

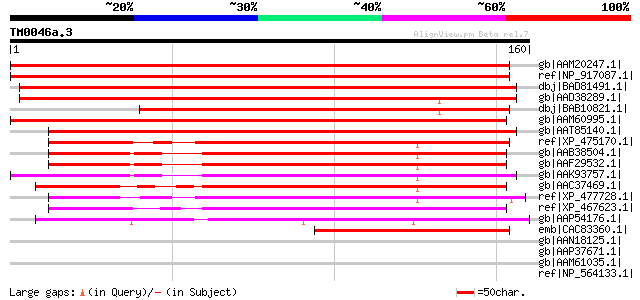
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0046a.3
(160 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM20247.1| putative plasma membrane associated protein [Arab... 253 1e-66
ref|NP_917087.1| putative plasma membrane associated protein [Or... 218 3e-56
dbj|BAD81491.1| putative ABA induced plasma membrane protein PM ... 193 2e-48
gb|AAD38289.1| putative ABA induced plasma membrane protein [Ory... 182 3e-45
dbj|BAB10821.1| unnamed protein product [Arabidopsis thaliana] g... 166 3e-40
gb|AAM60995.1| plasma membrane associated protein-like [Arabidop... 134 8e-31
gb|AAT85140.1| unknown protein [Oryza sativa (japonica cultivar-... 128 5e-29
ref|XP_475170.1| putative plasma membrane associated protein [Or... 115 4e-25
gb|AAB38504.1| ABA induced plasma membrane protein PM 19 [Tritic... 113 2e-24
gb|AAF29532.1| plasma membrane associated protein [Hordeum vulgare] 112 3e-24
gb|AAK93757.1| unknown protein [Arabidopsis thaliana] gi|1350755... 112 3e-24
gb|AAC37469.1| a Lea protein with hydrophobic domain, high pI va... 107 8e-23
ref|XP_477728.1| putative plasma membrane associated protein [Or... 107 1e-22
ref|XP_467623.1| putative ABA-induced plasma membrane protein [O... 107 1e-22
gb|AAP54176.1| unknown protein [Oryza sativa (japonica cultivar-... 82 4e-15
emb|CAC83360.1| putative hydrophobic LEA-like protein [Pinus pin... 75 8e-13
gb|AAN18125.1| At1g76670/F28O16_4 [Arabidopsis thaliana] gi|1849... 43 0.003
gb|AAP37671.1| At5g40240 [Arabidopsis thaliana] gi|10177511|dbj|... 43 0.003
gb|AAM61035.1| unknown [Arabidopsis thaliana] 42 0.004
ref|NP_564133.1| transporter-related [Arabidopsis thaliana] gi|2... 42 0.004
>gb|AAM20247.1| putative plasma membrane associated protein [Arabidopsis thaliana]
gi|18377741|gb|AAL67020.1| putative plasma membrane
associated protein [Arabidopsis thaliana]
gi|15218953|ref|NP_174245.1| AWPM-19-like membrane
family protein [Arabidopsis thaliana]
gi|12323516|gb|AAG51728.1| plasma membrane associated
protein, putative; 66162-66952 [Arabidopsis thaliana]
gi|25354594|pir||B86418 probable plasma membrane
associated protein F15D2.10 - Arabidopsis thaliana
Length = 158
Score = 253 bits (646), Expect = 1e-66
Identities = 121/154 (78%), Positives = 140/154 (90%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MA EQ+KP+A+ LL LNFCMYVIVLGIGGWAMNRAID GF +GP L+LPAHFSPI+FPMG
Sbjct: 1 MAGEQIKPVASGLLVLNFCMYVIVLGIGGWAMNRAIDHGFEVGPNLELPAHFSPIYFPMG 60
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
NA+TGFFV FAL+AGVVGAAS ISG++HIRSWT SLP+AA+ AT+AWTLT+LAMGFAWK
Sbjct: 61 NAATGFFVIFALLAGVVGAASTISGLSHIRSWTVGSLPAAATAATIAWTLTVLAMGFAWK 120
