

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.6
(396 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
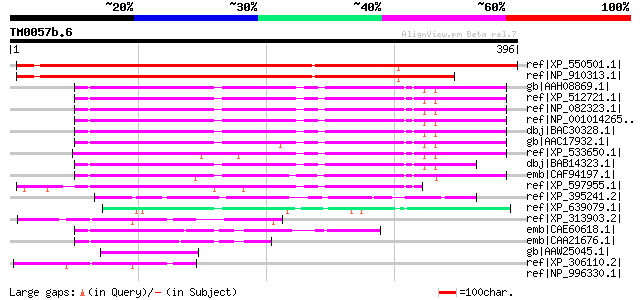
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_550501.1| unknown protein [Oryza sativa (japonica cultiva... 402 e-110
ref|NP_910313.1| Similar to Homo sapiens chromosome 19, cosmid F... 337 3e-91
gb|AAH08869.1| FLJ12886 protein [Homo sapiens] gi|12052738|emb|C... 137 8e-31
ref|XP_512721.1| PREDICTED: similar to hypothetical protein [Pan... 137 8e-31
ref|NP_082323.1| hypothetical protein LOC71997 [Mus musculus] gi... 136 1e-30
ref|NP_001014265.1| hypothetical protein LOC365215 [Rattus norve... 135 2e-30
dbj|BAC30328.1| unnamed protein product [Mus musculus] 133 8e-30
gb|AAC17932.1| F17127_1 [Homo sapiens] gi|10092659|ref|NP_061981... 127 8e-28
ref|XP_533650.1| PREDICTED: similar to hypothetical protein [Can... 125 3e-27
dbj|BAB14323.1| unnamed protein product [Homo sapiens] 118 3e-25
emb|CAF94197.1| unnamed protein product [Tetraodon nigroviridis] 115 3e-24
ref|XP_597955.1| PREDICTED: similar to RIKEN cDNA 1500002O20, pa... 99 3e-19
ref|XP_395241.2| PREDICTED: hypothetical protein XP_395241 [Apis... 91 6e-17
ref|XP_639079.1| hypothetical protein DDB0185525 [Dictyostelium ... 71 5e-11
ref|XP_313903.2| ENSANGP00000021895 [Anopheles gambiae str. PEST... 57 1e-06
emb|CAE60618.1| Hypothetical protein CBG04259 [Caenorhabditis br... 57 1e-06
emb|CAA21676.1| Hypothetical protein Y54E2A.2 [Caenorhabditis el... 54 8e-06
gb|AAW25045.1| unknown [Schistosoma japonicum] 54 8e-06
ref|XP_306110.2| ENSANGP00000015543 [Anopheles gambiae str. PEST... 50 2e-04
ref|NP_996330.1| CG3857-PA [Drosophila melanogaster] gi|45446781... 43 0.015
>ref|XP_550501.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|55295893|dbj|BAD67761.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|55296185|dbj|BAD67903.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 412
Score = 402 bits (1032), Expect = e-110
Identities = 212/394 (53%), Positives = 276/394 (69%), Gaps = 9/394 (2%)
Query: 6 PSPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHRSRLPSV-ASLNLLSDSWDFHID 64
P P PKILLAKP L P G G RSR + SL+L+SD+W+ H D
Sbjct: 25 PPPPPKILLAKPPL----PPPSSSGADDDGGGGGGAGRSRQATQPGSLSLVSDAWEVHTD 80
Query: 65 RFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAMARHC 124
+ LP+LTEN DF VIGVIGP GVGKSTIMNELYG+D SSPGMLPPFATQ+EE +AMA+HC
Sbjct: 81 KILPYLTENNDFMVIGVIGPTGVGKSTIMNELYGYDGSSPGMLPPFATQTEEIKAMAKHC 140
Query: 125 STGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSAELAHEIMGIQL 184
+ GI+ RIS ER+ILLDTQPVFS SVL +MM+PDGSS + +LSG+ +SA+LAHE+MGIQL
Sbjct: 141 TAGIDIRISNERVILLDTQPVFSPSVLIDMMKPDGSSAIPILSGDPLSADLAHELMGIQL 200
Query: 185 AVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNK 244
V LASVC+ILLVVSEG++D MW L+LTVDLLKH IPDPSLL SS +Q ++ DN+
