

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052a.3
(242 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
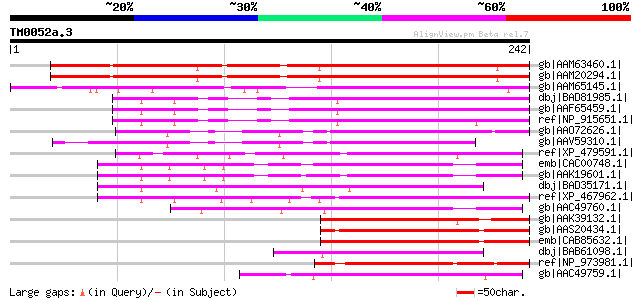
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM63460.1| unknown [Arabidopsis thaliana] 197 3e-49
gb|AAM20294.1| unknown protein [Arabidopsis thaliana] gi|1817562... 196 4e-49
gb|AAM65145.1| G-box binding factor, GBF4 [Arabidopsis thaliana]... 190 3e-47
dbj|BAD81985.1| OSE2 [Oryza sativa (japonica cultivar-group)] gi... 133 5e-30
gb|AAF65459.1| OSE2 [Oryza sativa] 132 6e-30
ref|NP_915651.1| putative DNA binding protein [Oryza sativa (jap... 124 3e-27
gb|AAO72626.1| OSE2-like protein [Oryza sativa (japonica cultiva... 114 3e-24
gb|AAV59310.1| unknown protein [Oryza sativa (japonica cultivar-... 96 7e-19
ref|XP_479591.1| putative bZIP protein DPBF3 [Oryza sativa (japo... 91 2e-17
emb|CAC00748.1| promoter-binding factor-like protein [Arabidopsi... 86 1e-15
gb|AAK19601.1| bZIP protein DPBF3 [Arabidopsis thaliana] 84 3e-15
dbj|BAD35171.1| putative bZIP transcription factor [Oryza sativa... 81 3e-14
ref|XP_467962.1| putative bZIP transcription factor ABI5 [Oryza ... 80 6e-14
gb|AAC49760.1| Dc3 promoter-binding factor-2 [Helianthus annuus]... 80 6e-14
gb|AAK39132.1| bZIP transcription factor 6 [Phaseolus vulgaris] 78 2e-13
gb|AAS20434.1| AREB-like protein [Lycopersicon esculentum] 78 2e-13
emb|CAB85632.1| putative ripening-related bZIP protein [Vitis vi... 78 2e-13
dbj|BAB61098.1| phi-2 [Nicotiana tabacum] 77 3e-13
ref|NP_973981.1| ABA-responsive element-binding protein 1 (AREB1... 77 4e-13
gb|AAC49759.1| Dc3 promoter-binding factor-1 [Helianthus annuus]... 77 5e-13
>gb|AAM63460.1| unknown [Arabidopsis thaliana]
Length = 315
Score = 197 bits (500), Expect = 3e-49
Identities = 120/231 (51%), Positives = 157/231 (67%), Gaps = 14/231 (6%)
Query: 20 SSPPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEIIAGDHHQCKEEIPDEMMTLE 79
+SP P + T ++ +DH T T ++VD+VW+E+++G+ KEE +E+MTLE
Sbjct: 91 ASPAPMEITTTTASDVVDHGGGTETTRG-GKSVDEVWREMVSGEGKGMKEETSEEIMTLE 149
Query: 80 DFLVKAG-----AVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGF 134
DFL KA AV ED +VK+ P+T + + FQ ++ VEG S+ F
Sbjct: 150 DFLAKAAVEDETAVTASAEDLDVKI--PVTNYGFDHSAPPHNPFQMIDKVEG---SIVAF 204
Query: 135 GNGVEVVEG-TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAV 193
GNG++V G RGKR R +VE LDKAA QRQRRMIKNRESAARSRERKQAYQVELE+LA
Sbjct: 205 GNGLDVYGGGARGKRARVMVEPLDKAAAQRQRRMIKNRESAARSRERKQAYQVELEALAA 264
Query: 194 KLEEENDKLLKEKAERTKERFKQLMEEVIPVVE--KRRPPRPLRRINSMQW 242
KLEEEN+ L KE ++ KER+++LME VIPVVE K++PPR LRRI S++W
Sbjct: 265 KLEEENELLSKEIEDKRKERYQKLMEFVIPVVEKPKQQPPRFLRRIRSLEW 315
>gb|AAM20294.1| unknown protein [Arabidopsis thaliana] gi|18175622|gb|AAL59898.1|
unknown protein [Arabidopsis thaliana]
