

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.1
(332 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
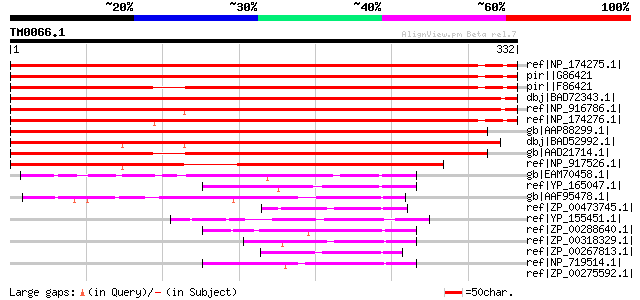
ref|NP_174275.1| expressed protein [Arabidopsis thaliana] 537 e-151
pir||G86421 F1N18.13 protein - Arabidopsis thaliana gi|9972363|g... 491 e-137
pir||F86421 hypothetical protein F1N18.14 - Arabidopsis thaliana... 488 e-137
dbj|BAD72343.1| unknown protein [Oryza sativa (japonica cultivar... 484 e-135
ref|NP_916786.1| P0003E08.5 [Oryza sativa (japonica cultivar-gro... 477 e-133
ref|NP_174276.1| hypothetical protein [Arabidopsis thaliana] 473 e-132
gb|AAP88299.1| At2g42950 [Arabidopsis thaliana] gi|26450698|dbj|... 471 e-131
dbj|BAD52992.1| hypothetical protein [Oryza sativa (japonica cul... 453 e-126
gb|AAD21714.1| hypothetical protein [Arabidopsis thaliana] gi|20... 428 e-118
ref|NP_917526.1| P0518F01.15 [Oryza sativa (japonica cultivar-gr... 338 1e-91
gb|EAM70458.1| Mg2+ transporter protein, CorA-like [Desulfuromon... 62 2e-08
ref|YP_165047.1| magnesium transporter, CorA family [Silicibacte... 62 2e-08
gb|AAF95478.1| conserved hypothetical protein [Vibrio cholerae O... 52 2e-05
ref|ZP_00473745.1| Mg2+ transporter protein, CorA-like [Chromoha... 49 2e-04
ref|YP_155451.1| Mg/Co transporter [Idiomarina loihiensis L2TR] ... 49 2e-04
ref|ZP_00288640.1| COG0598: Mg2+ and Co2+ transporters [Magnetoc... 49 2e-04
ref|ZP_00318329.1| COG0598: Mg2+ and Co2+ transporters [Microbul... 48 4e-04
ref|ZP_00267813.1| COG0598: Mg2+ and Co2+ transporters [Rhodospi... 47 8e-04
ref|NP_719514.1| magnesium transporter, putative [Shewanella one... 47 8e-04
ref|ZP_00275592.1| COG0598: Mg2+ and Co2+ transporters [Ralstoni... 46 0.001
>ref|NP_174275.1| expressed protein [Arabidopsis thaliana]
Length = 540
Score = 537 bits (1383), Expect = e-151
Identities = 268/332 (80%), Positives = 301/332 (89%), Gaps = 6/332 (1%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP++ EDD+P+V RSWQA+NFLV+VMHIKG+V+ NVLGI
Sbjct: 214 VPVRVAGGLLFELLGQSVGDPVISEDDVPVVFRSWQAKNFLVSVMHIKGNVTNTNVLGIT 273
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EV+ELL GGYNVPRTVHEVIA LACRLSRWDDRLFRKSIFG ADEIELKFMNRRNYEDL
Sbjct: 274 EVEELLYAGGYNVPRTVHEVIAHLACRLSRWDDRLFRKSIFGAADEIELKFMNRRNYEDL 333
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF IILNQEIRKLS QVIRVKWSLHAREEI+FELLQHL+G AR LL+G+RK+TREM+E
Sbjct: 334 NLFSIILNQEIRKLSRQVIRVKWSLHAREEIIFELLQHLRGNIARHLLDGLRKNTREMLE 393
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV QS+VRAWLQD+SLRV+HNLAVF G G+VLTII GLFGINVDGIP
Sbjct: 394 EQEAVRGRLFTIQDVMQSSVRAWLQDKSLRVSHNLAVFGGCGLVLTIIVGLFGINVDGIP 453
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA++TPYAFGLFT ++V +GA+L+VVGLVYLGLKKPI EE+VEVRKLELQ++VK+FQHEA
Sbjct: 454 GAQNTPYAFGLFTFLMVLLGAILIVVGLVYLGLKKPITEEQVEVRKLELQDVVKIFQHEA 513