Sbjct: 201 GVFLASVCNILLVVSEGINDLSMWDLILTVDLLKHNIPDPSLLTSSTTQDKE--NKNDNQ 258
Query: 245 LSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISS-- 302
+Y+A FVH +L +Q+F+P + L++ L ++FK SSF + P+ SS
Sbjct: 259 SGIEDYIADLCFVHARLREQDFSPSKLMVLKRVLEKHFKSSSFSIGSSGATPQVSDSSVP 318
Query: 303 SMVHDSPMDSNVPSLFAIPFKKKDENPRAQHGSYISALWKLRDQILSMKSPSFTRPVSER 362
S + + SN ++ +P + D + ++ + S L LRDQILS S SF++ ++ER
Sbjct: 319 SSMKIEDLSSNQQDIYLLPLRTPDNSTNFEYRTCPSMLGMLRDQILSRPSRSFSKNLTER 378
Query: 363 EWLKNSAKIWEQVKNSPTMLEYCRTLQQSGMFRR 396
+WL++SAKIW+ VK SP +LEYC+TLQ SG FR+
Sbjct: 379 DWLRSSAKIWDMVKKSPVILEYCKTLQDSGFFRK 412
>ref|NP_910313.1| Similar to Homo sapiens chromosome 19, cosmid F17127; hypothetical
59 kDa protein (AC004780) [Oryza sativa (japonica
cultivar-group)]
Length = 377
Score = 337 bits (865), Expect = 3e-91
Identities = 182/345 (52%), Positives = 236/345 (67%), Gaps = 9/345 (2%)
Query: 6 PSPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHRSRLPSV-ASLNLLSDSWDFHID 64
P P PKILLAKP L P G G RSR + SL+L+SD+W+ H D
Sbjct: 25 PPPPPKILLAKPPL----PPPSSSGADDDGGGGGGAGRSRQATQPGSLSLVSDAWEVHTD 80
Query: 65 RFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAMARHC 124
+ LP+LTEN DF VIGVIGP GVGKSTIMNELYG+D SSPGMLPPFATQ+EE +AMA+HC
Sbjct: 81 KILPYLTENNDFMVIGVIGPTGVGKSTIMNELYGYDGSSPGMLPPFATQTEEIKAMAKHC 140
Query: 125 STGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSAELAHEIMGIQL 184
+ GI+ RIS ER+ILLDTQPVFS SVL +MM+PDGSS + +LSG+ +SA+LAHE+MGIQL
Sbjct: 141 TAGIDIRISNERVILLDTQPVFSPSVLIDMMKPDGSSAIPILSGDPLSADLAHELMGIQL 200
Query: 185 AVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNK 244
V LASVC+ILLVVSEG++D MW L+LTVDLLKH IPDPSLL SS +Q ++ DN+
Sbjct: 201 GVFLASVCNILLVVSEGINDLSMWDLILTVDLLKHNIPDPSLLTSSTTQDKE--NKNDNQ 258
Query: 245 LSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISS-- 302
+Y+A FVH +L +Q+F+P + L++ L ++FK SSF + P+ SS
Sbjct: 259 SGIEDYIADLCFVHARLREQDFSPSKLMVLKRVLEKHFKSSSFSIGSSGATPQVSDSSVP 318
Query: 303 SMVHDSPMDSNVPSLFAIPFKKKDENPRAQHGSYISALWKLRDQI 347
S + + SN ++ +P + D + ++ + S L LRDQ+
Sbjct: 319 SSMKIEDLSSNQQDIYLLPLRTPDNSTNFEYRTCPSMLGMLRDQV 363
>gb|AAH08869.1| FLJ12886 protein [Homo sapiens] gi|12052738|emb|CAB66541.1|
hypothetical protein [Homo sapiens]
Length = 520
Score = 137 bits (344), Expect = 8e-31
Identities = 104/358 (29%), Positives = 171/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRTYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSSSG ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSSGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +DS V +LF +PF + + EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDSEV-NLFLVPFMDSEAESENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ V+ S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVRKSSALAEYSRLL 519
>ref|XP_512721.1| PREDICTED: similar to hypothetical protein [Pan troglodytes]
Length = 636
Score = 137 bits (344), Expect = 8e-31