gi|9758567|dbj|BAB09068.1| unnamed protein product
[Arabidopsis thaliana] gi|28317385|tpe|CAD29862.1| TPA:
basic leucine zipper transcription factor [Arabidopsis
thaliana] gi|15240086|ref|NP_199221.1| bZIP
transcription factor family protein [Arabidopsis
thaliana]
Length = 315
Score = 196 bits (499), Expect = 4e-49
Identities = 119/231 (51%), Positives = 157/231 (67%), Gaps = 14/231 (6%)
Query: 20 SSPPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEIIAGDHHQCKEEIPDEMMTLE 79
+SP P + T ++ +DH T T ++VD++W+E+++G+ KEE +E+MTLE
Sbjct: 91 ASPAPMEITTTTASDVVDHGGGTETTRG-GKSVDEIWREMVSGEGKGMKEETSEEIMTLE 149
Query: 80 DFLVKAG-----AVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGF 134
DFL KA AV ED +VK+ P+T + + FQ ++ VEG S+ F
Sbjct: 150 DFLAKAAVEDETAVTASAEDLDVKI--PVTNYGFDHSAPPHNPFQMIDKVEG---SIVAF 204
Query: 135 GNGVEVVEG-TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAV 193
GNG++V G RGKR R +VE LDKAA QRQRRMIKNRESAARSRERKQAYQVELE+LA
Sbjct: 205 GNGLDVYGGGARGKRARVMVEPLDKAAAQRQRRMIKNRESAARSRERKQAYQVELEALAA 264
Query: 194 KLEEENDKLLKEKAERTKERFKQLMEEVIPVVE--KRRPPRPLRRINSMQW 242
KLEEEN+ L KE ++ KER+++LME VIPVVE K++PPR LRRI S++W
Sbjct: 265 KLEEENELLSKEIEDKRKERYQKLMEFVIPVVEKPKQQPPRFLRRIRSLEW 315
>gb|AAM65145.1| G-box binding factor, GBF4 [Arabidopsis thaliana]
gi|15219608|ref|NP_171893.1| G-box binding factor 4
(GBF4) [Arabidopsis thaliana]
gi|1169863|sp|P42777|GBF4_ARATH G-box binding factor 4
(AtbZIP40) gi|403418|gb|AAA18414.1| GBF4
gi|4204292|gb|AAD10673.1| GBF4 [Arabidopsis thaliana]
Length = 270
Score = 190 bits (482), Expect = 3e-47
Identities = 130/289 (44%), Positives = 169/289 (57%), Gaps = 66/289 (22%)
Query: 1 MASSKMMMSSSKSLDHQQPSSPPPSSMPQFTTTNFI------DHA-----------DSTL 43
MAS K+M SS+ L + SS SS P +++ + DH+ D T
Sbjct: 1 MASFKLMSSSNSDLSRRNSSSA--SSSPSIRSSHHLRPNPHADHSRISFAYGGGVNDYTF 58
Query: 44 TDPYVP--------------------RTVDDVWQEIIAGDHH--QCKEEIPDEMMTLEDF 81
P ++VDDVW+EI++G+ KEE P+++MTLEDF
Sbjct: 59 ASDSKPFEMAIDVDRSIGDRNSVNNGKSVDDVWKEIVSGEQKTIMMKEEEPEDIMTLEDF 118
Query: 82 LVKAGAVDGDDEDDEVKMSMPLTERLS--GVFSLD-----GSSFQGMEGVEGGSASVGGF 134
L KA +G ++ +VK+ TERL+ G ++ D SSFQ +EG GG
Sbjct: 119 LAKAEMDEGASDEIDVKIP---TERLNNDGSYTFDFPMQRHSSFQMVEGSMGGGV----- 170
Query: 135 GNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVK 194
TRGKRGR ++E +DKAA QRQ+RMIKNRESAARSRERKQAYQVELE+LA K
Sbjct: 171 ---------TRGKRGRVMMEAMDKAAAQRQKRMIKNRESAARSRERKQAYQVELETLAAK 221
Query: 195 LEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPP-RPLRRINSMQW 242
LEEEN++LLKE E TKER+K+LME +IPV EK RPP RPL R +S++W
Sbjct: 222 LEEENEQLLKEIEESTKERYKKLMEVLIPVDEKPRPPSRPLSRSHSLEW 270
>dbj|BAD81985.1| OSE2 [Oryza sativa (japonica cultivar-group)]
gi|1753085|gb|AAB39320.1| leucine zipper protein
gi|7489466|pir||T04353 DNA binding protein - rice
Length = 217
Score = 133 bits (334), Expect = 5e-30