Query: 301 ETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
ETHAQ+R RNNL PTAGD F D DY+L+
Sbjct: 514 ETHAQLR----RNNLSPTAGDVF--DADYILI 539
>pir||G86421 F1N18.13 protein - Arabidopsis thaliana gi|9972363|gb|AAG10613.1|
Unknown protein [Arabidopsis thaliana]
Length = 533
Score = 491 bits (1264), Expect = e-137
Identities = 246/332 (74%), Positives = 279/332 (83%), Gaps = 6/332 (1%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRV GGLLFELLGQS GDP++ EDD+P+V RSWQA+NFLV+VMHIKG+VS+ NVLGI
Sbjct: 207 VPVRVDGGLLFELLGQSMGDPVIGEDDVPVVFRSWQAKNFLVSVMHIKGNVSKSNVLGIT 266
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EV+ELL G YNVPRT+HEVIA LACRLSRWDDRLFRKSIFG ADEIELKFMNRRN+EDL
Sbjct: 267 EVEELLYAGSYNVPRTIHEVIAHLACRLSRWDDRLFRKSIFGAADEIELKFMNRRNHEDL 326
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF IILNQEIRKL+ Q IRVKWSLHAREEI+ ELLQHL+G R LLEG+R +TREM+E
Sbjct: 327 NLFSIILNQEIRKLARQTIRVKWSLHAREEIILELLQHLRGNIPRHLLEGLRNNTREMLE 386
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQD QS +R+WLQD+SL +HNLA+F G G+VLTII GLF +N+DG+P
Sbjct: 387 EQEAVRGRLFTIQDNIQSNIRSWLQDQSLNGSHNLAIFGGCGLVLTIILGLFSVNLDGVP 446
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
G +HTPYAF LF+ LV IG VL+ GL YLG KKPI EE VE RKLELQ +VK+FQHEA
Sbjct: 447 GVKHTPYAFVLFSVFLVLIGIVLIAFGLRYLGPKKPITEEHVEARKLELQNVVKIFQHEA 506
Query: 301 ETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
ETHAQ+R RNNL PTAGD F D DY L+
Sbjct: 507 ETHAQLR----RNNLSPTAGDVF--DADYFLI 532
>pir||F86421 hypothetical protein F1N18.14 - Arabidopsis thaliana
gi|9972364|gb|AAG10614.1| Unknown protein [Arabidopsis
thaliana]
Length = 520
Score = 488 bits (1257), Expect = e-137
Identities = 249/332 (75%), Positives = 282/332 (84%), Gaps = 26/332 (7%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP++ EDD+P+V RSWQA+NFLV+VMHIKG+V+ NVLGI
Sbjct: 214 VPVRVAGGLLFELLGQSVGDPVISEDDVPVVFRSWQAKNFLVSVMHIKGNVTNTNVLGIT 273
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EV+ELL GGYNVPRTVHEVIA LACRLSRWDDR RNYEDL
Sbjct: 274 EVEELLYAGGYNVPRTVHEVIAHLACRLSRWDDR--------------------RNYEDL 313
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF IILNQEIRKLS QVIRVKWSLHAREEI+FELLQHL+G AR LL+G+RK+TREM+E
Sbjct: 314 NLFSIILNQEIRKLSRQVIRVKWSLHAREEIIFELLQHLRGNIARHLLDGLRKNTREMLE 373
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV QS+VRAWLQD+SLRV+HNLAVF G G+VLTII GLFGINVDGIP
Sbjct: 374 EQEAVRGRLFTIQDVMQSSVRAWLQDKSLRVSHNLAVFGGCGLVLTIIVGLFGINVDGIP 433
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA++TPYAFGLFT ++V +GA+L+VVGLVYLGLKKPI EE+VEVRKLELQ++VK+FQHEA
Sbjct: 434 GAQNTPYAFGLFTFLMVLLGAILIVVGLVYLGLKKPITEEQVEVRKLELQDVVKIFQHEA 493
Query: 301 ETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
ETHAQ+R RNNL PTAGD F D DY+L+
Sbjct: 494 ETHAQLR----RNNLSPTAGDVF--DADYILI 519
>dbj|BAD72343.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 558