Query: 121 EVELRIRNARLKTMEAFLIILSATQLFYIAAIHG 154
E+EL+ RNA+L+TMEAFLIILS TQL YIAA+HG
Sbjct: 121 EIELQGRNAKLRTMEAFLIILSVTQLIYIAAVHG 154
>ref|NP_917087.1| putative plasma membrane associated protein [Oryza sativa (japonica
cultivar-group)] gi|15408768|dbj|BAB64168.1| plasma
membrane associated protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 156
Score = 218 bits (556), Expect = 3e-56
Identities = 106/154 (68%), Positives = 129/154 (82%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MAN +KP+A LLL LNFCMYVIV +GGWA+N AI G+ IG + LPA+FSPI+FPMG
Sbjct: 1 MANAGLKPVAGLLLVLNFCMYVIVAAVGGWAINHAIHTGYFIGSGMALPANFSPIYFPMG 60
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
NA+TGFFV FA+IAGVVGAA+A++G +H+R+W+ ESLP+AAS +AWTLTLLAMG A K
Sbjct: 61 NAATGFFVIFAVIAGVVGAAAALAGFHHVRAWSHESLPAAASSGFIAWTLTLLAMGLAVK 120
Query: 121 EVELRIRNARLKTMEAFLIILSATQLFYIAAIHG 154
E++L RNARLKTME+F IILSATQLFY+ AIHG
Sbjct: 121 EIDLHGRNARLKTMESFTIILSATQLFYLLAIHG 154
>dbj|BAD81491.1| putative ABA induced plasma membrane protein PM 19 [Oryza sativa
(japonica cultivar-group)]
Length = 171
Score = 193 bits (490), Expect = 2e-48
Identities = 93/153 (60%), Positives = 118/153 (76%)
Query: 4 EQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNAS 63
E +KP+A LLL LN CMYVI+ IGGWA+N +ID+GFI+GP L+LPAHF P+FFP+GN +
Sbjct: 3 ETLKPVALLLLILNLCMYVILAIIGGWAVNISIDRGFILGPGLRLPAHFHPMFFPIGNWA 62
Query: 64 TGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVE 123
TGFFV F+L+AGVVG AS + G +HIR W SL AA+ +AW LT+LAMG A +E+
Sbjct: 63 TGFFVVFSLLAGVVGIASGLVGFSHIRHWNYYSLQPAATTGLLAWALTVLAMGLACQEIS 122
Query: 124 LRIRNARLKTMEAFLIILSATQLFYIAAIHGAA 156
L RNA+L TMEAF I+L+ATQLFY+ AIH +
Sbjct: 123 LDRRNAKLGTMEAFTIVLTATQLFYVLAIHSGS 155
>gb|AAD38289.1| putative ABA induced plasma membrane protein [Oryza sativa
(japonica cultivar-group)] gi|34904644|ref|NP_913669.1|
putative ABA-induced plasma membrane protein [Oryza
sativa (japonica cultivar-group)]
gi|13486855|dbj|BAB40086.1| putative ABA-induced plasma
membrane protein [Oryza sativa (japonica
cultivar-group)]
Length = 189
Score = 182 bits (461), Expect = 3e-45
Identities = 93/171 (54%), Positives = 118/171 (68%), Gaps = 18/171 (10%)
Query: 4 EQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNAS 63
E +KP+A LLL LN CMYVI+ IGGWA+N +ID+GFI+GP L+LPAHF P+FFP+GN +
Sbjct: 3 ETLKPVALLLLILNLCMYVILAIIGGWAVNISIDRGFILGPGLRLPAHFHPMFFPIGNWA 62
Query: 64 TGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVE 123
TGFFV F+L+AGVVG AS + G +HIR W SL AA+ +AW LT+LAMG A +E+
Sbjct: 63 TGFFVVFSLLAGVVGIASGLVGFSHIRHWNYYSLQPAATTGLLAWALTVLAMGLACQEIS 122
Query: 124 LRIRNARL------------------KTMEAFLIILSATQLFYIAAIHGAA 156
