

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0044a.9
(464 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
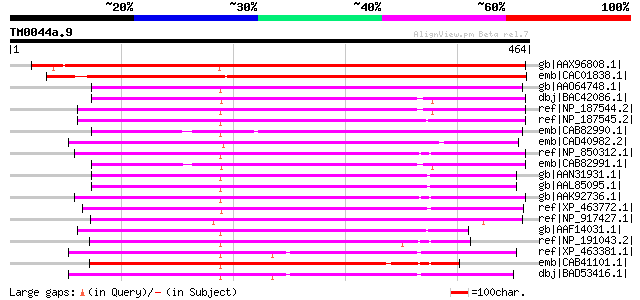
Score E
Sequences producing significant alignments: (bits) Value
gb|AAX96808.1| Transmembrane amino acid transporter protein [Ory... 512 e-143
emb|CAC01838.1| putative protein [Arabidopsis thaliana] gi|15237... 510 e-143
gb|AAO64748.1| At5g02170/T7H20_220 [Arabidopsis thaliana] gi|208... 283 1e-74
dbj|BAC42086.1| unknown protein [Arabidopsis thaliana] gi|306795... 282 1e-74
ref|NP_187544.2| amino acid transporter family protein [Arabidop... 272 1e-71
ref|NP_187545.2| amino acid transporter family protein [Arabidop... 272 2e-71
emb|CAB82990.1| putative protein [Arabidopsis thaliana] gi|11358... 261 2e-68
emb|CAD40982.2| OSJNBa0072F16.7 [Oryza sativa (japonica cultivar... 261 4e-68
ref|NP_850312.1| amino acid transporter family protein [Arabidop... 258 2e-67
emb|CAB82991.1| putative protein [Arabidopsis thaliana] gi|11358... 258 2e-67
gb|AAN31931.1| unknown protein [Arabidopsis thaliana] 258 2e-67
gb|AAL85095.1| unknown protein [Arabidopsis thaliana] gi|1453270... 258 2e-67
gb|AAK92736.1| unknown protein [Arabidopsis thaliana] 258 4e-67
ref|XP_463772.1| putative amino acid transport protein [Oryza sa... 252 2e-65
ref|NP_917427.1| P0712E02.23 [Oryza sativa (japonica cultivar-gr... 244 4e-63
gb|AAF14031.1| hypothetical protein [Arabidopsis thaliana] 241 5e-62
ref|NP_191043.2| amino acid transporter family protein [Arabidop... 238 2e-61
ref|XP_463381.1| P0025A05.3 [Oryza sativa (japonica cultivar-gro... 236 1e-60
emb|CAB41101.1| putative protein [Arabidopsis thaliana] gi|74863... 235 2e-60
dbj|BAD53416.1| amino acid transporter-like [Oryza sativa (japon... 234 3e-60
>gb|AAX96808.1| Transmembrane amino acid transporter protein [Oryza sativa
(japonica cultivar-group)]
Length = 483
Score = 512 bits (1318), Expect = e-143
Identities = 270/457 (59%), Positives = 334/457 (73%), Gaps = 16/457 (3%)
Query: 20 DSGHGANLGVQ-LANCNSC----------VEESQLCKCDHINNIEESAVARDTNVDAEHD 68
+S HGA L +Q L C VE + CKC + E +A R V+A H
Sbjct: 22 ESVHGAQLALQRLTAARRCGGGGDAACVDVEAGKPCKCGE-EHTEAAAAGRVAAVEAVHH 80
Query: 69 AKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLK 128
K SS AHSV+NMVGMLIGLGQLSTPYA+E GGWAS LL+ LGVMC YT HL+GKCL
Sbjct: 81 GKPTSSFAHSVINMVGMLIGLGQLSTPYALENGGWASVFLLVGLGVMCAYTAHLIGKCLD 140
Query: 129 KNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHL-- 186
+P ++Y DIG AFG KGR +A+ IY++IF +LVSYTISL DNL V HL
Sbjct: 141 DDPASKTYQDIGERAFGGKGRVVASAFIYLEIFFALVSYTISLSDNLPLVFAGAASHLHL 200
Query: 187 --AKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRV 244
+L+A Q+LTV AVLVALPSLWLRDLS+ISFLS G++MS++IF +V A FGGV +
Sbjct: 201 PWVRLTATQLLTVAAVLVALPSLWLRDLSTISFLSFAGIVMSLLIFGTVVCAAAFGGVGL 260
Query: 245 NHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIM 304
IP LRL IP++SG+Y+FSY GHIVFPN++ AMKDPS FT+VS+ SF VTALYT +
Sbjct: 261 GGYIPALRLERIPAVSGLYMFSYAGHIVFPNIHAAMKDPSAFTRVSVASFAVVTALYTAL 320
Query: 305 GYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSS 364
++GA MFGP V+SQ+TLSMP VT+IALWATVLTP+TKYALEFAP AIQLE LP++
Sbjct: 321 AFVGASMFGPSVSSQITLSMPPGLAVTRIALWATVLTPVTKYALEFAPFAIQLERHLPAA 380
Query: 365 MSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICR 424
MS R + ++RG +GS+ LL+ILALAL+VPYF++VLSLTGSLVSVAIS+IFPCAFY+KI
Sbjct: 381 MSPRARTLVRGGVGSAALLLILALALSVPYFQYVLSLTGSLVSVAISIIFPCAFYLKIRW 440
Query: 425 GQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
G++S+P LN ++I G +L VVGT SS+ LV+ I
Sbjct: 441 GRVSRPAVALNAAMIAAGVVLAVVGTASSATSLVQSI 477
>emb|CAC01838.1| putative protein [Arabidopsis thaliana]
gi|15237401|ref|NP_197176.1| amino acid transporter
family protein [Arabidopsis thaliana]
gi|11357814|pir||T51506 hypothetical protein F5E19_80 -
Arabidopsis thaliana
Length = 426
Score = 510 bits (1313), Expect = e-143
Identities = 260/429 (60%), Positives = 325/429 (75%), Gaps = 10/429 (2%)
Query: 34 CNSCVEESQLCKCDHINNIEESAVARDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLS 93
C +CVEE++ C+C+H + E + A NSS HSV+NMVGMLIGLGQLS
Sbjct: 3 CVACVEENKGCECEHEKPVRELVLEA---------ASENSSFLHSVINMVGMLIGLGQLS 53
Query: 94 TPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAA 153
PYAVE GGW S LLI+ G++ TYT H+LGKC+++NPK +SY DIG AFG GR +