Score = 484 bits (1246), Expect = e-135
Identities = 243/332 (73%), Positives = 281/332 (84%), Gaps = 2/332 (0%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP EE+DIPIVLRSWQAQNFLVT MH+KG S INVLG+
Sbjct: 228 VPVRVAGGLLFELLGQSVGDPNREEEDIPIVLRSWQAQNFLVTAMHVKGPSSNINVLGVT 287
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EVQELLS GG PR+ HEVIA L RLSRWDDRLFRK +FG ADEIELKF+NRRN+EDL
Sbjct: 288 EVQELLSAGGSQTPRSAHEVIAHLIGRLSRWDDRLFRKYVFGEADEIELKFVNRRNHEDL 347
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NL IILNQEIR+L+TQVIRVKWSLHAREEI+ ELL+HL+G R +L+ IRK TREM+E
Sbjct: 348 NLVSIILNQEIRRLATQVIRVKWSLHAREEIIIELLRHLRGNTTRVILDSIRKDTREMLE 407
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV QSTVRAWLQDRSLR+THNLA+F G GMVL+IITGLFGINVDGIP
Sbjct: 408 EQEAVRGRLFTIQDVMQSTVRAWLQDRSLRITHNLAIFGGGGMVLSIITGLFGINVDGIP 467
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA++TPYAFGLF +L F+G VL+ VG++YLGL+ P+ EKV+VRKLELQ+LV FQHEA
Sbjct: 468 GAQNTPYAFGLFAGLLFFVGFVLIGVGILYLGLQNPVTNEKVKVRKLELQDLVSAFQHEA 527
Query: 301 ETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
E H +VR+ +SR++ P + A +VDY+L+
Sbjct: 528 EQHGKVREGLSRHSSSPKSSSA--SNVDYVLI 557
>ref|NP_916786.1| P0003E08.5 [Oryza sativa (japonica cultivar-group)]
Length = 565
Score = 477 bits (1228), Expect = e-133
Identities = 243/339 (71%), Positives = 281/339 (82%), Gaps = 9/339 (2%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP EE+DIPIVLRSWQAQNFLVT MH+KG S INVLG+
Sbjct: 228 VPVRVAGGLLFELLGQSVGDPNREEEDIPIVLRSWQAQNFLVTAMHVKGPSSNINVLGVT 287
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMN------- 113
EVQELLS GG PR+ HEVIA L RLSRWDDRLFRK +FG ADEIELKF+N
Sbjct: 288 EVQELLSAGGSQTPRSAHEVIAHLIGRLSRWDDRLFRKYVFGEADEIELKFVNSSKQPFR 347
Query: 114 RRNYEDLNLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRK 173
RRN+EDLNL IILNQEIR+L+TQVIRVKWSLHAREEI+ ELL+HL+G R +L+ IRK
Sbjct: 348 RRNHEDLNLVSIILNQEIRRLATQVIRVKWSLHAREEIIIELLRHLRGNTTRVILDSIRK 407
Query: 174 STREMIEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFG 233
TREM+EEQEAVRGRLFTIQDV QSTVRAWLQDRSLR+THNLA+F G GMVL+IITGLFG
Sbjct: 408 DTREMLEEQEAVRGRLFTIQDVMQSTVRAWLQDRSLRITHNLAIFGGGGMVLSIITGLFG 467
Query: 234 INVDGIPGAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELV 293
INVDGIPGA++TPYAFGLF +L F+G VL+ VG++YLGL+ P+ EKV+VRKLELQ+LV
Sbjct: 468 INVDGIPGAQNTPYAFGLFAGLLFFVGFVLIGVGILYLGLQNPVTNEKVKVRKLELQDLV 527
Query: 294 KMFQHEAETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
FQHEAE H +VR+ +SR++ P + A +VDY+L+
Sbjct: 528 SAFQHEAEQHGKVREGLSRHSSSPKSSSA--SNVDYVLI 564
>ref|NP_174276.1| hypothetical protein [Arabidopsis thaliana]
Length = 570
Score = 473 bits (1216), Expect = e-132
Identities = 246/369 (66%), Positives = 279/369 (74%), Gaps = 43/369 (11%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRV GGLLFELLGQS GDP++ EDD+P+V RSWQA+NFLV+VMHIKG+VS+ NVLGI