L RNA+L TMEAF I+L+ATQLFY+ AIH +
Sbjct: 123 LDRRNAKLINSFRMVCELKPWEFGIQGTMEAFTIVLTATQLFYVLAIHSGS 173
>dbj|BAB10821.1| unnamed protein product [Arabidopsis thaliana]
gi|15237455|ref|NP_199465.1| AWPM-19-like membrane
family protein [Arabidopsis thaliana]
Length = 138
Score = 166 bits (419), Expect = 3e-40
Identities = 85/120 (70%), Positives = 98/120 (80%), Gaps = 6/120 (5%)
Query: 41 IIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSA 100
I G + LPAHFSPI FPMGNA+TGFF+ FALIAGV GAAS ISGI+H++SWT SLP+A
Sbjct: 16 IQGADYSLPAHFSPIHFPMGNAATGFFIMFALIAGVAGAASVISGISHLQSWTTTSLPAA 75
Query: 101 ASVATMAWTLTLLAMGFAWKEVELRIRNARL------KTMEAFLIILSATQLFYIAAIHG 154
S AT+AW+LTLLAMGF KE+EL +RNARL +TMEAFLIILSATQL YIAAI+G
Sbjct: 76 VSAATIAWSLTLLAMGFGCKEIELGMRNARLVSKPLMRTMEAFLIILSATQLLYIAAIYG 135
>gb|AAM60995.1| plasma membrane associated protein-like [Arabidopsis thaliana]
gi|15239619|ref|NP_197398.1| AWPM-19-like membrane
family protein [Arabidopsis thaliana]
Length = 171
Score = 134 bits (337), Expect = 8e-31
Identities = 70/153 (45%), Positives = 99/153 (63%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MA+ K A +LL LN +Y ++ I WA+N I++ L LPA PI+FP+G
Sbjct: 1 MASGGSKSAAFMLLMLNLGLYFVITIIASWAVNHGIERTRESASTLSLPAKIFPIYFPVG 60
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
N +TGFFV F LIAGVVG A++++GI ++ W + +L SAA+ + ++W+LTLLAMG A K
Sbjct: 61 NMATGFFVIFTLIAGVVGMATSLTGIINVLQWDSPNLHSAAASSLISWSLTLLAMGLACK 120
Query: 121 EVELRIRNARLKTMEAFLIILSATQLFYIAAIH 153
E+ + A L+T+E II+SATQL AIH
Sbjct: 121 EINIGWTEANLRTLEVLTIIVSATQLLCTGAIH 153
>gb|AAT85140.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 169
Score = 128 bits (322), Expect = 5e-29
Identities = 64/144 (44%), Positives = 93/144 (64%)
Query: 13 LLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFAL 72
LL LN +YV+V I GWA+N +ID+ F + P PI+FP+GN +TGFFV FAL
Sbjct: 13 LLFLNLVLYVVVAVIAGWAINYSIDESFNSLQGVSPPVRLFPIYFPIGNLATGFFVIFAL 72
Query: 73 IAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVELRIRNARLK 132
+AGVVG +++++G++ + S+ SAA+ + + WTLTLLAMG A KE+ + R L+
Sbjct: 73 LAGVVGVSTSLTGLHDVSQGYPASMMSAAAASIVTWTLTLLAMGLACKEISIGWRPPSLR 132
Query: 133 TMEAFLIILSATQLFYIAAIHGAA 156
+E F IIL+ TQL ++H A
Sbjct: 133 ALETFTIILAGTQLLCAGSLHAGA 156
>ref|XP_475170.1| putative plasma membrane associated protein [Oryza sativa (japonica
cultivar-group)] gi|47777422|gb|AAT38056.1| putative
plasma membrane associated protein [Oryza sativa
(japonica cultivar-group)]
Length = 173
Score = 115 bits (288), Expect = 4e-25
Identities = 63/143 (44%), Positives = 89/143 (62%), Gaps = 14/143 (9%)
Query: 13 LLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFAL 72
LL LN MY+IV+G W +N I+ E P GN +T +F+ FA+