Identities = 104/358 (29%), Positives = 171/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 296 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRTYVF 354
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 355 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 409
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 410 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 464
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSSSG ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 465 HESSSSSGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 519
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +DS V +LF +PF + + EN PRA
Sbjct: 520 GTLSMLQCNVFPGLPPDF-LDSEV-NLFLVPFMDSEAESENPPRAGPGSSPLFSLLPGYR 577
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ V+ S + EY R L
Sbjct: 578 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVRKSSALAEYSRLL 635
>ref|NP_082323.1| hypothetical protein LOC71997 [Mus musculus]
gi|21707691|gb|AAH34211.1| RIKEN cDNA 1500002O20 [Mus
musculus] gi|12836835|dbj|BAB23829.1| unnamed protein
product [Mus musculus]
Length = 520
Score = 136 bits (342), Expect = 1e-30
Identities = 103/358 (28%), Positives = 172/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSS+G ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +D+ V +LF +PF + ++EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ VK S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVKKSSALAEYSRLL 519
>ref|NP_001014265.1| hypothetical protein LOC365215 [Rattus norvegicus]
gi|56268844|gb|AAH87051.1| Hypothetical LOC365215
[Rattus norvegicus]
Length = 520
Score = 135 bits (340), Expect = 2e-30
Identities = 103/358 (28%), Positives = 172/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SS++G ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSAAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ +I + D +D+ V +LF +PF + ++EN PRA
Sbjct: 404 GTLSMLQCNIFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ VK S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVKKSSALAEYSRLL 519
>dbj|BAC30328.1| unnamed protein product [Mus musculus]
Length = 520
Score = 133 bits (335), Expect = 8e-30
Identities = 102/358 (28%), Positives = 171/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSS+G ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +D+ V +LF +PF + ++EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + + + W +A+IW+ VK S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTGKNWFHYAARIWDGVKKSSALAEYSRLL 519
>gb|AAC17932.1| F17127_1 [Homo sapiens] gi|10092659|ref|NP_061981.1| hypothetical
protein LOC56006 [Homo sapiens]
Length = 528
Score = 127 bits (318), Expect = 8e-28
Identities = 105/377 (27%), Positives = 172/377 (44%), Gaps = 57/377 (15%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 169 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRTYVF 227