Identities = 90/216 (41%), Positives = 119/216 (54%), Gaps = 47/216 (21%)
Query: 49 PRTVDDVWQEII-AGDHHQCKEEIPDEM-------------------MTLEDFLVKAGAV 88
PRT ++VW+EI AG +P MTLEDFL + GAV
Sbjct: 27 PRTAEEVWKEITGAGVAAAAGGVVPPAAAAAAAPAVVAGAGAGTGAEMTLEDFLAREGAV 86
Query: 89 DGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKR 148
++DE ++ P S+ +G V GF NG EV G G R
Sbjct: 87 ----KEDEAVVTDP-------------SAAKGQV--------VMGFLNGAEVTGGVTGGR 121
Query: 149 GRP--VVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEK 206
R +++ +D+AA QRQ+RMIKNRESAARSRERKQAY ELESL +LEEEN K+ KE+
Sbjct: 122 SRKRHLMDPMDRAAMQRQKRMIKNRESAARSRERKQAYIAELESLVTQLEEENAKMFKEQ 181
Query: 207 AERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
E+ ++R K+L E V+PV+ ++ R LRR NSM+W
Sbjct: 182 EEQHQKRLKELKEMVVPVIIRKTSARDLRRTNSMEW 217
>gb|AAF65459.1| OSE2 [Oryza sativa]
Length = 217
Score = 132 bits (333), Expect = 6e-30
Identities = 90/216 (41%), Positives = 119/216 (54%), Gaps = 47/216 (21%)
Query: 49 PRTVDDVWQEII-AGDHHQCKEEIPDEM-------------------MTLEDFLVKAGAV 88
PRT ++VW+EI AG +P MTLEDFL + GAV
Sbjct: 27 PRTAEEVWKEITGAGVAAAAGVGLPPAAAAAAAPAVVAGAGAGTGAEMTLEDFLAREGAV 86
Query: 89 DGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKR 148
++DE ++ P S+ +G V GF NG EV G G R
Sbjct: 87 ----KEDEAVVTDP-------------SAAKGQV--------VMGFLNGAEVTGGVTGGR 121
Query: 149 GRP--VVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEK 206
R +++ +D+AA QRQ+RMIKNRESAARSRERKQAY ELESL +LEEEN K+ KE+
Sbjct: 122 SRKRHLMDPMDRAAMQRQKRMIKNRESAARSRERKQAYIAELESLVTQLEEENAKMFKEQ 181
Query: 207 AERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
E+ ++R K+L E V+PV+ ++ R LRR NSM+W
Sbjct: 182 EEQHQKRLKELKEMVVPVIIRKTSARDLRRTNSMEW 217
>ref|NP_915651.1| putative DNA binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 278
Score = 124 bits (310), Expect = 3e-27
Identities = 90/229 (39%), Positives = 119/229 (51%), Gaps = 60/229 (26%)
Query: 49 PRTVDDVWQEII-AGDHHQCKEEIPDEM-------------------MTLEDFLVKAGAV 88
PRT ++VW+EI AG +P MTLEDFL + GAV
Sbjct: 75 PRTAEEVWKEITGAGVAAAAGGVVPPAAAAAAAPAVVAGAGAGTGAEMTLEDFLAREGAV 134
Query: 89 DGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKR 148
++DE ++ P S+ +G V GF NG EV G G R
Sbjct: 135 ----KEDEAVVTDP-------------SAAKGQV--------VMGFLNGAEVTGGVTGGR 169
Query: 149 GRP--VVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEK 206
R +++ +D+AA QRQ+RMIKNRESAARSRERKQAY ELESL +LEEEN K+ KE+
Sbjct: 170 SRKRHLMDPMDRAAMQRQKRMIKNRESAARSRERKQAYIAELESLVTQLEEENAKMFKEQ 229
Query: 207 AERTKERFKQ-------------LMEEVIPVVEKRRPPRPLRRINSMQW 242
E+ ++R K+ L E V+PV+ ++ R LRR NSM+W
Sbjct: 230 EEQHQKRLKEPSYRNIQYRTQYWLKEMVVPVIIRKTSARDLRRTNSMEW 278
>gb|AAO72626.1| OSE2-like protein [Oryza sativa (japonica cultivar-group)]
Length = 179
Score = 114 bits (284), Expect = 3e-24
