

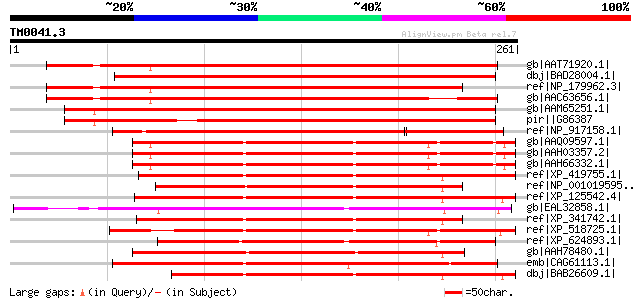
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.3
(261 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAT71920.1| At2g23820 [Arabidopsis thaliana] gi|58331781|gb|A... 324 2e-87
dbj|BAD28004.1| putative metal-dependent phosphohydrolase HD dom... 305 6e-82
ref|NP_179962.3| metal-dependent phosphohydrolase HD domain-cont... 290 2e-77
gb|AAC63656.1| unknown protein [Arabidopsis thaliana] gi|2541216... 289 5e-77
gb|AAM65251.1| unknown [Arabidopsis thaliana] gi|21280857|gb|AAM... 282 8e-75
pir||G86387 hypothetical protein F28B23.16 - Arabidopsis thalian... 258 1e-67
ref|NP_917158.1| P0039G05.13 [Oryza sativa (japonica cultivar-gr... 214 1e-54
gb|AAQ09597.1| NS5ATP2 [Homo sapiens] 174 2e-42
gb|AAH03357.2| C6orf74 protein [Homo sapiens] 174 2e-42
gb|AAH66332.1| C6orf74 protein [Homo sapiens] 174 2e-42
ref|XP_419755.1| PREDICTED: similar to chromosome 6 open reading... 170 3e-41
ref|NP_001019595.1| hypothetical protein LOC554129 [Danio rerio]... 169 9e-41
ref|XP_125542.4| PREDICTED: hypothetical protein LOC69692 [Mus m... 169 9e-41
gb|EAL32858.1| GA10728-PA [Drosophila pseudoobscura] 167 2e-40
ref|XP_341742.1| PREDICTED: similar to chromosome 6 open reading... 167 4e-40
ref|XP_518725.1| PREDICTED: similar to C6orf74 protein [Pan trog... 167 4e-40
ref|XP_624893.1| PREDICTED: similar to GA10728-PA [Apis mellifera] 166 5e-40
gb|AAH78480.1| MGC85244 protein [Xenopus laevis] 164 2e-39
emb|CAG61113.1| unnamed protein product [Candida glabrata CBS138... 162 7e-39
dbj|BAB26609.1| unnamed protein product [Mus musculus] 162 7e-39
>gb|AAT71920.1| At2g23820 [Arabidopsis thaliana] gi|58331781|gb|AAW70388.1|
At2g23820 [Arabidopsis thaliana]
gi|42570897|ref|NP_973522.1| metal-dependent
phosphohydrolase HD domain-containing protein
[Arabidopsis thaliana]
Length = 257
Score = 324 bits (830), Expect = 2e-87
Identities = 165/238 (69%), Positives = 188/238 (78%), Gaps = 9/238 (3%)
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS------AS 73
S + + +RF SP+R H S SP S + S P +A PS AS
Sbjct: 21 SFTRSHIRFTYSAAGASSPNRAIH---CMASDSPQSGDGSVSSPPNVAAVPSSSSSSSAS 77
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
SAIDFLS+C RLKTT RAGWI++DV+DPESIADHMYRM LMALI++DIPGV+RDKC+KMA
Sbjct: 78 SAIDFLSLCTRLKTTPRAGWIKRDVKDPESIADHMYRMGLMALISSDIPGVNRDKCMKMA 137
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSS 193
IVHDIAEAIVGDITP+ G+ KEEKNR E EAL+HMCK+LGGG RAKE+AELW EYE NSS
Sbjct: 138 IVHDIAEAIVGDITPSCGISKEEKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSS 197
Query: 194 PEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKK 251
PEAK VKD DKVE+ILQALEYE +QGKDL+EFFQSTAGKFQT IGKAWASEIVSRR+K
Sbjct: 198 PEAKVVKDFDKVELILQALEYEQDQGKDLEEFFQSTAGKFQTNIGKAWASEIVSRRRK 255
>dbj|BAD28004.1| putative metal-dependent phosphohydrolase HD domain-containing
protein [Oryza sativa (japonica cultivar-group)]
Length = 228
Score = 305 bits (782), Expect = 6e-82