Sbjct: 12 LLVLNLIMYLIVIGFASWNLNHYING------ETNHPG-------VAGNGATFYFLVFAI 58
Query: 73 IAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVEL-RIRNARL 131
+AGVVGAAS ++G++H+RSW A SL + A+ A +AW +T LA G A KE+ + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRSWGAHSLAAGAASALIAWAITALAFGLACKEIHIGGYRGWRL 118
Query: 132 KTMEAFLIILSATQLFYIAAIHG 154
+ +EAF+IIL+ TQL Y+A +HG
Sbjct: 119 RVLEAFVIILAFTQLLYVAMLHG 141
>gb|AAB38504.1| ABA induced plasma membrane protein PM 19 [Triticum aestivum]
gi|7489659|pir||T06978 ABA-induced plasma membrane
protein PM 19 - wheat
Length = 182
Score = 113 bits (282), Expect = 2e-24
Identities = 60/142 (42%), Positives = 89/142 (62%), Gaps = 14/142 (9%)
Query: 13 LLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFAL 72
LL LN MY++V+G W +N I+ G P + GN +T +F+ FA+
Sbjct: 12 LLVLNLIMYIVVIGFASWNLNHFIN-GLTNRPGVG------------GNGATFYFLVFAI 58
Query: 73 IAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVEL-RIRNARL 131
+AGVVGAAS ++G++H+R+W +SL ++AS A +AW +T LA G A KE+ + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRTWRGDSLATSASSALVAWAITALAFGLACKEIHIGGYRGWRL 118
Query: 132 KTMEAFLIILSATQLFYIAAIH 153
+ +EAF+IIL TQL Y+ A+H
Sbjct: 119 RVLEAFVIILMFTQLLYVLALH 140
>gb|AAF29532.1| plasma membrane associated protein [Hordeum vulgare]
Length = 181
Score = 112 bits (281), Expect = 3e-24
Identities = 60/142 (42%), Positives = 90/142 (63%), Gaps = 14/142 (9%)
Query: 13 LLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFAL 72
LL LN MY+IV+G W +N I+ G P + GN +T +F+ FA+
Sbjct: 12 LLVLNLIMYIIVIGFASWNLNHFIN-GITNRPGVG------------GNGATFYFLVFAI 58
Query: 73 IAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVEL-RIRNARL 131
+AGVVGAAS ++G++H+R+W +SL ++AS + +AW +T LA G A KE+ + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRTWRGDSLATSASSSLVAWAITALAFGLACKEIHVGGYRGWRL 118
Query: 132 KTMEAFLIILSATQLFYIAAIH 153
+ +EAF+IIL+ TQL Y+ A+H
Sbjct: 119 RVLEAFVIILAFTQLLYVLALH 140
>gb|AAK93757.1| unknown protein [Arabidopsis thaliana] gi|13507555|gb|AAK28640.1|
unknown protein [Arabidopsis thaliana]
gi|18390408|ref|NP_563710.1| AWPM-19-like membrane
family protein [Arabidopsis thaliana]
gi|2494121|gb|AAB80630.1| Strong similarity to Triticum
ABA induced membrane protein (gb|U80037). EST gb|Z27032
comes from this gene. [Arabidopsis thaliana]
gi|25354593|pir||B86178 hypothetical protein [imported]
- Arabidopsis thaliana
Length = 186
Score = 112 bits (280), Expect = 3e-24
Identities = 66/157 (42%), Positives = 92/157 (58%), Gaps = 14/157 (8%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MA + IA LL LN MY+IVLG W +N+ I+ G P G
Sbjct: 1 MATTVGRNIAAPLLFLNLVMYLIVLGFASWCLNKYIN-GQTNHPSFG------------G 47