Identities = 79/203 (38%), Positives = 112/203 (54%), Gaps = 36/203 (17%)
Query: 50 RTVDDVWQEIIAGDHHQCKEEIP---------DEMMTLEDFLVKAGAVDGDDEDDEVKMS 100
RT ++VW+EI + P MTLEDFL + EDD
Sbjct: 3 RTAEEVWKEISSSGGLSAPAPAPAAGAAGRGGGPEMTLEDFLAR--------EDDP---- 50
Query: 101 MPLTERLSGVFSLDGSSFQGMEGV-EGGSASVGGFGNGVEVVEGTRGKRGRPVVENLDKA 159
+++G+ G V EG + GG G G G RG++ R +++ D+A
Sbjct: 51 --------RATAVEGNMVVGFPNVTEGVGTAGGGRGGG----GGGRGRK-RTLMDPADRA 97
Query: 160 AQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLME 219
A QRQ+RMIKNRESAARSRERKQAY ELE+ +LEEE+ +LL+E+ E+ ++R K++ E
Sbjct: 98 AMQRQKRMIKNRESAARSRERKQAYIAELEAQVAELEEEHAQLLREQEEKNQKRLKEIKE 157
Query: 220 EVIPVVEKRRPPRPLRRINSMQW 242
+ + VV R+ + LRR NSM+W
Sbjct: 158 QAVAVV-IRKKTQDLRRTNSMEW 179
>gb|AAV59310.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|50931561|ref|XP_475308.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|46981289|gb|AAT07607.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 247
Score = 96.3 bits (238), Expect = 7e-19
Identities = 72/207 (34%), Positives = 106/207 (50%), Gaps = 45/207 (21%)
Query: 21 SPPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEIIAGDHHQCKEEIP-------- 72
+PPP+ + A + + + RT ++VW+EI + P
Sbjct: 72 APPPAG----------EAAGAPVAEVAARRTAEEVWKEISSSGGLSAPAPAPAAGAAGRG 121
Query: 73 -DEMMTLEDFLVKAGAVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGV-EGGSAS 130
MTLEDFL + EDD +++G+ G V EG +
Sbjct: 122 GGPEMTLEDFLAR--------EDDP------------RATAVEGNMVVGFPNVTEGVGTA 161
Query: 131 VGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELES 190
GG G G G RG++ R +++ D+AA QRQ+RMIKNRESAARSRERKQAY ELE+
Sbjct: 162 GGGRGGG----GGGRGRK-RTLMDPADRAAMQRQKRMIKNRESAARSRERKQAYIAELEA 216
Query: 191 LAVKLEEENDKLLKEKAERTKERFKQL 217
+LEEE+ +LL+E+ E+ ++R K++
Sbjct: 217 QVAELEEEHAQLLREQEEKNQKRLKEV 243
>ref|XP_479591.1| putative bZIP protein DPBF3 [Oryza sativa (japonica
cultivar-group)] gi|50509142|dbj|BAD30282.1| putative
bZIP protein DPBF3 [Oryza sativa (japonica
cultivar-group)] gi|33146493|dbj|BAC79602.1| putative
bZIP protein DPBF3 [Oryza sativa (japonica
cultivar-group)]
Length = 269
Score = 91.3 bits (225), Expect = 2e-17
Identities = 83/215 (38%), Positives = 104/215 (47%), Gaps = 36/215 (16%)
Query: 50 RTVDDVWQEI---IAGDHHQCKEEIPDEMMTLEDFLVKAG-AVDGDDEDDEVKMSM---- 101
+TVD+VW++I G HH P MTLEDFL +AG AVDG
Sbjct: 63 KTVDEVWRDIQGASTGRHHAT----PMGEMTLEDFLSRAGVAVDGAASAAGAHWLRGHYP 118
Query: 102 ----PLTERLSGVFSLDGSSFQGMEGVEG--GSASVGGFGNGVEVVEGTRGKRGRPVVEN 155
P T L V G S ++GV V GF + V V RG VVE
Sbjct: 119 PPPPPTTTTLQYV----GGSGAVVDGVYNRVDGHGVAGFLSQVGVAGRKRGGGVDGVVE- 173
Query: 156 LDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKA-------- 207
K ++RQ+RMIKNRESAARSR RKQAY ELE+ +LEEEN +L + KA
Sbjct: 174 --KTVERRQKRMIKNRESAARSRARKQAYTNELENKISRLEEENQRLREHKAVADFSTFP 231
