

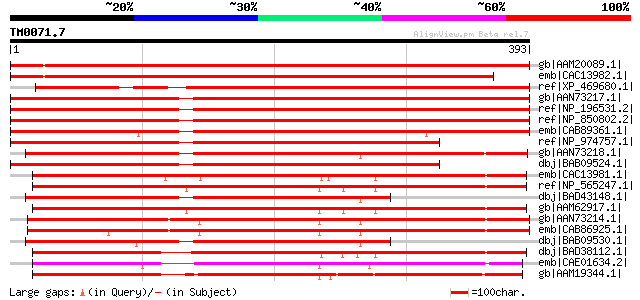
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0071.7
(393 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM20089.1| unknown protein [Arabidopsis thaliana] gi|1752932... 598 e-170
emb|CAC13982.1| putative magnesium transporter [Arabidopsis thal... 573 e-162
ref|XP_469680.1| putative CorA-like Mg2+ transporter protein [O... 563 e-159
gb|AAN73217.1| MRS2-7 [Arabidopsis thaliana] 546 e-154
ref|NP_196531.2| magnesium transporter CorA-like family protein ... 544 e-153
ref|NP_850802.2| magnesium transporter CorA-like family protein ... 544 e-153
emb|CAB89361.1| putative protein [Arabidopsis thaliana] gi|11282... 523 e-147
ref|NP_974757.1| magnesium transporter CorA-like family protein ... 452 e-126
gb|AAN73218.1| MRS2-8 [Arabidopsis thaliana] 446 e-124
dbj|BAB09524.1| unnamed protein product [Arabidopsis thaliana] 432 e-120
emb|CAC13981.1| putative magnesium transporter [Arabidopsis thal... 330 5e-89
ref|NP_565247.1| magnesium transporter CorA-like family protein ... 330 5e-89
dbj|BAD43148.1| unnamed protein product [Arabidopsis thaliana] 330 5e-89
gb|AAM62917.1| unknown [Arabidopsis thaliana] gi|25360983|gb|AAN... 328 2e-88
gb|AAN73214.1| MRS2-4 [Arabidopsis thaliana] gi|16209702|gb|AAL1... 328 2e-88
emb|CAB86925.1| putative protein [Arabidopsis thaliana] gi|11282... 322 2e-86
dbj|BAB09530.1| unnamed protein product [Arabidopsis thaliana] 320 4e-86
dbj|BAD38112.1| magnesium transporter CorA-like [Oryza sativa (j... 318 2e-85
emb|CAE01634.2| OSJNBa0029H02.17 [Oryza sativa (japonica cultiva... 298 2e-79
gb|AAM19344.1| hypothetical protein [Arabidopsis thaliana] gi|20... 287 3e-76
>gb|AAM20089.1| unknown protein [Arabidopsis thaliana] gi|17529326|gb|AAL38890.1|
unknown protein [Arabidopsis thaliana]
gi|21536628|gb|AAM60960.1| putative magnesium
transporter [Arabidopsis thaliana]
gi|10178059|dbj|BAB11423.1| unnamed protein product
[Arabidopsis thaliana] gi|25360813|gb|AAN73212.1| MRS2-2
[Arabidopsis thaliana] gi|30698045|ref|NP_851269.1|
magnesium transporter CorA-like family protein (MRS2-2)
[Arabidopsis thaliana]
Length = 394
Score = 598 bits (1542), Expect = e-170
Identities = 312/395 (78%), Positives = 342/395 (85%), Gaps = 3/395 (0%)
Query: 1 MARDGSVLPADPQAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRI 60
MA++G ++PADP AV VKKKT + SW L DA GQ LDVDKY IMHRVQIHARDLRI
Sbjct: 1 MAQNGYLVPADPSAVVTVKKKTPQA-SWALIDATGQSEPLDVDKYEIMHRVQIHARDLRI 59
Query: 61 LDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSS 120
LDP LSYPSTILGRE+AIVLNLEHIKAIIT+EEVLLRDP+DENV+PVVEEL+RRLP ++
Sbjct: 60 LDPNLSYPSTILGRERAIVLNLEHIKAIITSEEVLLRDPSDENVIPVVEELRRRLPVGNA 119
Query: 121 LHQQ-QGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H QGDG+E +Q+D + +EDESPFEFRALEVALEAICSFLAART ELE AAYPAL
Sbjct: 120 SHNGGQGDGKEIAGAQNDGDTGDEDESPFEFRALEVALEAICSFLAARTAELETAAYPAL 179
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKAGS-SS 238
DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK S SS
Sbjct: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKLSSASS 239
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKL 298
P+S G NW+ SPTIGSKISRASRASLATV DENDVEELEMLLEAYFMQID T N+L
Sbjct: 240 PISSIGEPNWYTTSPTIGSKISRASRASLATVHGDENDVEELEMLLEAYFMQIDSTLNRL 299
Query: 299 TTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDN 358
TTLREYIDDTEDYINIQLDNHRNQLIQLEL LSSGTVCLS YSLVA IFGMNIPY+WND
Sbjct: 300 TTLREYIDDTEDYINIQLDNHRNQLIQLELVLSSGTVCLSMYSLVAGIFGMNIPYTWNDG 359
Query: 359 HGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
HG+MFK+VV ++G ++F++I+ YAR KGLVGS
Sbjct: 360 HGYMFKYVVGLTGTLCVVVFVIIMSYARYKGLVGS 394
>emb|CAC13982.1| putative magnesium transporter [Arabidopsis thaliana]
gi|30698047|ref|NP_201261.2| magnesium transporter
CorA-like family protein (MRS2-2) [Arabidopsis thaliana]
Length = 378
Score = 573 bits (1476), Expect = e-162
Identities = 299/368 (81%), Positives = 322/368 (87%), Gaps = 3/368 (0%)
Query: 1 MARDGSVLPADPQAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRI 60
MA++G ++PADP AV VKKKT + SW L DA GQ LDVDKY IMHRVQIHARDLRI
Sbjct: 1 MAQNGYLVPADPSAVVTVKKKTPQA-SWALIDATGQSEPLDVDKYEIMHRVQIHARDLRI 59
Query: 61 LDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSS 120