Sbjct: 207 VPVRVDGGLLFELLGQSMGDPVIGEDDVPVVFRSWQAKNFLVSVMHIKGNVSKSNVLGIT 266
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDD--------------------------- 93
EV+ELL G YNVPRT+HEVIA LACRLSRWDD
Sbjct: 267 EVEELLYAGSYNVPRTIHEVIAHLACRLSRWDDSVVNSIDKDTRRSLEIACSSDTWDETN 326
Query: 94 ----------RLFRKSIFGVADEIELKFMNRRNYEDLNLFVIILNQEIRKLSTQVIRVKW 143
RLFRKSIFG ADEIELKFMNRRN+EDLNLF IILNQEIRKL+ Q IRVKW
Sbjct: 327 IKVLVLVRCGRLFRKSIFGAADEIELKFMNRRNHEDLNLFSIILNQEIRKLARQTIRVKW 386
Query: 144 SLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIEEQEAVRGRLFTIQDVTQSTVRAW 203
SLHAREEI+ ELLQHL+G R LLEG+R +TREM+EEQEAVRGRLFTIQD QS +R+W
Sbjct: 387 SLHAREEIILELLQHLRGNIPRHLLEGLRNNTREMLEEQEAVRGRLFTIQDNIQSNIRSW 446
Query: 204 LQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIPGAEHTPYAFGLFTAILVFIGAVL 263
LQD+SL +HNLA+F G G+VLTII GLF +N+DG+PG +HTPYAF LF+ LV IG VL
Sbjct: 447 LQDQSLNGSHNLAIFGGCGLVLTIILGLFSVNLDGVPGVKHTPYAFVLFSVFLVLIGIVL 506
Query: 264 VVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEAETHAQVRKNVSRNNLPPTAGDAF 323
+ GL YLG KKPI EE VE RKLELQ +VK+FQHEAETHAQ+R RNNL PTAGD F
Sbjct: 507 IAFGLRYLGPKKPITEEHVEARKLELQNVVKIFQHEAETHAQLR----RNNLSPTAGDVF 562
Query: 324 RRDVDYLLL 332
D DY L+
Sbjct: 563 --DADYFLI 569
>gb|AAP88299.1| At2g42950 [Arabidopsis thaliana] gi|26450698|dbj|BAC42458.1|
unknown protein [Arabidopsis thaliana]
gi|30689197|ref|NP_181823.2| expressed protein
[Arabidopsis thaliana]
Length = 501
Score = 471 bits (1211), Expect = e-131
Identities = 235/313 (75%), Positives = 266/313 (84%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQSAGDP ++EDDIPIVLRSWQAQNFLVT +H+KG I+VLGI
Sbjct: 167 VPVRVAGGLLFELLGQSAGDPFIQEDDIPIVLRSWQAQNFLVTALHVKGFALNISVLGIT 226
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EVQE+L GG +PRTVHE+IA LACRL+RWDDRLFRK IFG ADE+EL FMN+R YED
Sbjct: 227 EVQEMLIAGGACIPRTVHELIAHLACRLARWDDRLFRKYIFGAADEVELMFMNKRLYEDP 286
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF ILNQEIR+LSTQVIRVKWSLHAREEIVFELLQ LKG + LLEGI+KSTR+MI
Sbjct: 287 NLFTTILNQEIRRLSTQVIRVKWSLHAREEIVFELLQQLKGNRTKDLLEGIKKSTRDMIN 346
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV Q+TVRAWLQD+SL VTHNL +F GVG+++TI+TGLFGINVDGIP
Sbjct: 347 EQEAVRGRLFTIQDVMQNTVRAWLQDQSLTVTHNLGIFGGVGLLITIVTGLFGINVDGIP 406
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA+ P AF LF+ +L G VLVV GL+YLGLK+P+AEE VE RKLEL E+VK FQ EA
Sbjct: 407 GAKDFPQAFALFSVVLFVSGLVLVVAGLIYLGLKEPVAEENVETRKLELDEMVKKFQQEA 466
Query: 301 ETHAQVRKNVSRN 313
E+HAQV K V +N
Sbjct: 467 ESHAQVCKKVPQN 479
>dbj|BAD52992.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 575
Score = 453 bits (1165), Expect = e-126
Identities = 223/326 (68%), Positives = 272/326 (83%), Gaps = 5/326 (1%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP +E+DIPIVLR+WQAQNFL+T +H+KGS +NV+G+