Query: 208 ---ERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
+ K Q +E V+ +V + P + LRR S
Sbjct: 232 SCVDFLKAFLTQKLEPVMQIVPQPEPKQQLRRTTS 266
>emb|CAC00748.1| promoter-binding factor-like protein [Arabidopsis thaliana]
gi|20148683|gb|AAM10232.1| promoter-binding factor-like
protein [Arabidopsis thaliana]
gi|17064744|gb|AAL32526.1| promoter-binding factor-like
protein [Arabidopsis thaliana]
gi|15230146|ref|NP_191244.1| ABA-responsive
element-binding protein 3 (AREB3) [Arabidopsis thaliana]
gi|11358714|pir||T51273 promoter-binding factor-like
protein - Arabidopsis thaliana
gi|9967421|dbj|BAB12406.1| ABA-responsive element
binding protein 3 (AREB3) [Arabidopsis thaliana]
Length = 297
Score = 85.5 bits (210), Expect = 1e-15
Identities = 76/233 (32%), Positives = 108/233 (45%), Gaps = 56/233 (24%)
Query: 42 TLTDPYVPRTVDDVWQEII----AGDHHQCKEEIPD-EMMTLEDFLVKAGAVD----GDD 92
TL +TVD+VW++I G H+ +++ P MTLED L+KAG V G +
Sbjct: 83 TLPRDLSKKTVDEVWKDIQQNKNGGSAHERRDKQPTLGEMTLEDLLLKAGVVTETIPGSN 142
Query: 93 EDDEVK--------------------------MSMPLTERLSGVFSLDGSSFQGMEGVEG 126
D V SMP + D M+ +
Sbjct: 143 HDGPVGGGSAGSGAGLGQNITQVGPWIQYHQLPSMPQPQAFMPYPVSD------MQAMVS 196
Query: 127 GSASVGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQV 186
S+ +GG + T G++ E ++K ++RQ+RMIKNRESAARSR RKQAY
Sbjct: 197 QSSLMGGLSD-----TQTPGRKRVASGEVVEKTVERRQKRMIKNRESAARSRARKQAYTH 251
Query: 187 ELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
ELE +LEEEN++L K+K +E+++P V P R LRR +S
Sbjct: 252 ELEIKVSRLEEENERLRKQKE----------VEKILPSVPPPDPKRQLRRTSS 294
>gb|AAK19601.1| bZIP protein DPBF3 [Arabidopsis thaliana]
Length = 297
Score = 84.0 bits (206), Expect = 3e-15
Identities = 77/233 (33%), Positives = 107/233 (45%), Gaps = 56/233 (24%)
Query: 42 TLTDPYVPRTVDDVWQEII----AGDHHQCKEEIPD-EMMTLEDFLVKAGAVD----GDD 92
TL +TVD+VW++I G H+ +++ P MTLED L+KAG V G +
Sbjct: 83 TLPRDLSKKTVDEVWKDIQQNKNGGSAHERRDKQPTLGEMTLEDLLLKAGVVTETIPGSN 142
Query: 93 EDDEVK--------------------------MSMPLTERLSGVFSLDGSSFQGMEGVEG 126
D V SMP + D M+ +
Sbjct: 143 HDGPVGGGSAGSGAGLGQNITQVGPWIQYHQLPSMPQPQAFMPYPVSD------MQAMVS 196
Query: 127 GSASVGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQV 186
S+ +GG + + G + VVE K ++RQ+RMIKNRESAARSR RKQAY
Sbjct: 197 QSSLMGGLSD--TQIPGRKRVASGEVVE---KTVERRQKRMIKNRESAARSRARKQAYTH 251
Query: 187 ELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
ELE +LEEEN++L K+K +E+++P V P R LRR +S
Sbjct: 252 ELEIKVSRLEEENERLRKQKE----------VEKILPSVPPPDPKRQLRRTSS 294
>dbj|BAD35171.1| putative bZIP transcription factor [Oryza sativa (japonica
cultivar-group)] gi|51090510|dbj|BAD35712.1| putative
bZIP transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 324
Score = 80.9 bits (198), Expect = 3e-14
Identities = 68/218 (31%), Positives = 101/218 (46%), Gaps = 38/218 (17%)