Sbjct: 54 MPYAVESGGWMSIFLLISFGILTTYTSHILGKCIRRNPKSKSYSDIGYSAFGRHGRLIVC 113
Query: 154 ILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALPSLWLRDLS 213
+ IY++IFM+LVSYTISLHDN++ + A + LT AV +ALPSLW+RDLS
Sbjct: 114 LFIYLEIFMALVSYTISLHDNISAAFPATFSNHGHFPAAK-LTAVAVAIALPSLWIRDLS 172
Query: 214 SISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVF 273
SISFLSS G+LMS +IF SV TA+FGGV + +IP LRL NIP++SGIY+FS+GGHIVF
Sbjct: 173 SISFLSSGGILMSAIIFGSVVYTAIFGGVIDDGKIPVLRLENIPTVSGIYLFSFGGHIVF 232
Query: 274 PNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKI 333
PNLY +MKDPSKFTKVSI+SF TVTALY + GAKMFGP VNSQ+TLS+P +VTKI
Sbjct: 233 PNLYTSMKDPSKFTKVSIVSFATVTALYGALAITGAKMFGPSVNSQITLSLPKHLVVTKI 292
Query: 334 ALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVP 393
ALWATVLTPMTKYALEFAP+AIQLE LPS+M+ RTK++ RG++GS+LLLVILALALTVP
Sbjct: 293 ALWATVLTPMTKYALEFAPLAIQLERSLPSTMTDRTKLVARGLMGSALLLVILALALTVP 352
Query: 394 YFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISS 453
YF +VLSLTGSLVSV I++ P AFY+KIC ++K N+ + G +LGV+G+ S
Sbjct: 353 YFGYVLSLTGSLVSVTIAVTLPSAFYLKICWDGMTKFTRAANLGFVVLGCVLGVLGSFES 412
Query: 454 SKLLVEKIL 462
SKLLV++++
Sbjct: 413 SKLLVKELV 421
>gb|AAO64748.1| At5g02170/T7H20_220 [Arabidopsis thaliana]
gi|20856761|gb|AAM26683.1| AT5g02170/T7H20_220
[Arabidopsis thaliana] gi|22326569|ref|NP_195837.2|
amino acid transporter family protein [Arabidopsis
thaliana]
Length = 526
Score = 283 bits (723), Expect = 1e-74
Identities = 148/387 (38%), Positives = 229/387 (58%), Gaps = 2/387 (0%)
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
+ + SV+N + +L G+ L+ PYAV++GGW +L + G++ YT LL +CL+ +P +
Sbjct: 136 TFSQSVLNGINVLCGVALLTMPYAVKEGGWLGLFILFSFGIITFYTGILLKRCLENSPGI 195
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KLSA 191
+Y DIG AFG GR L +IL+Y++++ S V Y I + DNL+ + L++ L +
Sbjct: 196 HTYPDIGQAAFGTTGRILVSILLYVELYASCVEYIIMMSDNLSRMFPNTSLYINGFSLDS 255
Query: 192 PQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPFL 251
Q+ + L+ LP++WL+DLS +S+LS+ GV+ S+++ + + GV + L
Sbjct: 256 TQVFAITTTLIVLPTVWLKDLSLLSYLSAGGVISSILLALCLFWAGSVDGVGFHISGQAL 315
Query: 252 RLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKM 311
+ NIP GIY F +G H VFPN+Y +MK+PSKF V ++SF T Y + G M
Sbjct: 316 DITNIPVAIGIYGFGFGSHSVFPNIYSSMKEPSKFPTVLLISFAFCTLFYIAVAVCGFTM 375
Query: 312 FGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKM 371
FG + SQ TL+MP +KIA+W V+TPMTKYAL PV + LE +PSS
Sbjct: 376 FGDAIQSQFTLNMPPHFTSSKIAVWTAVVTPMTKYALTITPVMLSLEELIPSSSRKMRSK 435
Query: 372 ILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPL 431
+ + + L+L L +ALTVP+F V +L GS +++ I+LIFPC YI I +G+++
Sbjct: 436 GVSMLFRTILVLSTLVVALTVPFFATVAALIGSFIAMLIALIFPCLCYISIMKGRLTNFQ 495
Query: 432 FVLNISIITFGFLLGVVGTISSSKLLV 458
+ I I+ G + G GT S+ L+
Sbjct: 496 IGICILIVIIGIVSGCCGTYSAIARLI 522
>dbj|BAC42086.1| unknown protein [Arabidopsis thaliana] gi|30679549|ref|NP_195838.2|
amino acid transporter family protein [Arabidopsis
thaliana]
Length = 550
Score = 282 bits (722), Expect = 1e-74
Identities = 150/394 (38%), Positives = 234/394 (59%), Gaps = 11/394 (2%)
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
S + SV+N +L GLG ++ PYA+++ GW +L+ GV+ YT L+ +CL+ +P +
Sbjct: 160 SFSQSVLNGTNVLCGLGLITMPYAIKESGWLGLPILLFFGVITCYTGVLMKRCLESSPGI 219
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAK---LS 190
++Y DIG AFG GRF+ +IL+Y++++ + V Y I + DNL+ + L +A L
Sbjct: 220 QTYPDIGQAAFGITGRFIISILLYVELYAACVEYIIMMSDNLSGLFPNVSLSIASGISLD 279
Query: 191 APQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPF 250
+PQI + L+ LP++WL+DLS +S+LS GVL S+++ + + G+ +
Sbjct: 280 SPQIFAILTTLLVLPTVWLKDLSLLSYLSVGGVLASILLGICLFWVGAVDGIGFHATGRV 339
Query: 251 LRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAK 310
L N+P GI+ F Y GH VFPN+Y +MKDPS+F V ++ F+ T LY + G
Sbjct: 340 FDLSNLPVTIGIFGFGYSGHSVFPNIYSSMKDPSRFPLVLVICFSFCTVLYIAVAVCGYT 399
Query: 311 MFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTK 370
MFG V SQ TL+MP +K+A+W V+TPMTKYAL P+ + LE +P++ K
Sbjct: 400 MFGEAVESQFTLNMPKHFFPSKVAVWTAVITPMTKYALTITPIVMSLEELIPTA-----K 454
Query: 371 MILRGI---IGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQI 427
M RG+ + L+ L +AL+VP+F V +L GS +++ ++LIFPC Y+ I +G++
Sbjct: 455 MRSRGVSILFRTMLVTSTLVVALSVPFFAIVAALIGSFLAMLVALIFPCLCYLSILKGKL 514
Query: 428 SKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