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 228 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 282
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHL-------------------M 211
E+ +Q+A L +VCH+++VV + D ++ L +
Sbjct: 283 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRLWDLGCKCKSNSHSPQTPRFL 342
Query: 212 LTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNF 271
T +++K P P SH SSSSG ++ G EY VF+ K ++F P+
Sbjct: 343 QTAEMVKPSTPSP-----SHESSSSSGSDE-----GTEYYPHLVFLQNKARREDFCPRKL 392
Query: 272 VQLRKTLIQYFKPSSFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN 328
Q+ + Q S + + ++ + D +DS V +LF +PF + + EN
Sbjct: 393 RQMHLMIDQLMAHSHLRYKGTLSMLQCNVFPGLPPDF-LDSEV-NLFLVPFMDSEAESEN 450
Query: 329 -PRA---------------QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKI 371
PRA H S+ S + KLR Q++SM P + + +E+ W +A+I
Sbjct: 451 PPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARI 510
Query: 372 WEQVKNSPTMLEYCRTL 388
W+ V+ S + EY R L
Sbjct: 511 WDGVRKSSALAEYSRLL 527
>ref|XP_533650.1| PREDICTED: similar to hypothetical protein [Canis familiaris]
Length = 912
Score = 125 bits (313), Expect = 3e-27
Identities = 107/388 (27%), Positives = 174/388 (44%), Gaps = 61/388 (15%)
Query: 50 ASLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPP 109
+++ L+D + H F +L + TD V+GV+G G GKS +M+ L
Sbjct: 536 SAIEPLTDLRESHAFIFPQYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYV 595
Query: 110 FATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSA-------------SVLAEMMR 156
F QS E + + ++GI+ I+ ERI+ LDTQ S VLA
Sbjct: 596 FRAQSAEMKERGGNQTSGIDFFITQERIVFLDTQVPTSTCLTLLHATLSSTPGVLATSPF 655
Query: 157 PDGSSTVSVLSGESMSAELAH----------------EIMGIQLAVLLASVCHILLVVSE 200
PD S++ + S+ L + E+ +Q+A L +VCH+++VV +
Sbjct: 656 PDTFSSLQPILSPSILDHLINNDRKLPPEYNLPHTYVEMQSLQIAAFLFTVCHVVIVVQD 715
Query: 201 GVHDDCMWHLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTK 260
D ++ + T +++K P P SH SSSSG ++ G EY VF+ K
Sbjct: 716 WFTDLSLYRFLQTAEMVKPSTPSP-----SHESSSSSGSDE-----GTEYYPHLVFLQNK 765
Query: 261 LHDQEFTPKNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAI 320
++F P+ Q+ + Q S + + ++ + D +DS V +LF +
Sbjct: 766 ARREDFCPRKLRQMHLMIDQLMAHSHLRYKGTLSMLQCNVFPGLPPDF-LDSEV-NLFLV 823
Query: 321 PF---KKKDEN-PRA---------------QHGSYISALWKLRDQILSMKSPSFTRPV-S 360
PF + + EN PRA H S+ S + KLR Q++SM P + + +
Sbjct: 824 PFMDSEAESENPPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSMARPQLSHTILT 883
Query: 361 EREWLKNSAKIWEQVKNSPTMLEYCRTL 388
E+ W +A+IW+ VK S + EY R L
Sbjct: 884 EKNWFHYAARIWDGVKKSSALAEYSRLL 911
>dbj|BAB14323.1| unnamed protein product [Homo sapiens]
Length = 495
Score = 118 bits (296), Expect = 3e-25
Identities = 95/334 (28%), Positives = 157/334 (46%), Gaps = 38/334 (11%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRTYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSSSG ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSSGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +DS V +LF +PF + + EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDSEV-NLFLVPFMDSEAESENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREW 364