Sbjct: 234 VPVRVAGGLLFELLGQSVGDPARDEEDIPIVLRAWQAQNFLITALHVKGSAPNVNVIGVT 293
Query: 61 EVQELLSGGGYN--VPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMN---RR 115
EVQELLS G P+ + EVIA LA RL+RWDDRL+RK +FG ADEIELKF+N RR
Sbjct: 294 EVQELLSACGSTGTAPKNIQEVIAHLASRLARWDDRLWRKYVFGAADEIELKFVNSVFRR 353
Query: 116 NYEDLNLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKST 175
EDL L +I NQ+IR+L+TQVIRVKWSLHAREEI+FELL++L G +SLLE I+K
Sbjct: 354 KQEDLKLLCMIFNQDIRRLATQVIRVKWSLHAREEIIFELLKYLGGSTTKSLLEAIKKDA 413
Query: 176 REMIEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGIN 235
R+MIEEQEAVRGRLFTIQDV QST+RAW Q++SLR+THNL +F G G+VL+II GLFGIN
Sbjct: 414 RQMIEEQEAVRGRLFTIQDVMQSTLRAWSQEKSLRITHNLTIFGGCGLVLSIIAGLFGIN 473
Query: 236 VDGIPGAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKM 295
VDGIPGAE+TPYAF LF+A+L +G +L++VG+VY GL+KPI++E+V+VRKLELQELV M
Sbjct: 474 VDGIPGAENTPYAFALFSALLFLVGLLLIIVGIVYFGLQKPISDEQVQVRKLELQELVSM 533
Query: 296 FQHEAETHAQVRKNVSRNNLPPTAGD 321
FQHEAETHA+V++ V R +LPP A D
Sbjct: 534 FQHEAETHARVKEGVLRTDLPPRAAD 559
>gb|AAD21714.1| hypothetical protein [Arabidopsis thaliana]
gi|20197871|gb|AAM15295.1| hypothetical protein
[Arabidopsis thaliana] gi|25372716|pir||C84860
hypothetical protein At2g42950 [imported] - Arabidopsis
thaliana
Length = 481
Score = 428 bits (1101), Expect = e-118
Identities = 220/313 (70%), Positives = 249/313 (79%), Gaps = 20/313 (6%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQSAGDP ++EDDIPIVLRSWQAQNFLVT +H+KG I+VLGI
Sbjct: 167 VPVRVAGGLLFELLGQSAGDPFIQEDDIPIVLRSWQAQNFLVTALHVKGFALNISVLGIT 226
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EVQE+L GG +PRTVHE+IA LACRL+RWDDR R YED
Sbjct: 227 EVQEMLIAGGACIPRTVHELIAHLACRLARWDDR--------------------RLYEDP 266
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF ILNQEIR+LSTQVIRVKWSLHAREEIVFELLQ LKG + LLEGI+KSTR+MI
Sbjct: 267 NLFTTILNQEIRRLSTQVIRVKWSLHAREEIVFELLQQLKGNRTKDLLEGIKKSTRDMIN 326
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV Q+TVRAWLQD+SL VTHNL +F GVG+++TI+TGLFGINVDGIP
Sbjct: 327 EQEAVRGRLFTIQDVMQNTVRAWLQDQSLTVTHNLGIFGGVGLLITIVTGLFGINVDGIP 386
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA+ P AF LF+ +L G VLVV GL+YLGLK+P+AEE VE RKLEL E+VK FQ EA
Sbjct: 387 GAKDFPQAFALFSVVLFVSGLVLVVAGLIYLGLKEPVAEENVETRKLELDEMVKKFQQEA 446
Query: 301 ETHAQVRKNVSRN 313
E+HAQV K V +N
Sbjct: 447 ESHAQVCKKVPQN 459
>ref|NP_917526.1| P0518F01.15 [Oryza sativa (japonica cultivar-group)]
Length = 506
Score = 338 bits (868), Expect = 1e-91
Identities = 171/286 (59%), Positives = 213/286 (73%), Gaps = 36/286 (12%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP +E+DIPIVLR+WQAQNFL+T +H+KGS +NV+G+
Sbjct: 234 VPVRVAGGLLFELLGQSVGDPARDEEDIPIVLRAWQAQNFLITALHVKGSAPNVNVIGVT 293
Query: 61 EVQELLSGGGYN--VPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYE 118