S L I II FG + G GT S+ L ++
Sbjct: 515 SNTQIGLCIFIIVFGVVSGCCGTYSAISRLANQM 548
>ref|NP_187544.2| amino acid transporter family protein [Arabidopsis thaliana]
Length = 524
Score = 272 bits (696), Expect = 1e-71
Identities = 151/407 (37%), Positives = 235/407 (57%), Gaps = 11/407 (2%)
Query: 61 TNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTC 120
+N D + S SV+N + +L G+ L+ PYAV++GGW +L++ ++ YT
Sbjct: 122 SNTDLSYGEPNFCSFPQSVLNGINVLCGISLLTMPYAVKEGGWLGLCILLSFAIITCYTG 181
Query: 121 HLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLH 180
LL +CL+ + LR+Y DIG AFG GR + +IL+YM++++ V Y I + DNL+ V
Sbjct: 182 ILLKRCLESSSDLRTYPDIGQAAFGFTGRLIISILLYMELYVCCVEYIIMMSDNLSRVFP 241
Query: 181 LKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV 238
L++ L +PQI + A L+ LP++WL+DLS +S+LS+ GV +S+++ + +
Sbjct: 242 NITLNIVGVSLDSPQIFAISATLIVLPTVWLKDLSLLSYLSAGGVFVSILLALCLFWVGS 301
Query: 239 FGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVT 298
GV + L L N+P GI+ F + GH V P++Y +MK+PSKF V ++SF
Sbjct: 302 VDGVGFHTGGKSLDLANLPVAIGIFGFGFSGHAVLPSIYSSMKEPSKFPLVLLISFGFCV 361
Query: 299 ALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLE 358
Y ++ G MFG + SQ TL+MP + +KIA+W V+ PMTKYAL P+ + LE
Sbjct: 362 FFYIVVAICGYSMFGEAIQSQFTLNMPQQYTASKIAVWTAVVVPMTKYALALTPIVLGLE 421
Query: 359 H-KLPSSMSGRTKMILRGI---IGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIF 414
LPS KM G+ I + L+L L +ALT P+F + +L GS +++ + IF
Sbjct: 422 ELMLPSE-----KMRSYGVSIFIKTILVLSTLVVALTFPFFAIMGALMGSFLAMLVDFIF 476
Query: 415 PCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
PC Y+ I +G++SK + + II G + G GT S+ LV K+
Sbjct: 477 PCLCYLSILKGRLSKTQIGICVFIIISGIVSGCCGTYSAIGRLVGKL 523
>ref|NP_187545.2| amino acid transporter family protein [Arabidopsis thaliana]
Length = 528
Score = 272 bits (695), Expect = 2e-71
Identities = 147/403 (36%), Positives = 232/403 (57%), Gaps = 3/403 (0%)
Query: 61 TNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTC 120
+N D + S SV+N + +L G+ L+ PYAV++GGW +L++ ++ YT
Sbjct: 122 SNTDLSYGEPNFCSFPQSVLNGINVLCGISLLTMPYAVKEGGWLGLCILLSFAIITCYTG 181
Query: 121 HLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLH 180
LL +CL+ + LR+Y DIG AFG GR + +IL+YM++++ V Y I + DNL+ V
Sbjct: 182 ILLKRCLESSSDLRTYPDIGQAAFGFTGRLIISILLYMELYVCCVEYIIMMSDNLSRVFP 241
Query: 181 LKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV 238
L++ L +PQI + A L+ LP++WL+DLS +S+LS+ GV +S+++ + +
Sbjct: 242 NITLNIVGVSLDSPQIFAISATLIVLPTVWLKDLSLLSYLSAGGVFVSILLALCLFWVGS 301
Query: 239 FGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVT 298
GV + L L N+P GI+ F + GH V P++Y +MK+PSKF V ++SF
Sbjct: 302 VDGVGFHTGGKALDLANLPVAIGIFGFGFSGHAVLPSIYSSMKEPSKFPLVLLISFGFCV 361
Query: 299 ALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLE 358
Y + G MFG + SQ TL+MP + +KIA+W V+ PMTKYAL P+ + LE
Sbjct: 362 FFYIAVAICGYSMFGEAIQSQFTLNMPQQYTASKIAVWTAVVVPMTKYALALTPIVLGLE 421
Query: 359 HKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAF 418
+P S R+ + I + L+L L +ALT P+F + +L GS ++ + IFPC
Sbjct: 422 ELMPPSEKMRSYGV-SIFIKTILVLSTLVVALTFPFFAIMGALMGSFLATLVDFIFPCLC 480
Query: 419 YIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
Y+ I +G++SK + + II G + G GT S+ LV ++
Sbjct: 481 YLSILKGRLSKTQIGICVFIIISGIVSGCCGTYSAIGRLVGEL 523
>emb|CAB82990.1| putative protein [Arabidopsis thaliana] gi|11358405|pir||T48238
hypothetical protein T7H20.220 - Arabidopsis thaliana
Length = 516
Score = 261 bits (668), Expect = 2e-68
Identities = 145/387 (37%), Positives = 220/387 (56%), Gaps = 12/387 (3%)
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
+ + SV+N + +L G+ L+ PYAV++GGW +L + G++ YT LL +CL+ +P +
Sbjct: 136 TFSQSVLNGINVLCGVALLTMPYAVKEGGWLGLFILFSFGIITFYTGILLKRCLENSPGI 195
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KLSA 191
+Y DIG AFG GR L + S V Y I + DNL+ + L++ L +
Sbjct: 196 HTYPDIGQAAFGTTGRILVSA--------SCVEYIIMMSDNLSRMFPNTSLYINGFSLDS 247
Query: 192 PQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPFL 251
Q+ + L+ LP++WL+DLS +S+LS GV+ S+++ + + GV + L
Sbjct: 248 TQVFAITTTLIVLPTVWLKDLSLLSYLS--GVISSILLALCLFWAGSVDGVGFHISGQAL 305
Query: 252 RLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKM 311
+ NIP GIY F +G H VFPN+Y +MK+PSKF V ++SF T Y + G M
Sbjct: 306 DITNIPVAIGIYGFGFGSHSVFPNIYSSMKEPSKFPTVLLISFAFCTLFYIAVAVCGFTM 365
Query: 312 FGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKM 371