H S+ S + KLR Q++SM P + + +E+ W
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNW 495
>emb|CAF94197.1| unnamed protein product [Tetraodon nigroviridis]
Length = 511
Score = 115 bits (287), Expect = 3e-24
Identities = 96/362 (26%), Positives = 164/362 (44%), Gaps = 42/362 (11%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GVIG G GKST+M+ L F
Sbjct: 168 SIKLVDDQMNW-CDSAMEYLRDQTDMLVVGVIGLQGAGKSTVMSLLSANTPEEDQRGYVF 226
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQ-----PVFSASVLAEMMRPDGSSTVSV 165
Q++E + + STGI+ I+ ER+I LDTQ P+ S S+L ++ D
Sbjct: 227 RAQAQEIKERGGNQSTGIDFFITQERVIFLDTQASNTHPILSPSILDHLINNDRK----- 281
Query: 166 LSGESMSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPS 225
L E E +Q+A L +VCH++++V + D ++ + T ++LK PS
Sbjct: 282 LPPEYNLPHTYVETQSLQIAAFLFTVCHVVILVQDWFTDVNLYRFLQTAEMLK-----PS 336
Query: 226 LLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPS 285
++SH + SSG + G EY VFV K +F P+N + + + S
Sbjct: 337 TPSTSHDSTGSSGTD-----DGAEYYPHIVFVQNKARLDDFCPRNLKNMHLVVDKLMAHS 391
Query: 286 SFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPFKKKDENPRAQ------------- 332
+ + +I + D + + V ++F +P + D +
Sbjct: 392 HLKYKGTLSMLDCNIFPGLGPDY-LTTEV-NMFLLPVQDSDSDDGPTKAAPGTYPLFSLL 449
Query: 333 -----HGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCR 386
H ++ + + KLR +IL+M + + +E+ W +A+IW+ VK S + EY R
Sbjct: 450 PGYKGHPTFSTTVSKLRSRILAMPRCQLSHTILTEKNWFHYAARIWDGVKKSSALSEYSR 509
Query: 387 TL 388
L
Sbjct: 510 LL 511
>ref|XP_597955.1| PREDICTED: similar to RIKEN cDNA 1500002O20, partial [Bos taurus]
Length = 482
Score = 98.6 bits (244), Expect = 3e-19
Identities = 95/359 (26%), Positives = 154/359 (42%), Gaps = 63/359 (17%)
Query: 6 PSPSP---KILLAKPGLVTGGPVAGK--FGRGAAGEDDSAQHRSRLPSVASLNLLSDSWD 60
P+P+P I+L KP GP A G+ + +H S+ L+ D +
Sbjct: 132 PAPAPLEKPIVLMKPREEGKGPAATTTVVGQAKLLPPERMKH--------SIKLVDDQMN 183
Query: 61 FHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAM 120
+ D + +L + TD V+GV+G G GKS +M+ L F QS E +
Sbjct: 184 W-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVFRAQSAEMKER 242
Query: 121 ARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPD---------------------- 158
+ ++GI+ I+ ERI+ LDTQP+ S S+L ++ D
Sbjct: 243 GGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRKLPPEYNLPHTYVEMQVRGPS 302
Query: 159 -------GSSTVSVLSGESMSAELAHEIMG--------IQLAVLLASVCHILLVVSEGVH 203
SS V V A + E++G +Q+A L +VCH+++VV +
Sbjct: 303 QAWGPGAESSWVWVCHFYLQRALVPRELVGEDGIKSRSLQIAAFLFTVCHVVIVVQDWFT 362
Query: 204 DDCMWHLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHD 263
D ++ + T +++K P P SH SSSSG E+ G EY VF+ K
Sbjct: 363 DLSLYRFLQTAEMVKPSTPSP-----SHESSSSSGSEE-----GAEYYPHLVFLQNKARR 412