Identities = 145/197 (73%), Positives = 177/197 (89%), Gaps = 1/197 (0%)
Query: 55 SSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLM 114
++S+++S PS + A P+++SAIDFL++C+RLKTTKRAGW+R+ VQ PES+ADHMYRM +M
Sbjct: 32 AASMSSSSPSPSPAAPASASAIDFLTLCYRLKTTKRAGWVRRGVQGPESVADHMYRMGVM 91
Query: 115 ALIAADIP-GVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLG 173
AL+AAD+P GV+RD+CVKMAIVHDIAEAIVGDITP+DGVPKEEK+R EQEALDHMC +LG
Sbjct: 92 ALVAADLPSGVNRDRCVKMAIVHDIAEAIVGDITPSDGVPKEEKSRREQEALDHMCSLLG 151
Query: 174 GGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKF 233
GG RA+E+ ELW EYE N++ EAK VKD DKVEMILQALEYE EQG DL+EFFQSTAGKF
Sbjct: 152 GGPRAEEIRELWMEYEQNATLEAKVVKDFDKVEMILQALEYEKEQGLDLEEFFQSTAGKF 211
Query: 234 QTEIGKAWASEIVSRRK 250
QT++GKAWA+E+ SRRK
Sbjct: 212 QTDVGKAWAAEVASRRK 228
>ref|NP_179962.3| metal-dependent phosphohydrolase HD domain-containing protein
[Arabidopsis thaliana]
Length = 245
Score = 290 bits (743), Expect = 2e-77
Identities = 148/220 (67%), Positives = 170/220 (77%), Gaps = 9/220 (4%)
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS------AS 73
S + + +RF SP+R H S SP S + S P +A PS AS
Sbjct: 21 SFTRSHIRFTYSAAGASSPNRAIH---CMASDSPQSGDGSVSSPPNVAAVPSSSSSSSAS 77
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
SAIDFLS+C RLKTT RAGWI++DV+DPESIADHMYRM LMALI++DIPGV+RDKC+KMA
Sbjct: 78 SAIDFLSLCTRLKTTPRAGWIKRDVKDPESIADHMYRMGLMALISSDIPGVNRDKCMKMA 137
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSS 193
IVHDIAEAIVGDITP+ G+ KEEKNR E EAL+HMCK+LGGG RAKE+AELW EYE NSS
Sbjct: 138 IVHDIAEAIVGDITPSCGISKEEKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSS 197
Query: 194 PEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKF 233
PEAK VKD DKVE+ILQALEYE +QGKDL+EFFQSTAG F
Sbjct: 198 PEAKVVKDFDKVELILQALEYEQDQGKDLEEFFQSTAGNF 237
>gb|AAC63656.1| unknown protein [Arabidopsis thaliana] gi|25412160|pir||D84629
hypothetical protein At2g23820 [imported] - Arabidopsis
thaliana
Length = 243
Score = 289 bits (740), Expect = 5e-77
Identities = 153/238 (64%), Positives = 174/238 (72%), Gaps = 23/238 (9%)
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS------AS 73
S + + +RF SP+R H S SP S + S P +A PS AS
Sbjct: 21 SFTRSHIRFTYSAAGASSPNRAIH---CMASDSPQSGDGSVSSPPNVAAVPSSSSSSSAS 77
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
SAIDFLS+C RLKTT RAGWI++DV+DPESIADHMYRM LMALI++DIPGV+RDKC+KMA
Sbjct: 78 SAIDFLSLCTRLKTTPRAGWIKRDVKDPESIADHMYRMGLMALISSDIPGVNRDKCMKMA 137
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSS 193
IVHDIAEAIVGDITP+ G+ KEEKNR E EAL+HMCK+LGGG RAKE+AELW EYE NSS
Sbjct: 138 IVHDIAEAIVGDITPSCGISKEEKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSS 197
Query: 194 PEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKK 251
PEAK VKD DKVE+ILQALEYE GKFQT IGKAWASEIVSRR+K
Sbjct: 198 PEAKVVKDFDKVELILQALEYEQ--------------GKFQTNIGKAWASEIVSRRRK 241
>gb|AAM65251.1| unknown [Arabidopsis thaliana] gi|21280857|gb|AAM45013.1| unknown