EVQELLS G P+ + EVIA LA RL+RWDDRL+RK +FG ADEIELKF+NR
Sbjct: 294 EVQELLSACGSTGTAPKNIQEVIAHLASRLARWDDRLWRKYVFGAADEIELKFVNR---- 349
Query: 119 DLNLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREM 178
EEI+FELL++L G +SLLE I+K R+M
Sbjct: 350 ------------------------------EEIIFELLKYLGGSTTKSLLEAIKKDARQM 379
Query: 179 IEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDG 238
IEEQEAVRGRLFTIQDV QST+RAW Q++SLR+THNL +F G G+VL+II GLFGINVDG
Sbjct: 380 IEEQEAVRGRLFTIQDVMQSTLRAWSQEKSLRITHNLTIFGGCGLVLSIIAGLFGINVDG 439
Query: 239 IPGAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEV 284
IPGAE+TPYAF LF+A+L +G +L++VG+VY GL+KPI++E+V++
Sbjct: 440 IPGAENTPYAFALFSALLFLVGLLLIIVGIVYFGLQKPISDEQVQL 485
>gb|EAM70458.1| Mg2+ transporter protein, CorA-like [Desulfuromonas acetoxidans DSM
684]
Length = 325
Score = 62.4 bits (150), Expect = 2e-08
Identities = 72/263 (27%), Positives = 120/263 (45%), Gaps = 30/263 (11%)
Query: 8 GLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIAEVQE-LL 66
G L L G + ED + LR W + ++T+ S + I ++Q+ +
Sbjct: 82 GALVGLRGVNKNPDEAMEDMVS--LRVWLDPDRIITL-------SNKPMASIRDIQQQFV 132
Query: 67 SGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDLNLFVII 126
G G P+T + ++L RL + ++ I DE+EL DL +
Sbjct: 133 QGRG---PKTTQGLFSELCDRLEWYVSQM----IDDYEDEVELI-----EQSDLAEDHLR 180
Query: 127 LNQEIRKLSTQVIRVKWSLHAREEIVFEL-LQHLKGIGARSL--LEGIRKSTREMIEEQE 183
+ I L +V+ + L + + + L ++ R+L L+ IR + +E+ +
Sbjct: 181 RREAISNLRRRVVTPRRYLAPQRDALSHLSVEPPSWFNERTLMHLKEIRNRLQRHMEDLD 240
Query: 184 AVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIPGAE 243
AVR R +Q+ Q+ V L R + +FA + + L+ TGLFG+N+ GIPGA
Sbjct: 241 AVRDRTVIVQEELQAQVSDQLNARMYIIN----IFAAIFLPLSFFTGLFGVNLGGIPGA- 295
Query: 244 HTPYAFGLFTAILVFIGAVLVVV 266
H+ AF LF L+ IG L+VV
Sbjct: 296 HSAVAFLLFCFCLLVIGVGLMVV 318
>ref|YP_165047.1| magnesium transporter, CorA family [Silicibacter pomeroyi DSS-3]
gi|56680687|gb|AAV97352.1| magnesium transporter, CorA
family [Silicibacter pomeroyi DSS-3]
Length = 322
Score = 62.0 bits (149), Expect = 2e-08
Identities = 48/144 (33%), Positives = 76/144 (52%), Gaps = 10/144 (6%)
Query: 127 LNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKS---TREMIEEQE 183
++ + L VI+++ LH + E + LL++ G+ + L +R++ TR IEE +
Sbjct: 178 MHDRLSALRLTVIKLRRYLHPQREALDALLRNEAGLFDSAGLVLLREAVDRTRRTIEELD 237
Query: 184 AVRGRLFTIQDVTQSTVRAWLQDRSL-RVTHNLAVFAGVGMVLTIITGLFGINVDGIPGA 242
A R RL +QD S Q R L R + L+V A V + L +TGLFG+N+ G+PG
Sbjct: 238 ATRDRLAAVQDQIASE-----QVRLLGRNSFVLSVVAAVFLPLGFLTGLFGVNIAGMPGT 292
Query: 243 EHTPYAFGLFTAILVFIGAVLVVV 266
TP AF L +G +V++
Sbjct: 293 -GTPLAFWLLAGASALLGLAIVLL 315
>gb|AAF95478.1| conserved hypothetical protein [Vibrio cholerae O1 biovar eltor
str. N16961] gi|15642332|ref|NP_231965.1| hypothetical
protein VC2334 [Vibrio cholerae O1 biovar eltor str.