FG + SQ TL+MP +KIA+W V+TPMTKYAL PV + LE +PSS
Sbjct: 366 FGDAIQSQFTLNMPPHFTSSKIAVWTAVVTPMTKYALTITPVMLSLEELIPSSSRKMRSK 425
Query: 372 ILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPL 431
+ + + L+L L +ALTVP+F V +L GS +++ I+LIFPC YI I +G+++
Sbjct: 426 GVSMLFRTILVLSTLVVALTVPFFATVAALIGSFIAMLIALIFPCLCYISIMKGRLTNFQ 485
Query: 432 FVLNISIITFGFLLGVVGTISSSKLLV 458
+ I I+ G + G GT S+ L+
Sbjct: 486 IGICILIVIIGIVSGCCGTYSAIARLI 512
>emb|CAD40982.2| OSJNBa0072F16.7 [Oryza sativa (japonica cultivar-group)]
gi|50924802|ref|XP_472750.1| OSJNBa0072F16.7 [Oryza
sativa (japonica cultivar-group)]
Length = 455
Score = 261 bits (666), Expect = 4e-68
Identities = 145/407 (35%), Positives = 234/407 (56%), Gaps = 8/407 (1%)
Query: 53 EESAVARDTNVDAEHDAKAN-SSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIA 111
++ V RD +A + ++ + N + L G+G LS PYA+ +GGW S +LL+A
Sbjct: 47 KQEPVERDHEAQCSPEADGDGATFVRTCFNGLNALSGVGLLSIPYALSEGGWLSLVLLLA 106
Query: 112 LGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISL 171
+ ++C YT LL +C+ +P +R Y DIG AFGAKGR + +Y ++++ + + I
Sbjct: 107 VAMVCCYTGLLLRRCMAASPAVRGYPDIGALAFGAKGRLAVSAFLYAELYLVAIGFLILE 166
Query: 172 HDNLTTVLHLKQLHLAKL--SAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVI 229
DNL + L + L S Q+ V +V LP+ WLR L+ ++++S+ GVL SVV+
Sbjct: 167 GDNLDKLFPGTSLAVGGLVVSGKQLFVVVVAVVILPTTWLRSLAVLAYVSASGVLASVVV 226
Query: 230 FVSVAATAVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKV 289
V AVF GV + + L + +P+ G+Y F Y GH +FP L +M++ KF++V
Sbjct: 227 VFCVLWAAVFDGVGFHGKGRMLNVSGLPTALGLYTFCYCGHAIFPTLCNSMQEKDKFSRV 286
Query: 290 SILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALE 349
++ F T Y M +G M+G +V SQVTL++P +I +K+A++ T++ P +KYAL
Sbjct: 287 LVICFVACTVNYGSMAILGYLMYGDDVKSQVTLNLPEGKISSKLAIYTTLINPFSKYALM 346
Query: 350 FAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVA 409
PVA +E KL + +++R +I S +++ ALTVP+F H+++L GSL+SV
Sbjct: 347 VTPVATAIEEKLLAGNKRSVNVLIRTLIVVSTVVI----ALTVPFFGHLMALVGSLLSVM 402
Query: 410 ISLIFPCAFYIKIC-RGQISKPLFVLNISIITFGFLLGVVGTISSSK 455
S++ PC Y+KI + + +L +II G L+ GT SS K
Sbjct: 403 ASMLLPCICYLKIFGLTRCGRGETLLIAAIIVLGSLVAATGTYSSLK 449
>ref|NP_850312.1| amino acid transporter family protein [Arabidopsis thaliana]
Length = 550
Score = 258 bits (660), Expect = 2e-67
Identities = 145/405 (35%), Positives = 230/405 (55%), Gaps = 4/405 (0%)
Query: 59 RDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTY 118
+ + V E NSS +V+N + +L G+G LSTPYA ++GGW ++L G++ Y
Sbjct: 146 KSSMVSHEIPMSRNSSYGQAVLNGLNVLCGVGILSTPYAAKEGGWLGLMILFVYGLLSFY 205
Query: 119 TCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTV 178
T LL CL L +Y DIG AFG GR +I++Y++++ V Y I DNL+++
Sbjct: 206 TGILLRYCLDSESDLETYPDIGQAAFGTTGRIFVSIVLYLELYACCVEYIILESDNLSSL 265
Query: 179 LHLKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAAT 236
L + +L A + + L LP++WLRDLS +S++S+ GV+ SV++ + +
Sbjct: 266 YPNAALSIGGFQLDARHLFALLTTLAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLFWI 325
Query: 237 AVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTT 296
+ V ++ + L L +P G+Y + Y GH VFPN+Y +M PS++ V + F
Sbjct: 326 GLVDEVGIHSKGTTLNLSTLPVAIGLYGYCYSGHAVFPNIYTSMAKPSQYPAVLLTCFGI 385
Query: 297 VTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQ 356
T +Y + MG MFG SQ TL++P I TKIA+W TV+ P TKYAL +PVA+
Sbjct: 386 CTLMYAGVAVMGYTMFGESTQSQFTLNLPQDLIATKIAVWTTVVNPFTKYALTISPVAMS 445
Query: 357 LEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPC 416
LE +PS R+ G I + L+ L + L +P+F V+SL GSL+++ ++LI P
Sbjct: 446 LEELIPSRHI-RSHWYAIG-IRTLLVFSTLLVGLAIPFFGLVMSLIGSLLTMLVTLILPP 503
Query: 417 AFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
A ++ I R +++ +L + II G + V+G+ S+ +VEK+
Sbjct: 504 ACFLSIVRRKVTPTQMMLCVLIIIVGAISSVIGSYSALSKIVEKL 548
>emb|CAB82991.1| putative protein [Arabidopsis thaliana] gi|11358406|pir||T48239
hypothetical protein T7H20.230 - Arabidopsis thaliana
Length = 543
Score = 258 bits (660), Expect = 2e-67
Identities = 144/394 (36%), Positives = 223/394 (56%), Gaps = 18/394 (4%)
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
S + SV+N +L GLG ++ PYA+++ GW +L+ GV+ YT L+ +CL+ +P +
Sbjct: 160 SFSQSVLNGTNVLCGLGLITMPYAIKESGWLGLPILLFFGVITCYTGVLMKRCLESSPGI 219
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAK---LS 190
++Y DIG AFG + ++ V Y I + DNL+ + L +A L