LDP LSYPSTILGRE+AIVLNLEHIKAIIT+EEVLLRDP+DENV+PVVEEL+RRLP ++
Sbjct: 60 LDPNLSYPSTILGRERAIVLNLEHIKAIITSEEVLLRDPSDENVIPVVEELRRRLPVGNA 119
Query: 121 LHQQ-QGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H QGDG+E +Q+D + +EDESPFEFRALEVALEAICSFLAART ELE AAYPAL
Sbjct: 120 SHNGGQGDGKEIAGAQNDGDTGDEDESPFEFRALEVALEAICSFLAARTAELETAAYPAL 179
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKAGS-SS 238
DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK S SS
Sbjct: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKLSSASS 239
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKL 298
P+S G NW+ SPTIGSKISRASRASLATV DENDVEELEMLLEAYFMQID T N+L
Sbjct: 240 PISSIGEPNWYTTSPTIGSKISRASRASLATVHGDENDVEELEMLLEAYFMQIDSTLNRL 299
Query: 299 TTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDN 358
TTLREYIDDTEDYINIQLDNHRNQLIQLEL LSSGTVCLS YSLVA IFGMNIPY+WND
Sbjct: 300 TTLREYIDDTEDYINIQLDNHRNQLIQLELVLSSGTVCLSMYSLVAGIFGMNIPYTWNDG 359
Query: 359 HGFMFKWV 366
HG+MFK+V
Sbjct: 360 HGYMFKYV 367
>ref|XP_469680.1| putative CorA-like Mg2+ transporter protein [Oryza sativa
(japonica cultivar-group)] gi|40539050|gb|AAR87307.1|
putative CorA-like Mg2+ transporter protein [Oryza
sativa (japonica cultivar-group)]
Length = 374
Score = 563 bits (1451), Expect = e-159
Identities = 296/375 (78%), Positives = 317/375 (83%), Gaps = 24/375 (6%)
Query: 20 KKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKAIV 79
KK +SRSWILFDA G+ +LD DKYAIMHRV I+ARDLRILDPLLSYPSTILGRE+AIV
Sbjct: 23 KKRGASRSWILFDAAGEERVLDADKYAIMHRVDINARDLRILDPLLSYPSTILGRERAIV 82
Query: 80 LNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTE 139
LNLE VLLRDP D+NV+PVVEEL+RRL S+ +QHD E
Sbjct: 83 LNLE----------VLLRDPLDDNVIPVVEELRRRLAPSSA-------------TQHDVE 119
Query: 140 AAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRKLKS 199
AEEDESPFEFRALEV LEAICSFL ARTTELE AAYPALDELTSKISSRNLDRVRKLKS
Sbjct: 120 GAEEDESPFEFRALEVTLEAICSFLGARTTELESAAYPALDELTSKISSRNLDRVRKLKS 179
Query: 200 AMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSGAANWFGGSPTIGSK 258
MTRL ARVQKVRDELEQLLDDDDDMADLYLSRK AG++SPVSGSG NWF SPTIGSK
Sbjct: 180 GMTRLNARVQKVRDELEQLLDDDDDMADLYLSRKLAGAASPVSGSGGPNWFPASPTIGSK 239
Query: 259 ISRASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYINIQLDN 318
ISRASRAS T+ +ENDVEELEMLLEAYFMQIDGT NKLTTLREYIDDTEDYINIQLDN
Sbjct: 240 ISRASRASAPTIHGNENDVEELEMLLEAYFMQIDGTLNKLTTLREYIDDTEDYINIQLDN 299
Query: 319 HRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWVVIVSGVFSAIMF 378
HRNQLIQLELFLSSGTVCLS YSLVA IFGMNIPY+WNDNHG++FKWVV+VSG+F A MF
Sbjct: 300 HRNQLIQLELFLSSGTVCLSLYSLVAGIFGMNIPYTWNDNHGYVFKWVVLVSGLFCAFMF 359
Query: 379 LLIIVYARKKGLVGS 393
+ I+ YAR KGLVGS
Sbjct: 360 VSIVAYARHKGLVGS 374
>gb|AAN73217.1| MRS2-7 [Arabidopsis thaliana]
Length = 386
Score = 546 bits (1407), Expect = e-154
Identities = 289/396 (72%), Positives = 325/396 (81%), Gaps = 13/396 (3%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 1 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 60
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEVL+RD +DENV+PV+EE QRRLP +
Sbjct: 61 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVLIRDSSDENVIPVLEEFQRRLPVGN 120
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE +AYPAL
Sbjct: 121 EEHGAHGDG----------DVGEEDESPFEFRALEVALEAICSFLAARTTELEKSAYPAL 170
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSS 238
DELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+RK G+SS
Sbjct: 171 DELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTRKLVGASS 230
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFD-ENDVEELEMLLEAYFMQIDGTFNK 297
VS S W+ SPTIGS ISRASR SLATVR D E DVEELEMLLEAYFMQID T NK
Sbjct: 231 SVSVSDEPIWYPTSPTIGSMISRASRMSLATVRGDDETDVEELEMLLEAYFMQIDSTLNK 290
Query: 298 LTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWND 357
LT LREYIDDTEDYINIQLDNHRNQLIQLEL LS+GTVC+S YS++A IFGMNIP +WN
Sbjct: 291 LTELREYIDDTEDYINIQLDNHRNQLIQLELMLSAGTVCVSVYSMIAGIFGMNIPNTWNH 350
Query: 358 NHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
+HG++FKWVV ++G F ++F++I+ YAR +GL+GS
Sbjct: 351 DHGYIFKWVVSLTGTFCIVLFVIILSYARFRGLIGS 386
>ref|NP_196531.2| magnesium transporter CorA-like family protein (MRS2-7)
[Arabidopsis thaliana]
Length = 386
Score = 544 bits (1401), Expect = e-153