Query: 264 QEFTPKNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF 322
++F P+ Q+ + Q S + + ++ + D +DS V +LF +PF
Sbjct: 413 EDFCPRKLRQMHLMIDQLMAHSHLRYKGTLSMLQCNVFPGLPPDF-LDSEV-NLFLMPF 469
>ref|XP_395241.2| PREDICTED: hypothetical protein XP_395241 [Apis mellifera]
Length = 359
Score = 90.9 bits (224), Expect = 6e-17
Identities = 81/299 (27%), Positives = 137/299 (45%), Gaps = 40/299 (13%)
Query: 67 LPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAMARHCST 126
L FL + DF V+G +G GVGKSTIM+ L + + G+ P T E+ A +C+
Sbjct: 100 LEFLYDQQDFLVVGCLGAQGVGKSTIMSLL--TCNIATGIFPIQDTSHHESGA---NCTI 154
Query: 127 GIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSAELAHEIMGIQLAV 186
GI+ ++ R+I LDTQP+ S S + D + + ++ E E+ Q A
Sbjct: 155 GIDFFVTKNRVIYLDTQPILSGSTIDYSTTYDQKKPTT----DFINIETNLELQSSQFAA 210
Query: 187 LLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNKLS 246
L SVCH+++ V + D + + T ++LK SS+S ++D
Sbjct: 211 FLFSVCHVIIFVQDWFVDPNLVRFLQTAEMLK--------------PSSTSNMDQDYV-- 254
Query: 247 GREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISSSMVH 306
EY VF+H K Q+FTP N ++++ + F SS ++ H+ H
Sbjct: 255 --EYYPHIVFLHNKAELQDFTPHNMEKVKEFYTKLFS-SSRLQIHSGLDMSNH------- 304
Query: 307 DSPMDSNVPSLFAIPFKKKDENPRAQHGSYISALWKLRDQILSMKSPSFT-RPVSEREW 364
M++ + F ++E Q+G + + KL+ +I + S T ++E+ W
Sbjct: 305 --SMEAGLNLFFIPRITNEEEFTMRQNGKKL--IEKLKTKIHGVNRNSMTPSTLTEKNW 359
>ref|XP_639079.1| hypothetical protein DDB0185525 [Dictyostelium discoideum]
gi|60467689|gb|EAL65708.1| hypothetical protein
DDB0185525 [Dictyostelium discoideum]
Length = 690
Score = 71.2 bits (173), Expect = 5e-11
Identities = 97/422 (22%), Positives = 164/422 (37%), Gaps = 120/422 (28%)
Query: 73 NTDFTVIGVIGPPGVGKSTIMNELY--GFDSS---------------------------- 102
NT FTVIGVIG GKST+++ L D+S
Sbjct: 182 NTQFTVIGVIGGQSCGKSTLLSSLIKRSHDNSQTQQQQQQTNDSTTTTTTTSNTTSTTTT 241
Query: 103 ----------SPGMLPPFATQSEENRAMARHCSTGIEPRISAE-RIILLDTQPVFSASVL 151
S F Q++ N + +H + GI+ +S E RII LDTQP+ S S++
Sbjct: 242 PSSSSSTTTVSNNNAKLFEIQTDNNFGLGKHQTIGIDLFVSEEERIIYLDTQPLQSLSII 301
Query: 152 AEMMRPDGSSTVSVLSGESMSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLM 211
EM+ + L + S E H + ++L V L S+C+++LVV E D + +
Sbjct: 302 CEMIEDNFH-----LPNQFNSYEQYHHMQCLRLIVYLLSICNVVLVVQENKSLDIPFCKL 356
Query: 212 LTVD--LLKHGIPDPSLLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQE---- 265
L +LK+ PD S+ SQ+ + +++ L E A +F+ KL + +
Sbjct: 357 LRSASLILKNKTPD----ISNPSQTGLPYNFENDIL---ESTARQLFIFNKLTEIDTNET 409
Query: 266 ----------FTPKNFVQ------------------------------------------ 273
PKN Q
Sbjct: 410 CFDKSLSVLFSNPKNLYQSFISNLSNNNNNNNSTNSSNNSNMSGSGSGSGDQQQQNTIGS 469
Query: 274 ---LRKTLIQYFKPSSFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPFKKKDENPR 330
L +F P +++E A + E + V+ S D N ++ +++ +
Sbjct: 470 KYQLMNHFDSFFIPDRYLQEIANNATTEILG---VNSS--DINPNTINGTKKRQQQQTNT 524