Sbjct: 220 QTYPDIGQAAFGITDSSIRGVV-------PCVEYIIMMSDNLSGLFPNVSLSIASGISLD 272
Query: 191 APQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPF 250
+PQI + L+ LP++WL+DLS +S+LS GVL S+++ + + G+ +
Sbjct: 273 SPQIFAILTTLLVLPTVWLKDLSLLSYLSVGGVLASILLGICLFWVGAVDGIGFHATGRV 332
Query: 251 LRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAK 310
L N+P GI+ F Y GH VFPN+Y +MKDPS+F V ++ F+ T LY + G
Sbjct: 333 FDLSNLPVTIGIFGFGYSGHSVFPNIYSSMKDPSRFPLVLVICFSFCTVLYIAVAVCGYT 392
Query: 311 MFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTK 370
MFG V SQ TL+MP +K+A+W V+TPMTKYAL P+ + LE +P++ K
Sbjct: 393 MFGEAVESQFTLNMPKHFFPSKVAVWTAVITPMTKYALTITPIVMSLEELIPTA-----K 447
Query: 371 MILRGI---IGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQI 427
M RG+ + L+ L +AL+VP+F V +L GS +++ ++LIFPC Y+ I +G++
Sbjct: 448 MRSRGVSILFRTMLVTSTLVVALSVPFFAIVAALIGSFLAMLVALIFPCLCYLSILKGKL 507
Query: 428 SKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
S L I II FG + G GT S+ L ++
Sbjct: 508 SNTQIGLCIFIIVFGVVSGCCGTYSAISRLANQM 541
>gb|AAN31931.1| unknown protein [Arabidopsis thaliana]
Length = 407
Score = 258 bits (660), Expect = 2e-67
Identities = 138/382 (36%), Positives = 218/382 (56%), Gaps = 4/382 (1%)
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
S+ ++ N + ++ G+G LSTPY V++ GWAS ++L+ V+C YT L+ C + +
Sbjct: 19 SVIQTIFNAINVMAGVGLLSTPYTVKEAGWASMVILLLFAVICCYTATLMKDCFENKTGI 78
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KLSA 191
+Y DIG AFG GR L +L+Y +++ V + I DNLT + L L +L +
Sbjct: 79 ITYPDIGEAAFGKYGRILICMLLYTELYSYCVEFIILEGDNLTGLFPGTSLDLLGFRLDS 138
Query: 192 PQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPFL 251
+ + L+ LP++WL+DL IS+LS+ GV+ + +I VSV GG+ +H +
Sbjct: 139 KHLFGILTALIVLPTVWLKDLRIISYLSAGGVIATALIAVSVFFLGTTGGIGFHHTGQAV 198
Query: 252 RLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKM 311
+ + IP GIY F Y GH VFPN+Y++M D +KF K I F LY + MG M
Sbjct: 199 KWNGIPFAIGIYGFCYSGHSVFPNIYQSMADKTKFNKAVITCFIICVLLYGGVAIMGYLM 258
Query: 312 FGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKM 371
FG SQ+TL+MP Q +K+A W TV++P TKYAL P+A +E LP MS
Sbjct: 259 FGEATLSQITLNMPQDQFFSKVAQWTTVVSPFTKYALLMNPLARSIEELLPERMSENIWC 318
Query: 372 ILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPL 431
L ++ ++L+ + A +P+F +++L GSL+S+ +++I P +IKI + ++
Sbjct: 319 FL--LLRTALVASSVCSAFLIPFFGLMMALIGSLLSILVAIIMPALCFIKIMGNKATRTQ 376
Query: 432 FVLNISIITFGFLLGVVGTISS 453
+L+ I+ G + G +GT SS
Sbjct: 377 MILSSIIVAIGVVSGTLGTYSS 398
>gb|AAL85095.1| unknown protein [Arabidopsis thaliana] gi|14532708|gb|AAK64155.1|
unknown protein [Arabidopsis thaliana]
gi|20197120|gb|AAD11993.2| expressed protein
[Arabidopsis thaliana] gi|18405629|ref|NP_030664.1|
amino acid transporter family protein [Arabidopsis
thaliana]
Length = 536
Score = 258 bits (660), Expect = 2e-67
Identities = 138/382 (36%), Positives = 218/382 (56%), Gaps = 4/382 (1%)
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
S+ ++ N + ++ G+G LSTPY V++ GWAS ++L+ V+C YT L+ C + +
Sbjct: 148 SVIQTIFNAINVMAGVGLLSTPYTVKEAGWASMVILLLFAVICCYTATLMKDCFENKTGI 207
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KLSA 191
+Y DIG AFG GR L +L+Y +++ V + I DNLT + L L +L +
Sbjct: 208 ITYPDIGEAAFGKYGRILICMLLYTELYSYCVEFIILEGDNLTGLFPGTSLDLLGFRLDS 267
Query: 192 PQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPFL 251
+ + L+ LP++WL+DL IS+LS+ GV+ + +I VSV GG+ +H +
Sbjct: 268 KHLFGILTALIVLPTVWLKDLRIISYLSAGGVIATALIAVSVFFLGTTGGIGFHHTGQAV 327
Query: 252 RLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKM 311
+ + IP GIY F Y GH VFPN+Y++M D +KF K I F LY + MG M
Sbjct: 328 KWNGIPFAIGIYGFCYSGHSVFPNIYQSMADKTKFNKAVITCFIICVLLYGGVAIMGYLM 387
Query: 312 FGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKM 371
FG SQ+TL+MP Q +K+A W TV++P TKYAL P+A +E LP MS
Sbjct: 388 FGEATLSQITLNMPQDQFFSKVAQWTTVVSPFTKYALLMNPLARSIEELLPERMSENIWC 447
Query: 372 ILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPL 431
L ++ ++L+ + A +P+F +++L GSL+S+ +++I P +IKI + ++
Sbjct: 448 FL--LLRTALVASSVCSAFLIPFFGLMMALIGSLLSILVAIIMPALCFIKIMGNKATRTQ 505
Query: 432 FVLNISIITFGFLLGVVGTISS 453
+L+ I+ G + G +GT SS
Sbjct: 506 MILSSIIVAIGVVSGTLGTYSS 527
>gb|AAK92736.1| unknown protein [Arabidopsis thaliana]
Length = 550
Score = 258 bits (658), Expect = 4e-67
Identities = 145/405 (35%), Positives = 230/405 (55%), Gaps = 4/405 (0%)