Identities = 288/396 (72%), Positives = 323/396 (80%), Gaps = 13/396 (3%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 1 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 60
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEVL+RD +DENV+PV+EE QRRLP +
Sbjct: 61 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVLIRDSSDENVIPVLEEFQRRLPVGN 120
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE AYPAL
Sbjct: 121 EAHGVHGDG----------DLGEEDESPFEFRALEVALEAICSFLAARTTELEKFAYPAL 170
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSS 238
DELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+RK G+SS
Sbjct: 171 DELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTRKLVGASS 230
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFD-ENDVEELEMLLEAYFMQIDGTFNK 297
VS S W+ SPTIGS ISRASR SL TVR D E DVEELEMLLEAYFMQID T NK
Sbjct: 231 SVSVSDEPIWYPTSPTIGSMISRASRVSLVTVRGDDETDVEELEMLLEAYFMQIDSTLNK 290
Query: 298 LTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWND 357
LT LREYIDDTEDYINIQLDNHRNQLIQLEL LS+GTVC+S YS++A IFGMNIP +WN
Sbjct: 291 LTELREYIDDTEDYINIQLDNHRNQLIQLELMLSAGTVCVSVYSMIAGIFGMNIPNTWNH 350
Query: 358 NHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
+HG++FKWVV ++G F ++F++I+ YAR +GL+GS
Sbjct: 351 DHGYIFKWVVSLTGTFCIVLFVIILSYARFRGLIGS 386
>ref|NP_850802.2| magnesium transporter CorA-like family protein (MRS2-7)
[Arabidopsis thaliana]
Length = 397
Score = 544 bits (1401), Expect = e-153
Identities = 288/396 (72%), Positives = 323/396 (80%), Gaps = 13/396 (3%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 12 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 71
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEVL+RD +DENV+PV+EE QRRLP +
Sbjct: 72 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVLIRDSSDENVIPVLEEFQRRLPVGN 131
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE AYPAL
Sbjct: 132 EAHGVHGDG----------DLGEEDESPFEFRALEVALEAICSFLAARTTELEKFAYPAL 181
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSS 238
DELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+RK G+SS
Sbjct: 182 DELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTRKLVGASS 241
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFD-ENDVEELEMLLEAYFMQIDGTFNK 297
VS S W+ SPTIGS ISRASR SL TVR D E DVEELEMLLEAYFMQID T NK
Sbjct: 242 SVSVSDEPIWYPTSPTIGSMISRASRVSLVTVRGDDETDVEELEMLLEAYFMQIDSTLNK 301
Query: 298 LTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWND 357
LT LREYIDDTEDYINIQLDNHRNQLIQLEL LS+GTVC+S YS++A IFGMNIP +WN
Sbjct: 302 LTELREYIDDTEDYINIQLDNHRNQLIQLELMLSAGTVCVSVYSMIAGIFGMNIPNTWNH 361
Query: 358 NHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
+HG++FKWVV ++G F ++F++I+ YAR +GL+GS
Sbjct: 362 DHGYIFKWVVSLTGTFCIVLFVIILSYARFRGLIGS 397
>emb|CAB89361.1| putative protein [Arabidopsis thaliana] gi|11282359|pir||T49929
hypothetical protein F17I14.120 - Arabidopsis thaliana
Length = 401
Score = 523 bits (1346), Expect = e-147
Identities = 285/411 (69%), Positives = 321/411 (77%), Gaps = 28/411 (6%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 1 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 60
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLL-------RDPTDENVVPVVEELQ 112
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEV + + +DENV+PV+EE Q
Sbjct: 61 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVRIISYWLVAKYSSDENVIPVLEEFQ 120
Query: 113 RRLPKVSSLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELE 172
RRLP + H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE
Sbjct: 121 RRLPVGNEAHGVHGDG----------DLGEEDESPFEFRALEVALEAICSFLAARTTELE 170
Query: 173 MAAYPALDELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSR 232
AYPALDELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+R
Sbjct: 171 KFAYPALDELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTR 230
Query: 233 K-AGSSSPVSGSGAANWFGGSPTIGSKISRASRASLATVR-FDENDVEELEMLLEAYFMQ 290
K G+SS VS S W+ SPTIGS ISRASR SL TVR DE DVEELEMLLEAYFMQ
Sbjct: 231 KLVGASSSVSVSDEPIWYPTSPTIGSMISRASRVSLVTVRGDDETDVEELEMLLEAYFMQ 290
Query: 291 IDGTFNKLTTLREYIDDTEDYINI--------QLDNHRNQLIQLELFLSSGTVCLSFYSL 342
ID T NKLT LREYIDDTEDYINI QLDNHRNQLIQLEL LS+GTVC+S YS+
Sbjct: 291 IDSTLNKLTELREYIDDTEDYINIQAIITVYLQLDNHRNQLIQLELMLSAGTVCVSVYSM 350