Query: 42 TLTDPYVPRTVDDVWQEII-------------AGDHHQCKEEIPDEMMTLEDFLVKAGAV 88
TL +TVD+VW++I+ A + + MTLE+FLV+AG V
Sbjct: 86 TLPRTLSQKTVDEVWRDIMGLGGSDDEDPAAAAAAAAPAQRQPTLGEMTLEEFLVRAGVV 145
Query: 89 DGDDEDD-----EVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFG-------- 135
D + + P + ++ L G+ GV G+A+
Sbjct: 146 REDMGQTIVLPPQAQALFPGSNVVAPAMQLANGMLPGVVGVAPGAAAAMTVAAPATPVVL 205
Query: 136 NGVEVVEGTRGKRGRPVVENLD------------KAAQQRQRRMIKNRESAARSRERKQA 183
NG+ VEG PV D K ++RQRRMIKNRESAARSR RKQA
Sbjct: 206 NGLGKVEGGDLSSLSPVPYPFDTALRVRKGPTVEKVVERRQRRMIKNRESAARSRARKQA 265
Query: 184 YQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEEV 221
Y +ELE+ KL+E+ +L K++ E +++ ++ME +
Sbjct: 266 YIMELEAEVAKLKEQKAELQKKQVEMIQKQNDEVMERI 303
>ref|XP_467962.1| putative bZIP transcription factor ABI5 [Oryza sativa (japonica
cultivar-group)] gi|46805743|dbj|BAD17130.1| putative
bZIP transcription factor ABI5 [Oryza sativa (japonica
cultivar-group)] gi|46806070|dbj|BAD17318.1| putative
bZIP transcription factor ABI5 [Oryza sativa (japonica
cultivar-group)]
Length = 357
Score = 79.7 bits (195), Expect = 6e-14
Identities = 81/263 (30%), Positives = 112/263 (41%), Gaps = 67/263 (25%)
Query: 42 TLTDPYVPRTVDDVWQEII------AGDHHQCKEEIPDEM-------MTLEDFLVKAGAV 88
TL +TVD+VW++++ A E P +TLE+FLV+AG V
Sbjct: 100 TLPRTLSQKTVDEVWRDMMCFGGGGASTAPAAAEPPPPAHRQQTLGEITLEEFLVRAGVV 159
Query: 89 DGDDEDDEV-----------------KMSMPLTERLSGVF-------SLDGSSFQGMEGV 124
D V + P+ S VF SL G G
Sbjct: 160 REDMSVPPVPPAPTPTAAAVPPPPPPQQQTPMLFGQSNVFPPMVPPLSLGNGLVSGAVGH 219
Query: 125 EGGSA----------SVGGFGNGVEVVEG---------------TRGKRGRPVVENLDKA 159
GG A S GFG +EG G RGR ++K
Sbjct: 220 GGGGAASLVSPVRPVSSNGFGK----MEGGDLSSLSPSPVPYVFKGGLRGRKA-PGIEKV 274
Query: 160 AQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLME 219
++RQRRMIKNRESAARSR+RKQAY +ELE+ KL+E ND+L K++ E +++ +++E
Sbjct: 275 VERRQRRMIKNRESAARSRQRKQAYMMELEAEVAKLKELNDELQKKQDEMLEQQKNEVLE 334
Query: 220 EVIPVVEKRRPPRPLRRINSMQW 242
+ V LRR + W
Sbjct: 335 RMSRQVGPTAKRICLRRTLTGPW 357
>gb|AAC49760.1| Dc3 promoter-binding factor-2 [Helianthus annuus]
gi|7489324|pir||T12622 Dc3 promoter-binding factor-2 -
common sunflower
Length = 174
Score = 79.7 bits (195), Expect = 6e-14
Identities = 63/181 (34%), Positives = 91/181 (49%), Gaps = 27/181 (14%)
Query: 76 MTLEDFLVKAGAV----------DGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVE 125
MTLEDFLVKAG V +G+ E E + + + + +Q V
Sbjct: 1 MTLEDFLVKAGVVAESSPGKVNEEGNLEPQETQWIGYQSHAVQQQNMIMAGHYQVQPSVT 60
Query: 126 --GGSASVGGFGNGVEVVEGTR-----GKRGRPVVENLDKAAQQRQRRMIKNRESAARSR 178
G S G+ + ++ G++ + ++K ++RQ+RMIKNRESAARSR
Sbjct: 61 VPGNSLMDVGYMSPTSLMGSLSDRHMSGRKRFASGDVMEKTVERRQKRMIKNRESAARSR 120
Query: 179 ERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRIN 238
RKQAY ELE+ +LEEEN++L +EK +E+VIP V +P LRR +