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
N +T FF+TF+++A VVG AS ++G NHIR W +SL +A + + +AW +T LAMG A K
Sbjct: 48 NGATPFFLTFSILAAVVGVASKLAGANHIRFWRNDSLAAAGASSIVAWAITALAMGLACK 107
Query: 121 EVEL-RIRNARLKTMEAFLIILSATQLFYIAAIHGAA 156
++ + R RLK +EAF+IIL+ TQL Y+ IH +
Sbjct: 108 QINIGGWRGWRLKMIEAFIIILTFTQLLYLMLIHAGS 144
>gb|AAC37469.1| a Lea protein with hydrophobic domain, high pI value (11.6); 15kD
protein; putative gi|7488688|pir||T07661 maturation
protein PM3 - soybean
Length = 181
Score = 107 bits (268), Expect = 8e-23
Identities = 63/146 (43%), Positives = 88/146 (60%), Gaps = 14/146 (9%)
Query: 9 IATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFV 68
+A LL LN MY IVLG W +NR FI G + P F GN +T FF+
Sbjct: 8 VAAPLLFLNLIMYFIVLGFASWCLNR-----FINGQT------YHPSFG--GNGATMFFL 54
Query: 69 TFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVEL-RIR 127
TF+++A V+G S + G NH+R+W ++SL SA + + +AW +T LA G A K++ L R
Sbjct: 55 TFSILAAVLGIVSKLLGGNHMRTWRSDSLASAGATSMVAWAVTALAFGLACKQIHLGGHR 114
Query: 128 NARLKTMEAFLIILSATQLFYIAAIH 153
RL+ +EAF+IIL+ TQL Y+ IH
Sbjct: 115 GWRLRVVEAFIIILTFTQLLYLILIH 140
>ref|XP_477728.1| putative plasma membrane associated protein [Oryza sativa (japonica
cultivar-group)] gi|4097340|gb|AAD10377.1| hydrophobic
LEA-like protein [Oryza sativa]
gi|34395385|dbj|BAC84545.1| putative plasma membrane
associated protein [Oryza sativa (japonica
cultivar-group)] gi|34394744|dbj|BAC84108.1| putative
plasma membrane associated protein [Oryza sativa
(japonica cultivar-group)]
Length = 173
Score = 107 bits (267), Expect = 1e-22
Identities = 61/154 (39%), Positives = 90/154 (57%), Gaps = 20/154 (12%)
Query: 13 LLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFAL 72
LL LN MY+IV+G W +N I + P GN +T +F+ FA+
Sbjct: 12 LLVLNLIMYLIVIGFASWNLNH------FINGQTNYPG-------VAGNGATFYFLVFAI 58
Query: 73 IAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVEL-RIRNARL 131
+AGVVGAAS ++G++H+R+W +SL + A+ + +AW +T LA G A KE+ + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRAWRHDSLATNAASSLIAWAITALAFGLACKEIHIGGHRGWRL 118
Query: 132 KTMEAFLIILSATQLFYIAAIH------GAAAYR 159
+ +EAF+IIL+ TQL Y+ +H G AYR
Sbjct: 119 RVLEAFVIILAFTQLLYVLMLHTGLFGGGDGAYR 152
>ref|XP_467623.1| putative ABA-induced plasma membrane protein [Oryza sativa
(japonica cultivar-group)] gi|46390474|dbj|BAD15935.1|
putative ABA-induced plasma membrane protein [Oryza
sativa (japonica cultivar-group)]
Length = 185
Score = 107 bits (266), Expect = 1e-22
Identities = 55/141 (39%), Positives = 83/141 (58%), Gaps = 14/141 (9%)
Query: 13 LLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFAL 72