Query: 343 VAAIFGMNIPYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
+A IFGMNIP +WN +HG++FKWVV ++G F ++F++I+ YAR +GL+GS
Sbjct: 351 IAGIFGMNIPNTWNHDHGYIFKWVVSLTGTFCIVLFVIILSYARFRGLIGS 401
>ref|NP_974757.1| magnesium transporter CorA-like family protein (MRS2-7)
[Arabidopsis thaliana]
Length = 338
Score = 452 bits (1163), Expect = e-126
Identities = 248/328 (75%), Positives = 266/328 (80%), Gaps = 13/328 (3%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 12 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 71
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEVL+RD +DENV+PV+EE QRRLP +
Sbjct: 72 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVLIRDSSDENVIPVLEEFQRRLPVGN 131
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE AYPAL
Sbjct: 132 EAHGVHGDG----------DLGEEDESPFEFRALEVALEAICSFLAARTTELEKFAYPAL 181
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSS 238
DELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+RK G+SS
Sbjct: 182 DELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTRKLVGASS 241
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVR-FDENDVEELEMLLEAYFMQIDGTFNK 297
VS S W+ SPTIGS ISRASR SL TVR DE DVEELEMLLEAYFMQID T NK
Sbjct: 242 SVSVSDEPIWYPTSPTIGSMISRASRVSLVTVRGDDETDVEELEMLLEAYFMQIDSTLNK 301
Query: 298 LTTLREYIDDTEDYINIQLDNHRNQLIQ 325
LT LREYIDDTEDYINIQLDNHRNQLIQ
Sbjct: 302 LTELREYIDDTEDYINIQLDNHRNQLIQ 329
>gb|AAN73218.1| MRS2-8 [Arabidopsis thaliana]
Length = 380
Score = 446 bits (1147), Expect = e-124
Identities = 245/385 (63%), Positives = 295/385 (75%), Gaps = 16/385 (4%)
Query: 13 QAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTIL 72
+ V V + QSS SWI DA G+ ++LDVDKY IMHRVQIHARDLRILDP L YPS IL
Sbjct: 6 ELVPVKRITPQSSWSWISIDATGKKTVLDVDKYVIMHRVQIHARDLRILDPNLFYPSAIL 65
Query: 73 GREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYV 132
GRE+AIVLNLEHIKAIITA+EVL++D +DEN++P +EE Q RL + H Q DG
Sbjct: 66 GRERAIVLNLEHIKAIITAKEVLIQDSSDENLIPTLEEFQTRLSVGNKAHGGQLDG---- 121
Query: 133 SSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
+ EEDES FEFRALEVALEAICSFLAART ELE +AYPALDELT K++SRNL
Sbjct: 122 ------DVVEEDESAFEFRALEVALEAICSFLAARTIELEKSAYPALDELTLKLTSRNLL 175
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSGA-ANWFG 250
RV KLKS+MTRLTA+VQK++DELEQLL+DD+DMA+LYLSRK AG+SSP SG NW+
Sbjct: 176 RVCKLKSSMTRLTAQVQKIKDELEQLLEDDEDMAELYLSRKLAGASSPAIDSGEHINWYP 235
Query: 251 GSPTIGSKISRASR--ASLATVRFDE-NDVEELEMLLEAYFMQIDGTFNKLTTLREYIDD 307
SPTIG+KISRA ATVR D+ NDVEE+EMLLEA+FMQID T NKLT LREY+D+
Sbjct: 236 TSPTIGAKISRAKSHLVRSATVRGDDKNDVEEVEMLLEAHFMQIDRTLNKLTELREYVDE 295
Query: 308 TEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWVV 367
TED++NIQLD+ RNQLI+ E+ L++G++C+S YS+V I GMNIP+ WN MFKWVV
Sbjct: 296 TEDFLNIQLDSSRNQLIKFEIILTAGSICVSVYSVVVGILGMNIPFPWNIKK-HMFKWVV 354
Query: 368 IVSGVFSAIMFLLIIVYARKKGLVG 392
+ AI+F+ I+ +AR K L G
Sbjct: 355 SGTATVCAILFVTIMSFARYKKLFG 379
>dbj|BAB09524.1| unnamed protein product [Arabidopsis thaliana]
Length = 413
Score = 432 bits (1112), Expect = e-120
Identities = 239/329 (72%), Positives = 263/329 (79%), Gaps = 14/329 (4%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 1 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 60
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEVL+RD +DENV+PV+EE QRRLP +
Sbjct: 61 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVLIRDSSDENVIPVLEEFQRRLPVGN 120
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE AYPAL
Sbjct: 121 EAHGVHGDG----------DLGEEDESPFEFRALEVALEAICSFLAARTTELEKFAYPAL 170
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSS 238
DELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+RK G+SS
Sbjct: 171 DELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTRKLVGASS 230
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVR-FDENDVEELEMLLEAYFMQIDGTFNK 297
VS S W+ SPTIGS ISRASR SL TVR DE DVEELEMLLEAYFMQID T NK
Sbjct: 231 SVSVSDEPIWYPTSPTIGSMISRASRVSLVTVRGDDETDVEELEMLLEAYFMQIDSTLNK 290
Query: 298 LTTLREYIDDTEDYINIQL-DNHRNQLIQ 325
LT LREYIDDTEDYINIQ+ N +++ ++
Sbjct: 291 LTELREYIDDTEDYINIQVTKNEKSEFLK 319
>emb|CAC13981.1| putative magnesium transporter [Arabidopsis thaliana]
gi|20148403|gb|AAM10092.1| unknown protein [Arabidopsis
thaliana] gi|25360797|gb|AAN73211.1| MRS2-1 [Arabidopsis
thaliana] gi|18394312|ref|NP_563988.1| magnesium