Sbjct: 121 ARKQAYTHELENKVSRLEEENERLRREKE----------VEKVIPWVPPPKPKYQLRRTS 170
Query: 239 S 239
S
Sbjct: 171 S 171
>gb|AAK39132.1| bZIP transcription factor 6 [Phaseolus vulgaris]
Length = 415
Score = 78.2 bits (191), Expect = 2e-13
Identities = 47/100 (47%), Positives = 63/100 (63%), Gaps = 8/100 (8%)
Query: 146 GKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
G RGR ++K ++RQRRMIKNRESAARSR RKQAY +ELE+ KL+EEN L K+
Sbjct: 321 GMRGRKSGGAVEKVIERRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENQGLQKK 380
Query: 206 KA---ERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
+A E K +FK++M +++ R LRR + W
Sbjct: 381 QAEIMEIQKNQFKEMMN-----LQREVKRRRLRRTQTGPW 415
Score = 37.4 bits (85), Expect = 0.37
Identities = 40/140 (28%), Positives = 65/140 (45%), Gaps = 14/140 (10%)
Query: 42 TLTDPYVPRTVDDVWQEIIA--GDHH-----QCKEEIPDEMMTLEDFLVKAGAVDGDDE- 93
TL +TVD+VW++I G H Q + MTLE+FLV+AG V D +
Sbjct: 94 TLPRTLSQKTVDEVWKDISKDYGGHGEPNLAQTPRQPTLREMTLEEFLVRAGVVREDAKP 153
Query: 94 DDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRGRPV- 152
+D V M + +G+ G FQ M V + +G N +V G + P+
Sbjct: 154 NDGVFMDLARAGNNNGL----GFEFQQMNKVAAATGLMGNRLNNDPLV-GLQSSANLPLN 208
Query: 153 VENLDKAAQQRQRRMIKNRE 172
V + ++QQ Q + ++++
Sbjct: 209 VNGVRSSSQQPQMQSPQSQQ 228
>gb|AAS20434.1| AREB-like protein [Lycopersicon esculentum]
Length = 447
Score = 78.2 bits (191), Expect = 2e-13
Identities = 46/97 (47%), Positives = 64/97 (65%), Gaps = 3/97 (3%)
Query: 146 GKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
G RGR ++K ++RQRRMIKNRESAARSR RKQAY +ELE+ KL+EEND+L K+
Sbjct: 354 GLRGRKY-STVEKVVERRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENDELQKK 412
Query: 206 KAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
+ E + + Q++E + ++K R LRR + W
Sbjct: 413 QEEMLEMQKNQVIE--MMNLQKGAKRRCLRRTQTGPW 447
>emb|CAB85632.1| putative ripening-related bZIP protein [Vitis vinifera]
Length = 447
Score = 77.8 bits (190), Expect = 2e-13
Identities = 45/97 (46%), Positives = 65/97 (66%), Gaps = 2/97 (2%)
Query: 146 GKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
G RGR ++K ++RQRRMIKNRESAARSR RKQAY +ELE+ KL+E+N++L K+
Sbjct: 353 GIRGRKCSGAVEKVIERRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEKNEELEKK 412
Query: 206 KAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
+AE + + Q+ME + +++ R LRR + W
Sbjct: 413 QAEMMEMQKNQVME--MMNLQREVKKRCLRRTLTGPW 447
>dbj|BAB61098.1| phi-2 [Nicotiana tabacum]
Length = 464
Score = 77.4 bits (189), Expect = 3e-13
Identities = 45/116 (38%), Positives = 68/116 (57%), Gaps = 18/116 (15%)
Query: 124 VEGGSASVGGFGNGVEVVEGT------------------RGKRGRPVVENLDKAAQQRQR 165
++GG + G NGV V+G G RGR + +K ++R++
Sbjct: 330 MQGGVMDMAGLHNGVTSVKGGSPGNLDPPSLSPSPYACGEGGRGRRSCTSFEKVVERRRK 389
Query: 166 RMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEEV 221