N16961] gi|11354697|pir||D82090 conserved hypothetical
protein VC2334 [imported] - Vibrio cholerae (strain
N16961 serogroup O1)
Length = 327
Score = 52.0 bits (123), Expect = 2e-05
Identities = 65/275 (23%), Positives = 118/275 (42%), Gaps = 47/275 (17%)
Query: 9 LLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFL-------------------VTVMHIKG 49
LL + + S D L+ ED P+ + QNFL V +++ G
Sbjct: 58 LLDQNVPSSVIDSLLAEDTRPL-FEQYDEQNFLLILRGVNLNDNADPADMLSVRILYYHG 116
Query: 50 ---SVSRINVLGIAEVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADE 106
S + I+ V+ L G P + E+I ++ L+ K+I D+
Sbjct: 117 ALISTRKTPSKAISTVRTALEAG--QGPSNLAELIVRVVDELN--------KNIDLYLDQ 166
Query: 107 IELKFMNRRNYEDLNLFVIILNQEIRKLSTQVIRVKWSL--HAREEIVFELLQHLKGIGA 164
IE + +L+ ++ ++ + KL + +++L + E+ QHL+ +
Sbjct: 167 IEERITQFDEELELSNELMQTHKSLLKLKRFIKPQQYALDDYLNSEVALAQSQHLRLRHS 226
Query: 165 RSLLEGIRKSTREMIEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMV 224
+ + I ++ + E E ++G L R + ++ + T+ +V A + +
Sbjct: 227 VNTVTRINETLDFFLSELEIIKGEL-----------RQYHAEKMNQNTYLFSVIAAIFLP 275
Query: 225 LTIITGLFGINVDGIPGAEHTPYAFGLFTAILVFI 259
+ +TGL G+NV GIPG E +P AFGLF A L I
Sbjct: 276 TSFLTGLLGVNVGGIPGTE-SPIAFGLFCAGLGLI 309
>ref|ZP_00473745.1| Mg2+ transporter protein, CorA-like [Chromohalobacter salexigens
DSM 3043] gi|67519016|gb|EAM22975.1| Mg2+ transporter
protein, CorA-like [Chromohalobacter salexigens DSM
3043]
Length = 324
Score = 49.3 bits (116), Expect = 2e-04
Identities = 33/95 (34%), Positives = 51/95 (52%), Gaps = 6/95 (6%)
Query: 166 SLLEGIRKSTREMIEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVL 225
SL E + +R +E+ A++ R +Q+ S L R + LA+ V + L
Sbjct: 223 SLRENANQLSR-YVEDFHAMQERALILQEQIWSEHNEQLNQRM----YMLAIITTVFLPL 277
Query: 226 TIITGLFGINVDGIPGAEHTPYAFGLFTAILVFIG 260
+ +TGL GINV GIPGA+ +PY F +F I + +G
Sbjct: 278 SFLTGLLGINVGGIPGAD-SPYGFSVFITITLLMG 311
>ref|YP_155451.1| Mg/Co transporter [Idiomarina loihiensis L2TR]
gi|56179180|gb|AAV81902.1| Mg/Co transporter [Idiomarina
loihiensis L2TR]
Length = 313
Score = 49.3 bits (116), Expect = 2e-04
Identities = 42/170 (24%), Positives = 85/170 (49%), Gaps = 30/170 (17%)
Query: 106 EIELKFMNRRNYEDLNLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGAR 165
+I+L ++RR LN F+ + KLST+ +V S + ++ E
Sbjct: 170 QIKLTALHRRLLR-LNRFIRPQLAALEKLSTETEKVM-STELHQWLINE----------- 216
Query: 166 SLLEGIRKSTREMIEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVL 225
R +T+ + E+ + + +++ ++ Q + ++ R T+ L+V AGV + L
Sbjct: 217 ------RDTTQRLFEQIDLMLEQVWMQREQLQQAIA----EKMNRNTYWLSVIAGVFLPL 266
Query: 226 TIITGLFGINVDGIPGAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKK 275
+ +TG+FGIN+ G+PG E A ++F GA++++ + +L L++
Sbjct: 267 SFLTGVFGINIGGMPGVESE-------GAFMIFCGALVIIAIVEFLLLRR 309
>ref|ZP_00288640.1| COG0598: Mg2+ and Co2+ transporters [Magnetococcus sp. MC-1]
Length = 326
Score = 48.9 bits (115), Expect = 2e-04
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 8/143 (5%)
Query: 127 LNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIEEQEAVR 186
L +E+ L +VI ++ L A ++I LL L +G + RE+ E +
Sbjct: 182 LREELLMLRRKVINLRRYL-APQKIALALLSKSP---PSWLNKGRTRQIREVEHELQRYL 237
Query: 187 GRLFTIQD---VTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIPGAE 243
G L + ++ V + + + R R + L++ G+ + L +TGL GINV G+PGA+