transporter CorA-like family protein (MRS2-1)
[Arabidopsis thaliana] gi|6587806|gb|AAF18497.1|
Contains similarity to gb|M82916 MRS2 protein from
Saccharomyces cerivisae. ESTs gb|N96043, gb|AI998651,
gb|AA585850, gb|T42027 come from this gene. [Arabidopsis
thaliana] gi|15451154|gb|AAK96848.1| Unknown protein
[Arabidopsis thaliana] gi|25352409|pir||G86294 T24D18.11
protein - Arabidopsis thaliana
Length = 442
Score = 330 bits (846), Expect = 5e-89
Identities = 194/399 (48%), Positives = 256/399 (63%), Gaps = 26/399 (6%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ Q RSWI D G +++VDK+ +M R + ARDLR+LDPL YPSTILGREKA
Sbjct: 43 LKKRGQGLRSWIRVDTSGNTQVMEVDKFTMMRRCDLPARDLRLLDPLFVYPSTILGREKA 102
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLP--KVSSLHQQQGDGQEYVSSQ 135
IV+NLE I+ IITA+EVLL + D V+ V ELQ+RL V + QQ+ S+
Sbjct: 103 IVVNLEQIRCIITADEVLLLNSLDNYVLRYVVELQQRLKTSSVGEMWQQENSQLSRRRSR 162
Query: 136 HDTEAAEE---DESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
A E D PFEFRALE+ALEA C+FL ++ +ELE+ AYP LDELTSKIS+ NL+
Sbjct: 163 SFDNAFENSSPDYLPFEFRALEIALEAACTFLDSQASELEIEAYPLLDELTSKISTLNLE 222
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKA--------GSSSPV---S 241
RVR+LKS + LT RVQKVRDE+EQL+DDD DMA++YL+ K G S + S
Sbjct: 223 RVRRLKSRLVALTRRVQKVRDEIEQLMDDDGDMAEMYLTEKKRRMEGSMYGDQSLLGYRS 282
Query: 242 GSGAANWFGGSPTIGSKISRASRASLATVRFDEN---------DVEELEMLLEAYFMQID 292
G + SP SR SL+ R + ++EELEMLLEAYF+ ID
Sbjct: 283 NDGLSVSAPVSPVSSPPDSRRLDKSLSIARSRHDSARSSEGAENIEELEMLLEAYFVVID 342
Query: 293 GTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIP 352
T NKLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN
Sbjct: 343 STLNKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNFE 402
Query: 353 YSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
+ + G F+WV+I++GV ++F + + + + L+
Sbjct: 403 IDFFNQPG-AFRWVLIITGVCGFVIFSAFVWFFKYRRLM 440
>ref|NP_565247.1| magnesium transporter CorA-like family protein (MGT1) (MRS2)
[Arabidopsis thaliana] gi|6503302|gb|AAF14678.1| Is a
member of PF|01544 CorA-like Mg2+ transporter protein
family. ESTs gb|Z48392 and gb|Z48391 come from this
gene. [Arabidopsis thaliana] gi|25352404|pir||H96841
hypothetical protein F23A5.26 [imported] - Arabidopsis
thaliana
Length = 443
Score = 330 bits (846), Expect = 5e-89
Identities = 192/400 (48%), Positives = 252/400 (63%), Gaps = 27/400 (6%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ Q +SWI D +++VDK+ +M R + ARDLR+LDPL YPSTILGREKA
Sbjct: 43 LKKRGQGLKSWIRVDTSANSQVIEVDKFTMMRRCDLPARDLRLLDPLFVYPSTILGREKA 102
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYV----- 132
IV+NLE I+ IITA+EVLL + D V+ V ELQ+RL S D E
Sbjct: 103 IVVNLEQIRCIITADEVLLLNSLDNYVLRYVVELQQRLKASSVTEVWNQDSLELSRRRSR 162
Query: 133 SSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
S + + + D PFEFRALEVALEA C+FL ++ +ELE+ AYP LDELTSKIS+ NL+
Sbjct: 163 SLDNVLQNSSPDYLPFEFRALEVALEAACTFLDSQASELEIEAYPLLDELTSKISTLNLE 222
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK---------AGSSSPVSGS 243
R R+LKS + LT RVQKVRDE+EQL+DDD DMA++YL+ K S PV +
Sbjct: 223 RARRLKSRLVALTRRVQKVRDEIEQLMDDDGDMAEMYLTEKKKRMEGSLYGDQSLPVYRT 282
Query: 244 GAANWFGG--SPTIGSKISRASRASLATVRFDEN----------DVEELEMLLEAYFMQI 291
SP SR SL+ VR + ++EELEMLLEAYF+ I
Sbjct: 283 NDCFSLSAPVSPVSSPPESRRLEKSLSIVRSRHDSARSSEDATENIEELEMLLEAYFVVI 342
Query: 292 DGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNI 351
D T NKLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN
Sbjct: 343 DSTLNKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNF 402
Query: 352 PYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
+ + G FKWV+ ++GV ++FL + Y +++ L+
Sbjct: 403 EIDFFEKPG-AFKWVLAITGVCGLVVFLAFLWYYKRRRLM 441
>dbj|BAD43148.1| unnamed protein product [Arabidopsis thaliana]
Length = 294
Score = 330 bits (846), Expect = 5e-89
Identities = 187/281 (66%), Positives = 218/281 (77%), Gaps = 15/281 (5%)
Query: 13 QAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTIL 72
+ V V + QSS SWI DA G+ ++LDVDKY IMHRVQIHARDLRILDP L YPS IL
Sbjct: 6 ELVPVKRITPQSSWSWISIDATGKKTVLDVDKYVIMHRVQIHARDLRILDPNLFYPSAIL 65
Query: 73 GREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYV 132
GRE+AIVLNLEHIKAIITA+EVL++D +DEN++P +EE Q RL + H Q DG