Query: 59 RDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTY 118
+ + V E NSS +V+N + +L G+G LSTPYA ++GGW ++L G++ Y
Sbjct: 146 KSSMVSHEIPMSRNSSYGQAVLNGLNVLCGVGILSTPYAAKEGGWLGLMILFVYGLLSFY 205
Query: 119 TCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTV 178
T LL CL L +Y DIG AFG GR +I++Y++++ V Y I DNL+++
Sbjct: 206 TGILLRYCLDSESDLETYPDIGQAAFGTTGRIFVSIVLYLELYACCVEYIILEIDNLSSL 265
Query: 179 LHLKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAAT 236
L + +L A + + L LP++WLRDLS +S++S+ GV+ SV++ + +
Sbjct: 266 YPNAALSIGGFQLDARHLFALLTTLAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLFWI 325
Query: 237 AVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTT 296
+ V ++ + L L +P G+Y + Y GH VFPN+Y +M PS++ V + F
Sbjct: 326 GLVDEVGIHSKGTTLNLSTLPVAIGLYGYCYSGHAVFPNIYTSMAKPSQYPAVLLTCFGI 385
Query: 297 VTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQ 356
T +Y + MG MFG SQ TL++P I TKIA+W TV+ P TKYAL +PVA+
Sbjct: 386 CTLMYAGVAVMGYTMFGESTQSQFTLNLPQDLIATKIAVWTTVVNPFTKYALTISPVAMS 445
Query: 357 LEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPC 416
LE +PS R+ G I + L+ L + L +P+F V+SL GSL+++ ++LI P
Sbjct: 446 LEELIPSRHI-RSHWYAIG-IRTLLVFSTLLVGLAIPFFGLVMSLIGSLLTMLVTLILPP 503
Query: 417 AFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
A ++ I R +++ +L + II G + V+G+ S+ +VEK+
Sbjct: 504 ACFLSIVRRKVTPTQMMLCVLIIIVGAISSVIGSYSALSKIVEKL 548
>ref|XP_463772.1| putative amino acid transport protein [Oryza sativa (japonica
cultivar-group)] gi|41053220|dbj|BAD08181.1| putative
amino acid transport protein [Oryza sativa (japonica
cultivar-group)]
Length = 571
Score = 252 bits (644), Expect = 2e-65
Identities = 137/396 (34%), Positives = 221/396 (55%), Gaps = 5/396 (1%)
Query: 66 EHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGK 125
E A S +VMN + +L G+G LSTPYA++QGGW ++L V+ YT LL +
Sbjct: 176 EVPAYQQCSYTQAVMNGINVLCGVGILSTPYAIKQGGWLGLVILCLFAVLAWYTGVLLRR 235
Query: 126 CLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLH 185
CL L +Y DIG+ AFG GR +I++Y++++ + Y I DNL+ + L
Sbjct: 236 CLDSKEGLETYPDIGHAAFGTTGRIAISIILYIELYACCIEYLILESDNLSKLFPNAHLT 295
Query: 186 LAK--LSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVR 243
+ L++ + L+ +P+ WLRDLS +S+LS+ GV+ S+++ V + V V
Sbjct: 296 IGSMTLNSHVFFAILTTLIVMPTTWLRDLSCLSYLSAGGVIASILVVVCLCWVGVVDHVG 355
Query: 244 VNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTI 303
++ L L IP G+Y + Y GH VFPN+Y ++K+ ++F + + L+
Sbjct: 356 FENKGTALNLPGIPIAIGLYGYCYSGHGVFPNIYSSLKNRNQFPSILFTCIGLSSILFAG 415
Query: 304 MGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPS 363
MG KMFG SQ TL++P +V+K+A+W TV P+TKYAL P+A+ LE LP
Sbjct: 416 AAVMGYKMFGESTESQFTLNLPENLVVSKVAVWTTVANPITKYALTITPLAMSLEELLPP 475
Query: 364 SMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKIC 423
+ +I+ + SSL++ L +AL+VP+F V++L GSL+++ ++ I PCA ++ I
Sbjct: 476 NQQKYANIIM---LRSSLVVSTLLIALSVPFFGLVMALVGSLLTMLVTYILPCACFLAIL 532
Query: 424 RGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVE 459
+ +++ II G VGT SS +++
Sbjct: 533 KRKVTWHQIAACSFIIVVGVCCACVGTYSSLSKIIQ 568
>ref|NP_917427.1| P0712E02.23 [Oryza sativa (japonica cultivar-group)]
gi|20160971|dbj|BAB89905.1| amino acid transporter-like
[Oryza sativa (japonica cultivar-group)]
gi|14588694|dbj|BAB61859.1| amino acid transporter-like
[Oryza sativa (japonica cultivar-group)]
Length = 460
Score = 244 bits (623), Expect = 4e-63
Identities = 125/390 (32%), Positives = 225/390 (57%), Gaps = 4/390 (1%)
Query: 73 SSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPK 132
+S S +N+ ++ G+G LS PYA+ QGGW S L +G +C YT +L+ +C++ +
Sbjct: 68 ASFGRSCLNLSNIISGIGMLSVPYALSQGGWLSLTLFTMVGAICFYTGNLIDRCMRVDRC 127
Query: 133 LRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLH--LKQLHLAKLS 190
+RSY DIG AFG+ GR ++IY+++++ +S+ I DNL +L + ++ ++
Sbjct: 128 VRSYPDIGYLAFGSYGRMAIGLVIYVELYLVAISFLILEGDNLDKLLPGIVVEILGYQVH 187
Query: 191 APQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPF 250
Q+ + A V LP+ WL++LS ++++S+VG++ SV + S+ V G
Sbjct: 188 GKQLFVLAAAAVILPTTWLKNLSMLAYVSAVGLVSSVALTASLVWAGVAGKGFHMEGSSL 247
Query: 251 LRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAK 310
L L +P+ +Y + GH VFP +Y +M F KV ++S + Y + +G
Sbjct: 248 LNLSELPTALSLYFVCFAGHGVFPTVYSSMNSKKDFPKVLLISLVLCSLNYAVTAVLGYL 307
Query: 311 MFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTK 370