Sbjct: 238 GDLESYRERAVVIREEINNQMNVRMNRAMYMLSLITGIFLPLGFLTGLLGINVGGMPGAD 297
Query: 244 HTPYAFGLFTAILVFIGAVLVVV 266
+ P+AF L +++ + A+ + V
Sbjct: 298 N-PWAFWLVCLLMIAVTAINLFV 319
>ref|ZP_00318329.1| COG0598: Mg2+ and Co2+ transporters [Microbulbifer degradans 2-40]
Length = 322
Score = 48.1 bits (113), Expect = 4e-04
Identities = 41/120 (34%), Positives = 59/120 (49%), Gaps = 12/120 (10%)
Query: 154 ELLQHLKGIGARSLLEGIRKSTRE-------MIEEQEAVRGRLFTIQDVTQSTVRAWLQD 206
E L L G A L E R RE ++E ++ R R +Q+ + + +
Sbjct: 201 EALSRLCGEQAPWLTETHRIGLRETSDHLLRLLENLDSARDRAAVVQEELVNHMSEQMNQ 260
Query: 207 RSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIPGAEHTPYAFGLFTAILVFIGAVLVVV 266
R + L++ A + + L +TGL GINV GIPGAE+ AF F AIL + A+ VVV
Sbjct: 261 RM----YVLSLVAAIFLPLGFLTGLLGINVGGIPGAENN-QAFFWFIAILCSLTALQVVV 315
>ref|ZP_00267813.1| COG0598: Mg2+ and Co2+ transporters [Rhodospirillum rubrum]
Length = 346
Score = 47.0 bits (110), Expect = 8e-04
Identities = 26/93 (27%), Positives = 49/93 (51%), Gaps = 5/93 (5%)
Query: 165 RSLLEGIRKSTREMIEEQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMV 224
R +L + +E+ +++R + ++D Q Q R R + + + AG+ +
Sbjct: 243 REILGAALATFTRYVEDLDSLREQAALLRDRIQQRA----QQRINRSIYKMTLLAGIFLP 298
Query: 225 LTIITGLFGINVDGIPGAEHTPYAFGLFTAILV 257
LT TGL G+N+ G+PGA+ +P+ F + +LV
Sbjct: 299 LTFFTGLLGVNLGGMPGAQ-SPFGFWILVGLLV 330
>ref|NP_719514.1| magnesium transporter, putative [Shewanella oneidensis MR-1]
gi|24350324|gb|AAN56958.1| magnesium transporter,
putative [Shewanella oneidensis MR-1]
Length = 321
Score = 47.0 bits (110), Expect = 8e-04
Identities = 37/143 (25%), Positives = 76/143 (52%), Gaps = 8/143 (5%)
Query: 127 LNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMI---EEQE 183
L +I +L Q + ++ L ++E ++L + +R++T +I E+ +
Sbjct: 177 LRTDIAELRRQTVALRRYLGPQKEAFAKMLSEQFVLFNEPEKLKMRETTNNLIRTIEDLD 236
Query: 184 AVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIPGAE 243
A+R R +VTQ + + ++ + + L++ + + + L +TGL G+N+ GIPGA+
Sbjct: 237 ALRDRA----NVTQEELLSQQSEQLNKRLYFLSLVSVIFLPLGFLTGLLGVNIGGIPGAD 292
Query: 244 HTPYAFGLFTAILVFIGAVLVVV 266
+ +AF F ILV + A+ +V+
Sbjct: 293 NN-FAFTSFCIILVSLVALQMVI 314
>ref|ZP_00275592.1| COG0598: Mg2+ and Co2+ transporters [Ralstonia metallidurans CH34]
Length = 346
Score = 46.2 bits (108), Expect = 0.001
Identities = 31/104 (29%), Positives = 53/104 (50%), Gaps = 7/104 (6%)
Query: 167 LLEGIRKSTREMIEEQEAVRGRLFTIQD---VTQSTVRAWLQDRSLRVTHNLAVFAGVGM 223
LLEG + RE EE V L + + + Q + A L +++ R L + + +
Sbjct: 238 LLEGDLRELRESTEEFSLVLNDLAALVERIKLLQEEIAAKLNEQTNRTLFTLTLVTVLAL 297
Query: 224 VLTIITGLFGINVDGIPGAEHTPYAFGLFTAILVFIGAVLVVVG 267
+ I+ G FG+NV GIP A+H P+ F + ++V + + V+ G
Sbjct: 298 PINIVAGFFGMNVGGIPLADH-PHGFWV---LVVLVASFTVLAG 337
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.141 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 520,635,478
Number of Sequences: 2540612
Number of extensions: 19976431
Number of successful extensions: 64636
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 64565
Number of HSP's gapped (non-prelim): 118
length of query: 332
length of database: 863,360,394
effective HSP length: 128
effective length of query: 204
effective length of database: 538,162,058
effective search space: 109785059832
effective search space used: 109785059832
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0066.1