Sbjct: 66 GRERAIVLNLEHIKAIITAKEVLIQDSSDENLIPTLEEFQTRLSVGNKAHGGQLDG---- 121
Query: 133 SSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
+ EEDES FEFRALEVALEAICSFLAART ELE +AYPALDELT K++SRNL
Sbjct: 122 ------DVVEEDESAFEFRALEVALEAICSFLAARTIELEKSAYPALDELTLKLTSRNLL 175
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSGA-ANWFG 250
RV KLKS+MTRLTA+VQK++DELEQLL+DD+DMA+LYLSRK AG+SSP SG NW+
Sbjct: 176 RVCKLKSSMTRLTAQVQKIKDELEQLLEDDEDMAELYLSRKLAGASSPAIDSGEHINWYP 235
Query: 251 GSPTIGSKISRASR--ASLATVRFDE-NDVEELEMLLEAYF 288
SPTIG+KISRA ATVR D+ NDVEE+EMLLEA+F
Sbjct: 236 TSPTIGAKISRAKSHLVRSATVRGDDKNDVEEVEMLLEAHF 276
>gb|AAM62917.1| unknown [Arabidopsis thaliana] gi|25360983|gb|AAN73219.1| MRS2-10
[Arabidopsis thaliana]
Length = 443
Score = 328 bits (841), Expect = 2e-88
Identities = 191/400 (47%), Positives = 252/400 (62%), Gaps = 27/400 (6%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ Q +SWI D +++VDK+ +M R + ARDLR+LDPL YPSTILGREKA
Sbjct: 43 LKKRGQGLKSWIRVDTSANSQVIEVDKFTMMRRCDLPARDLRLLDPLFVYPSTILGREKA 102
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYV----- 132
IV+NLE I+ IITA+EVLL + D V+ V ELQ+RL S D E
Sbjct: 103 IVVNLEQIRCIITADEVLLLNSLDNYVLRYVVELQQRLKASSVTEVWNQDSLELSRRRSR 162
Query: 133 SSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
S + + + D PFEFRALEVALEA C+FL ++ +ELE+ AYP LDELTSKIS+ NL+
Sbjct: 163 SLDNVFQNSSPDYLPFEFRALEVALEAACTFLDSQASELEIEAYPLLDELTSKISTLNLE 222
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK---------AGSSSPVSGS 243
R R+LKS + LT RVQKVRDE+EQL+DDD DMA++YL+ K S PV +
Sbjct: 223 RARRLKSRLVALTRRVQKVRDEIEQLMDDDGDMAEMYLTEKKKRMEGSLYGDQSLPVYRT 282
Query: 244 GAANWFGG--SPTIGSKISRASRASLATVRFDEN----------DVEELEMLLEAYFMQI 291
SP SR SL+ VR + ++EELEMLLEAYF+ I
Sbjct: 283 NDCFSLSAPVSPVSSPPESRRLEKSLSIVRSRHDSARSSEDATENIEELEMLLEAYFVVI 342
Query: 292 DGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNI 351
D T NKLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN
Sbjct: 343 DSTLNKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNF 402
Query: 352 PYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
+ + G FKWV+ ++GV ++FL + + +++ L+
Sbjct: 403 EIDFFEKPG-AFKWVLAITGVCGLVVFLAFLWFYKRRRLM 441
>gb|AAN73214.1| MRS2-4 [Arabidopsis thaliana] gi|16209702|gb|AAL14408.1|
AT3g58970/F17J16_20 [Arabidopsis thaliana]
gi|25090327|gb|AAN72277.1| At3g58970/F17J16_20
[Arabidopsis thaliana] gi|18411070|ref|NP_567076.1|
magnesium transporter CorA-like family protein
[Arabidopsis thaliana]
Length = 436
Score = 328 bits (840), Expect = 2e-88
Identities = 189/388 (48%), Positives = 252/388 (64%), Gaps = 10/388 (2%)
Query: 14 AVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILG 73
A KKKT +R W+ FD G +++ DK I+ R + ARDLRIL P+ S+ S IL
Sbjct: 51 ATGKAKKKTGGARLWMRFDRTGAMEVVECDKSTIIKRASVPARDLRILGPVFSHSSNILA 110
Query: 74 REKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVS 133
REKAIV+NLE IKAI+TAEEVLL DP V+P VE L+++ P+ + ++ V
Sbjct: 111 REKAIVVNLEVIKAIVTAEEVLLLDPLRPEVLPFVERLKQQFPQRNG-NENALQASANVQ 169
Query: 134 SQHDTEAAE--EDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNL 191
S D EAAE + E PFEF+ LE+ALE +CSF+ LE A+P LDELT +S+ NL
Sbjct: 170 SPLDPEAAEGLQSELPFEFQVLEIALEVVCSFVDKSVAALETEAWPVLDELTKNVSTENL 229
Query: 192 DRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK---AGSSSPVSGSGAANW 248
+ VR LKS +TRL ARVQKVRDELE LLDD++DMADLYL+RK + + A+N
Sbjct: 230 EYVRSLKSNLTRLLARVQKVRDELEHLLDDNEDMADLYLTRKWIQNQQTEAILAGTASNS 289
Query: 249 FGGSPTIGSKISRASR---ASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYI 305
S + R + AS+ T +E+DVE+LEMLLEAYFMQ+DG NK+ T+REYI
Sbjct: 290 IALPAHNTSNLHRLTSNRSASMVTSNTEEDDVEDLEMLLEAYFMQLDGMRNKILTVREYI 349
Query: 306 DDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKW 365
DDTEDY+NIQLDN RN+LIQL+L L+ + ++ +L+A++FGMNIP HG +F +
Sbjct: 350 DDTEDYVNIQLDNQRNELIQLQLTLTIASFAIAAETLLASLFGMNIPCPLYSIHG-VFGY 408
Query: 366 VVIVSGVFSAIMFLLIIVYARKKGLVGS 393
V ++F++ + YAR K L+GS
Sbjct: 409 FVWSVTALCIVLFMVTLGYARWKKLLGS 436
>emb|CAB86925.1| putative protein [Arabidopsis thaliana] gi|11282358|pir||T47779
hypothetical protein F17J16.20 - Arabidopsis thaliana
Length = 463
Score = 322 bits (824), Expect = 2e-86
Identities = 190/399 (47%), Positives = 253/399 (62%), Gaps = 21/399 (5%)
Query: 14 AVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILG 73