++G +V +QVTLS+P+ ++ T+IA+ T++TP+ KYAL PV I +E KL ++
Sbjct: 308 IYGEDVQAQVTLSLPTGKLYTRIAILTTLITPLAKYALVIQPVTIAIEEKLSATTDAEIN 367
Query: 371 MILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKI--CRGQIS 428
+ R + +++++ + LA TVP+F +++S GS ++V ++++FPC Y+KI RG +
Sbjct: 368 RLTRVLTSTAVVISTVVLACTVPFFGYLISFIGSSLNVTVAVLFPCLSYLKIYMSRGGVG 427
Query: 429 KPLFVLNISIITFGFLLGVVGTISSSKLLV 458
I I+ G + +VGT +S + ++
Sbjct: 428 CFEMAAIIGILVIGVCVAIVGTYTSLQQII 457
>gb|AAF14031.1| hypothetical protein [Arabidopsis thaliana]
Length = 478
Score = 241 bits (614), Expect = 5e-62
Identities = 129/352 (36%), Positives = 204/352 (57%), Gaps = 3/352 (0%)
Query: 61 TNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTC 120
+N D + S SV+N + +L G+ L+ PYAV++GGW +L++ ++ YT
Sbjct: 122 SNTDLSYGEPNFCSFPQSVLNGINVLCGISLLTMPYAVKEGGWLGLCILLSFAIITCYTG 181
Query: 121 HLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLH 180
LL +CL+ + LR+Y DIG AFG GR + +IL+YM++++ V Y I + DNL+ V
Sbjct: 182 ILLKRCLESSSDLRTYPDIGQAAFGFTGRLIISILLYMELYVCCVEYIIMMSDNLSRVFP 241
Query: 181 LKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV 238
L++ L +PQI + A L+ LP++WL+DLS +S+LS+ GV +S+++ + +
Sbjct: 242 NITLNIVGVSLDSPQIFAISATLIVLPTVWLKDLSLLSYLSAGGVFVSILLALCLFWVGS 301
Query: 239 FGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVT 298
GV + L L N+P GI+ F + GH V P++Y +MK+PSKF V ++SF
Sbjct: 302 VDGVGFHTGGKALDLANLPVAIGIFGFGFSGHAVLPSIYSSMKEPSKFPLVLLISFGFCV 361
Query: 299 ALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLE 358
Y + G MFG + SQ TL+MP + +KIA+W V+ PMTKYAL P+ + LE
Sbjct: 362 FFYIAVAICGYSMFGEAIQSQFTLNMPQQYTASKIAVWTAVVVPMTKYALALTPIVLGLE 421
Query: 359 HKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAI 410
+P S R+ + I + L+L L +ALT P+F + +L GS ++ +
Sbjct: 422 ELMPPSEKMRSYGV-SIFIKTILVLSTLVVALTFPFFAIMGALMGSFLATLV 472
>ref|NP_191043.2| amino acid transporter family protein [Arabidopsis thaliana]
Length = 546
Score = 238 bits (608), Expect = 2e-61
Identities = 132/350 (37%), Positives = 207/350 (58%), Gaps = 11/350 (3%)
Query: 72 NSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNP 131
+SS +V+N V +L G+G LSTPYAV++GGW I+L A G++C YT LL CL +P
Sbjct: 149 DSSFGQAVLNGVNVLCGVGILSTPYAVKEGGWLGLIILFAFGILCFYTGLLLRYCLDSHP 208
Query: 132 KLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KL 189
+++Y DIG+ AFG+ GR L ++++YM+++ V Y I DNL+++ L + L
Sbjct: 209 DVQTYPDIGHAAFGSTGRILVSVILYMELYAMSVEYIILEGDNLSSMFPNASLSIGGFHL 268
Query: 190 SAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIP 249
AP++ + L LP++WLRDLS +S++S+ GV+ SV++ + + + V ++ +
Sbjct: 269 DAPRLFALLTTLAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLFWVGLVDDVGIHSKGT 328
Query: 250 FLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGA 309
L L +P G+Y + Y GH VFPN+Y +M PS+F+ V + SF T +Y + MG
Sbjct: 329 PLNLATLPVSVGLYGYCYSGHGVFPNIYTSMAKPSQFSAVLLASFGICTLMYAGVAVMGY 388
Query: 310 KMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALE-------FAPVAIQLEHKLP 362
MFG SQ TL++P + +KIALW TV+ P TKY L A+ LE +P
Sbjct: 389 SMFGESTESQFTLNLPQDLVASKIALWTTVVNPFTKYPLSRNIQRLFIYLFAMSLEELIP 448
Query: 363 SSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISL 412
S+ G+++ I S+L + L + L +P+F V+SL GS +++ I L
Sbjct: 449 SNY-GKSRFYAIA-IRSALAISTLLVGLAIPFFGLVMSLIGSFLTMLIFL 496
>ref|XP_463381.1| P0025A05.3 [Oryza sativa (japonica cultivar-group)]
Length = 424
Score = 236 bits (602), Expect = 1e-60
Identities = 134/407 (32%), Positives = 232/407 (56%), Gaps = 12/407 (2%)
Query: 53 EESAVARDTNVDAEHDA-KANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIA 111
+E+ A D +V+A+ + +S + + +N+ + G+G LS PYAV QGGW S +L +
Sbjct: 16 KEADFADDDDVEAQLTSYHTGASFSRTCLNLTNAVSGIGVLSMPYAVSQGGWLSLLLFVL 75
Query: 112 LGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISL 171
+G +C YT L+ +C++ + + SY DIG +AFGA GR A +Y+++++ +S+ +
Sbjct: 76 VGAVCYYTGTLIERCMRADGSIASYPDIGQYAFGATGRRAVAFFMYVELYLVAISFLVLE 135
Query: 172 HDNLTTVLHLKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVI 229
DNL + + + +L Q+ V A V LP+ WL++L ++++S+ G++ SV +
Sbjct: 136 GDNLDKLFPGATMEILGYQLHGKQLFIVLAAAVILPTTWLKNLGMLAYVSAAGLIASVAL 195
Query: 230 FVSV--AATAVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFT 287
S+ A A G R ++ L L IP+ G+Y + GH VFP +Y +MK+ F+
Sbjct: 196 TASLIWAGVAETGFHRNSNT---LNLAGIPTSLGLYFVCFTGHAVFPTIYSSMKNSKHFS 252