Query: 331 AQHGSYISALWKLRDQILSMKSPS-FTRPVSEREWLKNSAKIWEQVKNSPTMLEYCRTLQ 389
++ ++ L +L M+S S FT +SER+WL+N+ +W +++ + Y L
Sbjct: 525 KLQPTFNESIEYLHKHLLLMRSVSKFTHEISERQWLENAQNLWNYLQDCDLLSSYNHQLS 584
Query: 390 QS 391
+S
Sbjct: 585 KS 586
>ref|XP_313903.2| ENSANGP00000021895 [Anopheles gambiae str. PEST]
gi|55240424|gb|EAA09364.2| ENSANGP00000021895 [Anopheles
gambiae str. PEST]
Length = 384
Score = 57.0 bits (136), Expect = 1e-06
Identities = 63/218 (28%), Positives = 92/218 (41%), Gaps = 41/218 (18%)
Query: 7 SPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHRSRLPSVASLNLLSDSWDFHIDRF 66
S +P+I + G + + K A D S P SL LL + + R
Sbjct: 37 STTPQIAVTSKGSESRHAASAKSLPATAAPDPY----SYTPMEGSLVLLKNHNAINY-RT 91
Query: 67 LPFLTE-NTDFTVIGVIGPPGVGKSTIMN----ELYGFDSSSPGMLPPFATQSEENRAMA 121
L FL E N D+ VIG IG PGVGKST++N LY D S F S N A
Sbjct: 92 LDFLHETNQDYVVIGTIGMPGVGKSTVLNLLNTNLYAPDQSDNRKETIFPVHSTLNVAGE 151
Query: 122 RHCSTGIEPRISAERIILLD-TQPVFSASVLAEMMRPDGSSTVSVLSGESMSAELAHEIM 180
+ I+ +R+ILLD ++PVF G + + +E
Sbjct: 152 NE----VRMHITEDRLILLDCSEPVF---------------------GHARKDFIQNEQD 186
Query: 181 GIQLAVLLASVCHILLVVSEGVHD-----DCMWHLMLT 213
++ ++L +CH++LVV E ++ MW M+T
Sbjct: 187 ELKKLMILLRICHVMLVVQEEYYNIRLMRTLMWAEMMT 224
>emb|CAE60618.1| Hypothetical protein CBG04259 [Caenorhabditis briggsae]
Length = 314
Score = 56.6 bits (135), Expect = 1e-06
Identities = 62/239 (25%), Positives = 102/239 (41%), Gaps = 30/239 (12%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+D + D LT + +F VI IGP G GKSTI++ L G +S F
Sbjct: 4 SVRFLNDIGEI-TDSISDLLTNSPNFNVISAIGPQGAGKSTILSMLAGNNSRQMYREYVF 62
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
S E +RH +T I+ ++ + I LD QP+ S S++ + + G + +
Sbjct: 63 RPVSREANEQSRHQTTQIDIYVTNHQ-IFLDCQPMHSFSIMEGLPKLRGG---RLDEATA 118
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
MS L +L L + H +LV+SE D + + T + ++ +
Sbjct: 119 MSDTL-------RLTAFLLFISHTVLVISETHFDKTIIDTIRTSEQIRPWL--------- 162
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVR 289
H+ KLS E M VF+ TK + P + + L F+ S +++
Sbjct: 163 --------HQFRPKLS-IERMTNLVFIKTKASSIDTAPSVIRERAQLLRLAFRDSKWLK 212
>emb|CAA21676.1| Hypothetical protein Y54E2A.2 [Caenorhabditis elegans]
gi|17537511|ref|NP_497058.1| gluconokinase like (43.8
kD) (2P34) [Caenorhabditis elegans]
gi|7510176|pir||T27143 hypothetical protein Y54E2A.2 -
Caenorhabditis elegans
Length = 385
Score = 53.9 bits (128), Expect = 8e-06
Identities = 48/154 (31%), Positives = 70/154 (45%), Gaps = 12/154 (7%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+D + D LT + +F VI IGP G GKST+++ L G +S F
Sbjct: 62 SVRFLTDFGEIS-DAISDLLTSSPNFNVISAIGPQGAGKSTLLSMLAGNNSRQMYREYVF 120
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
S E +RH + I+ I I LD QP++S S++ + + G +
Sbjct: 121 RPVSREANEQSRHQTIQIDIYI-VNHQIFLDCQPMYSFSIMEGLPKVRGG---RFDDSTA 176
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHD 204
MS L +L L V H +LVVSE +D
