sbjct: 137 GNKRYVGAHCVRPTTNLNEVHCHLLEAAAIHL 168
>gb|EAA77773.1| hypothetical protein FG09724.1 [Gibberella zeae PH-1]
gi|46136417|ref|XP_389900.1| hypothetical protein
FG09724.1 [Gibberella zeae PH-1]
Length = 275
Score = 89.0 bits (219), Expect = 9e-17
Identities = 52/163 (31%), Positives = 88/163 (53%), Gaps = 6/163 (3%)
Query: 2 FSVTQFDSSSG---GGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSF 58
F+ T + + G GG F S K + + L PVT+KQI +A ++ + F
Sbjct: 7 FTKTSYGAQGGDDSGGFFAGGSQQGSQGGGGKAYQDESLRPVTIKQILDAEEAY-AGADF 65
Query: 59 VINGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWIN-DAFDTQEVEEIMNGM 117
I+G +T +T +G V + T++ +DDGTG+I+ ++WI+ D D E ++
Sbjct: 66 KIDGSPVTQITFIGQVRSVQPQPTNITLKIDDGTGQIEVKKWIDVDKADDSEAGFELDS- 124
Query: 118 YVRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHL 160
++R+ G LKSF R V A +RPV +F+E+ +H ++ + HL
Sbjct: 125 HIRIWGRLKSFNNKRHVGAHVIRPVSDFNEVNYHMLEATYVHL 167
>emb|CAA21823.1| ssb2 [Schizosaccharomyces pombe]
Length = 247
Score = 87.8 bits (216), Expect = 2e-16
Identities = 58/187 (31%), Positives = 91/187 (48%), Gaps = 8/187 (4%)
Query: 23 DSSPAPQKGRESQG---LVPVTVKQINEASQSGDEKSSFVINGVELTNVTLVGMVFEKAE 79
D S P R+ L PVT+KQI ASQ + + F I+GVE+ VT VG++
Sbjct: 1 DQSSQPSVDRQQGAGNKLRPVTIKQILNASQVHAD-AEFKIDGVEVGQVTFVGVLRNIHA 59
Query: 80 RNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLKSFQGVRQVAAFSV 139
+ T+ + ++DGTG I+ R W + + E+ YVRV G++K F G +A+ +
Sbjct: 60 QTTNTTYQIEDGTGMIEVRHWEH----IDALSELATDTYVRVYGNIKIFSGKIYIASQYI 115
Query: 140 RPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASDSSLKTPVRNTSNGSQAPSSTP 199
R + + +E+ FHF++ I HL K + P +S N S+ A S
Sbjct: 116 RTIKDHNEVHFHFLEAIAVHLHFTQKANAVNGANAPGYGTSNALGYNNISSNGAANSLEQ 175
Query: 200 AYAQYGV 206
A+Y +
Sbjct: 176 KLAEYSL 182
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 385,239,401
Number of Sequences: 2540612
Number of extensions: 15692993
Number of successful extensions: 48155
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 47995
Number of HSP's gapped (non-prelim): 115
length of query: 224
length of database: 863,360,394
effective HSP length: 123
effective length of query: 101
effective length of database: 550,865,118
effective search space: 55637376918
effective search space used: 55637376918
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0072a.1