Query: 288 KVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYA 347
KV ++S + Y + +G ++G +V SQVTL++PS ++ TKIA+ T++ P+ KYA
Sbjct: 253 KVLLISSVLCSLNYGLTAVLGYMIYGDDVQSQVTLNLPSGKLYTKIAIVMTLVNPLAKYA 312
Query: 348 LEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVS 407
L AP+ +E +L + R R I +++L + +A TVP+F +++S GS +S
Sbjct: 313 LLVAPITAAVEERLSLT---RGSAPARVAISTAILASTVVVASTVPFFGYLMSFIGSFLS 369
Query: 408 VAISLIFPCAFYIKICRGQ-ISKPLFVLNISIITFGFLLGVVGTISS 453
V +++FPC Y+KI + I + V I+ G + V GT +S
Sbjct: 370 VMATVLFPCLCYLKIYKADGIHRTEMVAIAGILLLGVFVAVTGTYTS 416
>emb|CAB41101.1| putative protein [Arabidopsis thaliana] gi|7486337|pir||T06737
hypothetical protein F28P10.190 - Arabidopsis thaliana
Length = 571
Score = 235 bits (600), Expect = 2e-60
Identities = 128/333 (38%), Positives = 202/333 (60%), Gaps = 7/333 (2%)
Query: 72 NSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNP 131
+SS +V+N V +L G+G LSTPYAV++GGW I+L A G++C YT LL CL +P
Sbjct: 149 DSSFGQAVLNGVNVLCGVGILSTPYAVKEGGWLGLIILFAFGILCFYTGLLLRYCLDSHP 208
Query: 132 KLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KL 189
+++Y DIG+ AFG+ GR L ++++YM+++ V Y I DNL+++ L + L
Sbjct: 209 DVQTYPDIGHAAFGSTGRILVSVILYMELYAMSVEYIILEGDNLSSMFPNASLSIGGFHL 268
Query: 190 SAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIP 249
AP++ + L LP++WLRDLS +S++S+ GV+ SV++ + + + V ++ +
Sbjct: 269 DAPRLFALLTTLAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLFWVGLVDDVGIHSKGT 328
Query: 250 FLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGA 309
L L +P G+Y + Y GH VFPN+Y +M PS+F+ V + SF T +Y + MG
Sbjct: 329 PLNLATLPVSVGLYGYCYSGHGVFPNIYTSMAKPSQFSAVLLASFGICTLMYAGVAVMGY 388
Query: 310 KMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRT 369
MFG SQ TL++P + +KIALW T + YAL +PVA+ LE +PS+ G++
Sbjct: 389 SMFGESTESQFTLNLPQDLVASKIALWT---TKESTYALTLSPVAMSLEELIPSNY-GKS 444
Query: 370 KMILRGIIGSSLLLVILALALTVPYFEHVLSLT 402
+ I S+L + L + L +P+F ++LSLT
Sbjct: 445 RFYAIA-IRSALAISTLLVGLAIPFFVNLLSLT 476
>dbj|BAD53416.1| amino acid transporter-like [Oryza sativa (japonica
cultivar-group)]
Length = 631
Score = 234 bits (598), Expect = 3e-60
Identities = 133/404 (32%), Positives = 230/404 (56%), Gaps = 12/404 (2%)
Query: 53 EESAVARDTNVDAEHDA-KANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIA 111
+E+ A D +V+A+ + +S + + +N+ + G+G LS PYAV QGGW S +L +
Sbjct: 16 KEADFADDDDVEAQLTSYHTGASFSRTCLNLTNAVSGIGVLSMPYAVSQGGWLSLLLFVL 75
Query: 112 LGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISL 171
+G +C YT L+ +C++ + + SY DIG +AFGA GR A +Y+++++ +S+ +
Sbjct: 76 VGAVCYYTGTLIERCMRADGSIASYPDIGQYAFGATGRRAVAFFMYVELYLVAISFLVLE 135
Query: 172 HDNLTTVLHLKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVI 229
DNL + + + +L Q+ V A V LP+ WL++L ++++S+ G++ SV +
Sbjct: 136 GDNLDKLFPGATMEILGYQLHGKQLFIVLAAAVILPTTWLKNLGMLAYVSAAGLIASVAL 195
Query: 230 FVSV--AATAVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFT 287
S+ A A G R ++ L L IP+ G+Y + GH VFP +Y +MK+ F+
Sbjct: 196 TASLIWAGVAETGFHRNSNT---LNLAGIPTSLGLYFVCFTGHAVFPTIYSSMKNSKHFS 252
Query: 288 KVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYA 347
KV ++S + Y + +G ++G +V SQVTL++PS ++ TKIA+ T++ P+ KYA
Sbjct: 253 KVLLISSVLCSLNYGLTAVLGYMIYGDDVQSQVTLNLPSGKLYTKIAIVMTLVNPLAKYA 312
Query: 348 LEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVS 407
L AP+ +E +L + R R I +++L + +A TVP+F +++S GS +S
Sbjct: 313 LLVAPITAAVEERLSLT---RGSAPARVAISTAILASTVVVASTVPFFGYLMSFIGSFLS 369
Query: 408 VAISLIFPCAFYIKICRGQ-ISKPLFVLNISIITFGFLLGVVGT 450
V +++FPC Y+KI + I + V I+ G + V GT
Sbjct: 370 VMATVLFPCLCYLKIYKADGIHRTEMVAIAGILLLGVFVAVTGT 413
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 699,934,456
Number of Sequences: 2540612
Number of extensions: 27300024
Number of successful extensions: 95149
Number of sequences better than 10.0: 936
Number of HSP's better than 10.0 without gapping: 271
Number of HSP's successfully gapped in prelim test: 670
Number of HSP's that attempted gapping in prelim test: 93842
Number of HSP's gapped (non-prelim): 1177
length of query: 464
length of database: 863,360,394
effective HSP length: 132
effective length of query: 332
effective length of database: 527,999,610
effective search space: 175295870520
effective search space used: 175295870520
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0044a.9