Sbjct: 177 MSDTL-------RLTAFLLYVSHTVLVVSETHYD 203
>gb|AAW25045.1| unknown [Schistosoma japonicum]
Length = 193
Score = 53.9 bits (128), Expect = 8e-06
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query: 72 ENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGML-PPFATQSEENRAMARHCSTGIEP 130
EN+ F V+G IG G GKS+++N L S G+ PF Q+ ++ C+ GI
Sbjct: 65 ENSGFLVVGAIGLQGSGKSSVLNILANCISDDCGVFNAPFNVQTIKDVLTNSPCTGGINL 124
Query: 131 RISAERIILLDTQPVFS 147
I+ +R+ILLDTQP+ S
Sbjct: 125 YITQDRVILLDTQPLLS 141
>ref|XP_306110.2| ENSANGP00000015543 [Anopheles gambiae str. PEST]
gi|55246976|gb|EAA02587.3| ENSANGP00000015543 [Anopheles
gambiae str. PEST]
Length = 200
Score = 49.7 bits (117), Expect = 2e-04
Identities = 51/153 (33%), Positives = 68/153 (44%), Gaps = 15/153 (9%)
Query: 4 SEPSPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHR----SRLPSVASLNLLSDSW 59
S P +P IL + + G + A+ + A S P SL LL +
Sbjct: 26 STPRRTPTILASTTPQIAGTSKGSESRHAASAKSLPATAAPDPYSYTPMEGSLVLLKNHN 85
Query: 60 DFHIDRFLPFLTE-NTDFTVIGVIGPPGVGKSTIMN----ELYGFDSSSPGMLPPFATQS 114
+ R L FL E N D+ VIG IG PGVGKST++N LY D S F S
Sbjct: 86 AINY-RTLDFLHETNQDYVVIGTIGMPGVGKSTVLNLLNTNLYAPDQSDNRKETIFPVHS 144
Query: 115 EENRAMARHCSTGIEPRISAERIILLD-TQPVF 146
N A + I+ +R+ILLD ++PVF
Sbjct: 145 TINVAGENE----VRMHITEDRLILLDCSEPVF 173
>ref|NP_996330.1| CG3857-PA [Drosophila melanogaster] gi|45446781|gb|AAS65248.1|
CG3857-PA [Drosophila melanogaster]
gi|15291511|gb|AAK93024.1| GH24078p [Drosophila
melanogaster] gi|2661500|emb|CAA15683.1| EG:39E1.3
[Drosophila melanogaster] gi|41616158|tpg|DAA03173.1|
TPA: HDC17028 [Drosophila melanogaster]
Length = 487
Score = 43.1 bits (100), Expect = 0.015
Identities = 43/149 (28%), Positives = 65/149 (42%), Gaps = 37/149 (24%)
Query: 69 FLTENTDFTVIGVIGPPGVGKSTIMNEL-----YGFDSSSPGMLPP-----FATQ----- 113
F NTDFTVIGV+G GKST++N L +D P FAT+
Sbjct: 187 FHKTNTDFTVIGVLGGQSSGKSTLLNLLAAERSLDYDYYQHLFSPEADECIFATRHKLKP 246
Query: 114 SEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSA 173
+ +++ R + ++ I+ ER ILLDT P + P G +
Sbjct: 247 NNGQKSILRPRTETLQFFITRERHILLDTPP----------LMPVGKDSDH--------- 287
Query: 174 ELAHEIMGIQLAVLLASVCHILLVVSEGV 202
++ + L SVCHIL++V +G+
Sbjct: 288 ---QDLYSLGTMAQLLSVCHILILVIDGL 313
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 709,354,528
Number of Sequences: 2540612
Number of extensions: 30501948
Number of successful extensions: 107294
Number of sequences better than 10.0: 208
Number of HSP's better than 10.0 without gapping: 99
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 107094
Number of HSP's gapped (non-prelim): 250
length of query: 396
length of database: 863,360,394
effective HSP length: 130
effective length of query: 266
effective length of database: 533,080,834
effective search space: 141799501844
effective search space used: 141799501844
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0057b.6