LL +N M+ VLG+ GW++N+ ID + H GN S+G+ + F+L
Sbjct: 14 LLCVNLVMHAAVLGLAGWSLNKFIDG--------ETHHHLG------GNTSSGYLLVFSL 59
Query: 73 IAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVELRIRNARLK 132
+AGVVG S + + H+R+W E+L +AAS ++W LT L+ G A K + L R RL+
Sbjct: 60 MAGVVGVCSVLPRLLHVRAWRGETLAAAASTGLVSWALTALSFGLACKHITLGNRGRRLR 119
Query: 133 TMEAFLIILSATQLFYIAAIH 153
T+EAF+ IL+ TQL Y+ +H
Sbjct: 120 TLEAFIAILTLTQLLYLILLH 140
>gb|AAP54176.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|37535174|ref|NP_921889.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|22758326|gb|AAN05530.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 184
Score = 82.4 bits (202), Expect = 4e-15
Identities = 58/169 (34%), Positives = 80/169 (47%), Gaps = 21/169 (12%)
Query: 9 IATLLLGLNFCMYVIVLGIGGWAMNRAI-----DQGFIIGPELQLPAHFSPIFFPMGNAS 63
+A LL LN MYV +LG GWA+N +I D G G + P + +F AS
Sbjct: 11 VAGSLLLLNLLMYVFLLGFAGWALNSSIKNAGADVGVGWGEQPWSPYYRQSAWF----AS 66
Query: 64 TGFFVTFALIAGVVGAASAISGINHI----RSWTAESLPSAASVATMAWTLTLLAMGFAW 119
TFA +AG +G A+ S H SW + L +AAS+ T AW T LA G A
Sbjct: 67 RFHLATFAALAGALGVAAKASAAYHGGRSGASWRPQGLAAAASLGTAAWAATALAFGVAC 126
Query: 120 KEVE--------LRIRNARLKTMEAFLIILSATQLFYIAAIHGAAAYRR 160
+E+ R R++ +E + L+ TQL Y+ +H A A R
Sbjct: 127 REIHDAAAAGPAGAARGWRMRALEGLTVTLAFTQLLYVLLLHAAVAGER 175
>emb|CAC83360.1| putative hydrophobic LEA-like protein [Pinus pinaster]
Length = 93
Score = 74.7 bits (182), Expect = 8e-13
Identities = 39/60 (65%), Positives = 46/60 (76%)
Query: 95 ESLPSAASVATMAWTLTLLAMGFAWKEVELRIRNARLKTMEAFLIILSATQLFYIAAIHG 154
ESL S+AS A AW LTLLAMG A KE+ + RN++L T+E+FLIILS TQLFYI IHG
Sbjct: 1 ESLASSASSAMTAWGLTLLAMGLACKEIHVGGRNSKLITLESFLIILSGTQLFYILVIHG 60
>gb|AAN18125.1| At1g76670/F28O16_4 [Arabidopsis thaliana]
gi|18491195|gb|AAL69500.1| unknown protein [Arabidopsis
thaliana] gi|14532698|gb|AAK64150.1| unknown protein
[Arabidopsis thaliana] gi|46934766|emb|CAG18177.1|
UDP-galactose transporter [Arabidopsis thaliana]
gi|18411172|ref|NP_565138.1| transporter-related
[Arabidopsis thaliana] gi|16604380|gb|AAL24196.1|
At1g76670/F28O16_4 [Arabidopsis thaliana]
gi|25354481|pir||A96795 unknown protein F28O16.4
[imported] - Arabidopsis thaliana
gi|6143887|gb|AAF04433.1| unknown protein; 11341-9662
[Arabidopsis thaliana]
Length = 347
Score = 43.1 bits (100), Expect = 0.003
Identities = 28/96 (29%), Positives = 45/96 (46%), Gaps = 4/96 (4%)
Query: 24 VLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAI 83
V +G WAMN G I+ + + + S F TGF F + G+V A+ +
Sbjct: 12 VSDVGAWAMNVISSVGIIMANKQLMSS--SGFGFGFATTLTGFHFAFTALVGMVSNATGL 69
Query: 84 SGINHIRSWTA--ESLPSAASVATMAWTLTLLAMGF 117
S H+ W S+ + S+A M ++L L ++GF