A KKKT +R W+ FD G +++ DK I+ R + ARDLRIL P+ S+ S ILG
Sbjct: 67 ATGKAKKKTGGARLWMRFDRTGAMEVVECDKSTIIKRASVPARDLRILGPVFSHSSNILG 126
Query: 74 -----------REKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLH 122
REKAIV+NLE IKAI+TAEEVLL DP V+P VE L+++ P+ + +
Sbjct: 127 KNFDLLIPGLAREKAIVVNLEVIKAIVTAEEVLLLDPLRPEVLPFVERLKQQFPQRNG-N 185
Query: 123 QQQGDGQEYVSSQHDTEAAE--EDESPFEFRALEVALEAICSFLAARTTELEMAAYPALD 180
+ V S D EAAE + E PFEF+ LE+ALE +CSF+ LE A+P LD
Sbjct: 186 ENALQASANVQSPLDPEAAEGLQSELPFEFQVLEIALEVVCSFVDKSVAALETEAWPVLD 245
Query: 181 ELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK---AGSS 237
ELT +S+ NL+ VR LKS +TRL ARVQKVRDELE LLDD++DMADLYL+RK +
Sbjct: 246 ELTKNVSTENLEYVRSLKSNLTRLLARVQKVRDELEHLLDDNEDMADLYLTRKWIQNQQT 305
Query: 238 SPVSGSGAANWFGGSPTIGSKISRASR---ASLATVRFDENDVEELEMLLEAYFMQIDGT 294
+ A+N S + R + AS+ T +E+DVE+LEMLLEAYFMQ+DG
Sbjct: 306 EAILAGTASNSIALPAHNTSNLHRLTSNRSASMVTSNTEEDDVEDLEMLLEAYFMQLDGM 365
Query: 295 FNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYS 354
NK+ T+REYIDDTEDY+NIQLDN RN+LIQL+L L+ + ++ +L+A++FGMNIP
Sbjct: 366 RNKILTVREYIDDTEDYVNIQLDNQRNELIQLQLTLTIASFAIAAETLLASLFGMNIPCP 425
Query: 355 WNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
HG +F + V ++F++ + YAR K L+GS
Sbjct: 426 LYSIHG-VFGYFVWSVTALCIVLFMVTLGYARWKKLLGS 463
>dbj|BAB09530.1| unnamed protein product [Arabidopsis thaliana]
Length = 341
Score = 320 bits (821), Expect = 4e-86
Identities = 185/285 (64%), Positives = 216/285 (74%), Gaps = 19/285 (6%)
Query: 13 QAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTIL 72
+ V V + QSS SWI DA G+ ++LDVDKY IMHRVQIHARDLRILDP L YPS IL
Sbjct: 6 ELVPVKRITPQSSWSWISIDATGKKTVLDVDKYVIMHRVQIHARDLRILDPNLFYPSAIL 65
Query: 73 GREKAIVLNLEHIKAIITAEEV----LLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDG 128
GRE+AIVLNLEHIKAIITA+EV L++D +DEN++P +EE Q RL + H Q DG
Sbjct: 66 GRERAIVLNLEHIKAIITAKEVSLSVLIQDSSDENLIPTLEEFQTRLSVGNKAHGGQLDG 125
Query: 129 QEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISS 188
+ EEDES FEFRALEVALEAICSFLAART ELE +AYPALDELT K++S
Sbjct: 126 ----------DVVEEDESAFEFRALEVALEAICSFLAARTIELEKSAYPALDELTLKLTS 175
Query: 189 RNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSGA-A 246
RNL RV KLKS+MTRLTA+VQK++DELEQLL+DD+DMA+LYLSRK AG+SSP SG
Sbjct: 176 RNLLRVCKLKSSMTRLTAQVQKIKDELEQLLEDDEDMAELYLSRKLAGASSPAIDSGEHI 235
Query: 247 NWFGGSPTIGSKISRASR--ASLATVRFDE-NDVEELEMLLEAYF 288
NW+ SPTIG+KISRA ATVR D+ NDVEE+EMLLE +
Sbjct: 236 NWYPTSPTIGAKISRAKSHLVRSATVRGDDKNDVEEVEMLLEVVY 280
>dbj|BAD38112.1| magnesium transporter CorA-like [Oryza sativa (japonica
cultivar-group)]
Length = 436
Score = 318 bits (814), Expect = 2e-85
Identities = 191/398 (47%), Positives = 249/398 (61%), Gaps = 47/398 (11%)
Query: 18 VKKKTQSSRSWILFDA-DGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREK 76
+KK+ +RSWI +A L+VDK +M R ++ ARDLR+LDPL YPSTILGRE+
Sbjct: 60 LKKRGGGTRSWIRVEAATASVQTLEVDKATMMRRCELPARDLRLLDPLFVYPSTILGRER 119
Query: 77 AIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQH 136
AIV+NLE I+ +ITA+EVLL + D V+ ELQRRL
Sbjct: 120 AIVVNLEQIRCVITADEVLLLNSLDSYVLQYAAELQRRL--------------------- 158
Query: 137 DTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRK 196
+ AE DE PFEFRALE+ALEA CSFL A+ ELE+ AYP LDELTSKIS+ NL+RVR+
Sbjct: 159 -LQRAEGDELPFEFRALELALEAACSFLDAQAAELEIEAYPLLDELTSKISTLNLERVRR 217
Query: 197 LKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKA--------GSSSPVSGSGAANW 248
LKS + LT RVQKVRDE+EQL+DDD DMA++YLS K G S + + +
Sbjct: 218 LKSRLVALTRRVQKVRDEIEQLMDDDGDMAEMYLSEKKLRTEASFYGDQSMLGYNSVGDG 277
Query: 249 FG--------GSPTIGSKISRA---SRASLATVRFDEN----DVEELEMLLEAYFMQIDG 293
SPT K+ +A R+ +V+ +N ++ELEMLLEAYF+ ID
Sbjct: 278 TSFSAPVSPVSSPTESRKLEKAFSLCRSRHDSVKSSDNTATEHIQELEMLLEAYFVVIDS 337
Query: 294 TFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPY 353
T NKLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN
Sbjct: 338 TLNKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNFET 397
Query: 354 SWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
S F+WV+I++GV A +F + + + K L+
Sbjct: 398 SVFSIQN-AFQWVLIITGVIGAFIFCGFLWFFKYKRLM 434
>emb|CAE01634.2| OSJNBa0029H02.17 [Oryza sativa (japonica cultivar-group)]
gi|50926173|ref|XP_473061.1| OSJNBa0029H02.17 [Oryza