RMIKNRESAARSR+RKQAY +ELE+ KL+E +L K++AE +++ QL+E++
Sbjct: 390 RMIKNRESAARSRDRKQAYTLELETEVAKLKEIKQELQKKQAEFIEKQKNQLLEKM 445
>ref|NP_973981.1| ABA-responsive element-binding protein 1 (AREB1) [Arabidopsis
thaliana] gi|6739278|gb|AAF27180.1| abscisic acid
responsive elements-binding factor [Arabidopsis
thaliana] gi|9967417|dbj|BAB12404.1| ABA-responsive
element binding protein 1 (AREB1) [Arabidopsis thaliana]
Length = 416
Score = 77.0 bits (188), Expect = 4e-13
Identities = 48/100 (48%), Positives = 64/100 (64%), Gaps = 6/100 (6%)
Query: 143 GTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKL 202
G RG++ V +K ++RQRRMIKNRESAARSR RKQAY VELE+ KL+EEND+L
Sbjct: 323 GVRGRKSGTV----EKVVERRQRRMIKNRESAARSRARKQAYTVELEAEVAKLKEENDEL 378
Query: 203 LKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
+++A R E K E+ +++ P + LRR S W
Sbjct: 379 QRKQA-RIMEMQKNQETEMRNLLQ-GGPKKKLRRTESGPW 416
>gb|AAC49759.1| Dc3 promoter-binding factor-1 [Helianthus annuus]
gi|7489323|pir||T12621 Dc3 promoter-binding factor-1 -
common sunflower
Length = 378
Score = 76.6 bits (187), Expect = 5e-13
Identities = 59/163 (36%), Positives = 81/163 (49%), Gaps = 33/163 (20%)
Query: 108 SGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEV--------------------------V 141
+G + + G+ G GG A GG+G G+ + +
Sbjct: 215 AGNYQQQPPPYGGIGGNAGGVA--GGYGQGLGIGSPPSPVSSDGIATTQLDSGNQYALEM 272
Query: 142 EGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDK 201
G RG R R + ++K ++RQRRMIKNRESAARSR RKQAY VELE+ L+EEN +
Sbjct: 273 GGIRGGRKRIIDGPVEKVVERRQRRMIKNRESAARSRARKQAYTVELEAELNMLKEENAQ 332
Query: 202 LLKEKAERTKERFKQLMEE-----VIPVVEKRRPPRPLRRINS 239
L + AE ++R +Q EE V + R R LRR +S
Sbjct: 333 LKQALAEIERKRKQQFSEEIRMKGVTKCQKVRDKSRMLRRTSS 375
Score = 43.9 bits (102), Expect = 0.004
Identities = 26/63 (41%), Positives = 33/63 (52%), Gaps = 16/63 (25%)
Query: 42 TLTDPYVPRTVDDVWQEII---------AGDHHQCKEEIPDEM-------MTLEDFLVKA 85
TL P +TVD+VW EI + D++ C E++P MTLEDFLVKA
Sbjct: 78 TLPGPLSRKTVDEVWSEIQKTRQDHQQPSNDNNSCNEQVPGAQRQPTYGEMTLEDFLVKA 137
Query: 86 GAV 88
G V
Sbjct: 138 GVV 140
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 406,464,905
Number of Sequences: 2540612
Number of extensions: 17725846
Number of successful extensions: 107669
Number of sequences better than 10.0: 1917
Number of HSP's better than 10.0 without gapping: 900
Number of HSP's successfully gapped in prelim test: 1055
Number of HSP's that attempted gapping in prelim test: 103098
Number of HSP's gapped (non-prelim): 4624
length of query: 242
length of database: 863,360,394
effective HSP length: 124
effective length of query: 118
effective length of database: 548,324,506
effective search space: 64702291708
effective search space used: 64702291708
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0052a.3