Sbjct: 70 SASKHVPLWELLWFSIVANISIAAMNFSLMLNSVGF 105
>gb|AAP37671.1| At5g40240 [Arabidopsis thaliana] gi|10177511|dbj|BAB10905.1|
nodulin-like protein [Arabidopsis thaliana]
gi|42568214|ref|NP_198840.2| nodulin MtN21 family
protein [Arabidopsis thaliana]
Length = 368
Score = 42.7 bits (99), Expect = 0.003
Identities = 42/134 (31%), Positives = 64/134 (47%), Gaps = 16/134 (11%)
Query: 10 ATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVT 69
A L GL+F ++V I + + I G +LPA SP+FF +
Sbjct: 40 AATLRGLSFYVFVFYSYIVSTLL--LLPLSVIFGRSRRLPAAKSPLFFKI---------- 87
Query: 70 FALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVELRIRNA 129
+ G+VG S I+G I ++++ +L SA S T A+T T LA+ F ++V LR
Sbjct: 88 --FLLGLVGFMSQIAGCKGI-AYSSPTLASAISNLTPAFTFT-LAVIFRMEQVRLRSSAT 143
Query: 130 RLKTMEAFLIILSA 143
+ K + A L I A
Sbjct: 144 QAKIIGAILSISGA 157
>gb|AAM61035.1| unknown [Arabidopsis thaliana]
Length = 348
Score = 42.4 bits (98), Expect = 0.004
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 24 VLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAI 83
V +G WAMN G I+ + + + S F TGF + G+V A+ +
Sbjct: 13 VSDVGAWAMNVTSSVGIIMANKQLMSS--SGFGFSFATTLTGFHFALTALVGMVSNATGL 70
Query: 84 SGINHIRSWTA--ESLPSAASVATMAWTLTLLAMGF 117
S H+ W SL + S+A M ++L L ++GF
Sbjct: 71 SASKHVPLWELLWFSLVANISIAAMNFSLMLNSVGF 106
>ref|NP_564133.1| transporter-related [Arabidopsis thaliana] gi|25354485|pir||G86343
hypothetical protein T22I11.10 - Arabidopsis thaliana
gi|8886994|gb|AAF80654.1| Strong similarity to a
hypothetical protein F28O16.4 gi|6143887 from
Arabidopsis thaliana gb|AC010718. It contains a
integral membrane protein domain PF|00892
Length = 348
Score = 42.4 bits (98), Expect = 0.004
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 24 VLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAI 83
V +G WAMN G I+ + + + S F TGF + G+V A+ +
Sbjct: 13 VSDVGAWAMNVTSSVGIIMANKQLMSS--SGFGFSFATTLTGFHFALTALVGMVSNATGL 70
Query: 84 SGINHIRSWTA--ESLPSAASVATMAWTLTLLAMGF 117
S H+ W SL + S+A M ++L L ++GF
Sbjct: 71 SASKHVPLWELLWFSLVANISIAAMNFSLMLNSVGF 106
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.328 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 246,910,873
Number of Sequences: 2540612
Number of extensions: 9278261
Number of successful extensions: 31356
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 31308
Number of HSP's gapped (non-prelim): 63
length of query: 160
length of database: 863,360,394
effective HSP length: 117
effective length of query: 43
effective length of database: 566,108,790
effective search space: 24342677970
effective search space used: 24342677970
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0046a.3