sativa (japonica cultivar-group)]
Length = 428
Score = 298 bits (763), Expect = 2e-79
Identities = 185/404 (45%), Positives = 240/404 (58%), Gaps = 59/404 (14%)
Query: 18 VKKKTQSSRSWILFDA-DGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREK 76
+K++ RSW+ DA G ++V K A+M R+ + ARDLR+LDPL YPS ILGRE+
Sbjct: 46 LKRRGGGRRSWVRVDAATGASEAVEVAKPALMRRLDLPARDLRLLDPLFVYPSAILGRER 105
Query: 77 AIVLNLEHIKAIITAEEVL-LRDP--------TDENVVPVVEELQRRLPKVSSLHQQQGD 127
A+V NLE I+ IITA+E L LRDP T+E V V ELQRRL
Sbjct: 106 AVVCNLERIRCIITADEALILRDPDVAGGGAETEEAVRRYVAELQRRL------------ 153
Query: 128 GQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKIS 187
D+ PFEF ALEVALEA CSFL A+ ELE AYP LDELT+KIS
Sbjct: 154 ------------VDRADDLPFEFIALEVALEAACSFLDAQAVELEADAYPLLDELTTKIS 201
Query: 188 SRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK------------AG 235
+ NL+RVR+LKS + LT RVQKVRDE+EQL+DDD DMA++YL+ K A
Sbjct: 202 TLNLERVRRLKSKLVALTRRVQKVRDEIEQLMDDDGDMAEMYLTEKKRRMEASLLEEQAF 261
Query: 236 SSSPVSGSGAANWFGGSPTIGSKISRASRASLATVR----------FDENDVEELEMLLE 285
SG G++ SP SR L+ R + +EELEMLLE
Sbjct: 262 QGMGNSGFGSSFSAPVSPVSSPPASRRLEKELSFARSRHDSFKSADSSQYSIEELEMLLE 321
Query: 286 AYFMQIDGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAA 345
AYF+ ID T +KLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +V+
Sbjct: 322 AYFVVIDYTLSKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVSG 381
Query: 346 IFGMNIPYS-WNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKK 388
+FGMN +N H F+W ++++GV ++F I Y +K+
Sbjct: 382 VFGMNFEVDLFNVPHA--FEWTLVITGVCGLVIFCCFIWYFKKR 423
>gb|AAM19344.1| hypothetical protein [Arabidopsis thaliana]
gi|20465991|gb|AAM20217.1| unknown protein [Arabidopsis
thaliana] gi|17979301|gb|AAL49876.1| unknown protein
[Arabidopsis thaliana] gi|4406759|gb|AAD20070.1|
hypothetical protein [Arabidopsis thaliana]
gi|15227679|ref|NP_178460.1| magnesium transporter
CorA-like family protein (MRS2-5) [Arabidopsis thaliana]
gi|25352407|pir||F84450 hypothetical protein At2g03620
[imported] - Arabidopsis thaliana
Length = 421
Score = 287 bits (735), Expect = 3e-76
Identities = 169/398 (42%), Positives = 243/398 (60%), Gaps = 49/398 (12%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ QSSRSW+ D DG ++L++DK IM R + +RDLR+LDPL YPS+ILGRE+A
Sbjct: 41 LKKRGQSSRSWVKIDQDGNSAVLELDKATIMKRCSLPSRDLRLLDPLFIYPSSILGRERA 100
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHD 137
IV++LE I+ IITAEEV+L + D +VV EL +RL S H+
Sbjct: 101 IVVSLEKIRCIITAEEVILMNARDASVVQYQSELCKRL-----------------QSNHN 143
Query: 138 TEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRKL 197
+D+ PFEF+ALE+ LE C L A+ ELEM YP LDEL + IS+ NL+ VR+L
Sbjct: 144 LNV--KDDLPFEFKALELVLELSCLSLDAQVNELEMEVYPVLDELATNISTLNLEHVRRL 201
Query: 198 KSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK------------------------ 233
K + LT +VQKV DE+E L+DDDDDMA++YL+ K
Sbjct: 202 KGRLLTLTQKVQKVCDEIEHLMDDDDDMAEMYLTEKKERAEAHASEELEDNIGEDFESSG 261
Query: 234 -AGSSSPVS--GSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQ 290
S+PVS GS + N FG S I + ++ L++ EN +++LEMLLEAYF+
Sbjct: 262 IVSKSAPVSPVGSTSGN-FGKLQRAFSSIVGSHKSLLSSSSIGEN-IDQLEMLLEAYFVV 319
Query: 291 IDGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMN 350
+D T +KL++L+EYIDDTED INI+L N +NQLIQ +L L++ T + ++ V A+FGMN
Sbjct: 320 VDNTLSKLSSLKEYIDDTEDLINIKLGNVQNQLIQFQLLLTAATFVAAIFAAVTAVFGMN 379
Query: 351 IPYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKK 388
+ S N F++V++++G+ ++ ++Y + K
Sbjct: 380 LQDSVFQN-PTTFQYVLLITGIGCGFLYFGFVLYFKHK 416
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 630,297,723
Number of Sequences: 2540612
Number of extensions: 25884757
Number of successful extensions: 101419
Number of sequences better than 10.0: 300
Number of HSP's better than 10.0 without gapping: 106
Number of HSP's successfully gapped in prelim test: 195
Number of HSP's that attempted gapping in prelim test: 101013
Number of HSP's gapped (non-prelim): 425
length of query: 393
length of database: 863,360,394
effective HSP length: 130
effective length of query: 263
effective length of database: 533,080,834
effective search space: 140200259342
effective search space used: 140200259342
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0071.7