protein [Arabidopsis thaliana]
gi|15810417|gb|AAL07096.1| unknown protein [Arabidopsis
thaliana] gi|18395782|ref|NP_564240.1| metal-dependent
phosphohydrolase HD domain-containing protein
[Arabidopsis thaliana]
Length = 258
Score = 282 bits (721), Expect = 8e-75
Identities = 147/230 (63%), Positives = 170/230 (73%), Gaps = 8/230 (3%)
Query: 29 KSVTLSPHSPSRVF--------HMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLS 80
+S+ S HS SR F T + P S +S S S SS+IDFL+
Sbjct: 21 RSILASFHSSSRNFLFLGKPTPSSTIVSVRCQKPVSDGVSSMESMNHVASSVSSSIDFLT 80
Query: 81 ICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAE 140
+CHRLKTTKR GWI + + PESIADHMYRM+LMALIA D+ GVDR++C+KMAIVHDIAE
Sbjct: 81 LCHRLKTTKRKGWINQGINGPESIADHMYRMALMALIAGDLTGVDRERCIKMAIVHDIAE 140
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVK 200
AIVGDITP+DGVPKEEK+R E AL MC+VLGGG RA+E+ ELW EYE N+S EA VK
Sbjct: 141 AIVGDITPSDGVPKEEKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVK 200
Query: 201 DLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
D DKVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+EI +RRK
Sbjct: 201 DFDKVEMILQALEYEAEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRK 250
>pir||G86387 hypothetical protein F28B23.16 - Arabidopsis thaliana
gi|12321182|gb|AAG50684.1| hypothetical protein
[Arabidopsis thaliana]
Length = 248
Score = 258 bits (659), Expect = 1e-67
Identities = 140/230 (60%), Positives = 162/230 (69%), Gaps = 18/230 (7%)
Query: 29 KSVTLSPHSPSRVF--------HMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLS 80
+S+ S HS SR F T + P S +S S S SS+IDFL+
Sbjct: 21 RSILASFHSSSRNFLFLGKPTPSSTIVSVRCQKPVSDGVSSMESMNHVASSVSSSIDFLT 80
Query: 81 ICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAE 140
+CHRLK + PESIADHMYRM+LMALIA D+ GVDR++C+KMAIVHDIAE
Sbjct: 81 LCHRLK----------GINGPESIADHMYRMALMALIAGDLTGVDRERCIKMAIVHDIAE 130
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVK 200
AIVGDITP+DGVPKEEK+R E AL MC+VLGGG RA+E+ ELW EYE N+S EA VK
Sbjct: 131 AIVGDITPSDGVPKEEKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVK 190
Query: 201 DLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
D DKVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+EI +RRK
Sbjct: 191 DFDKVEMILQALEYEAEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRK 240
>ref|NP_917158.1| P0039G05.13 [Oryza sativa (japonica cultivar-group)]
gi|20805123|dbj|BAB92794.1| metal-dependent
phosphohydrolase HD domain-containing protein-like
[Oryza sativa (japonica cultivar-group)]
Length = 461
Score = 214 bits (546), Expect = 1e-54
Identities = 103/151 (68%), Positives = 125/151 (82%), Gaps = 2/151 (1%)
Query: 54 PSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSL 113
P+ + +PPSAA++ SASSAIDFL++CHRLKTTKR GWI ++ PESIADHMYRM+L
Sbjct: 146 PADAAAAAPPSAAAS--SASSAIDFLTLCHRLKTTKRKGWINHSIKGPESIADHMYRMAL 203
Query: 114 MALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLG 173
MALIA D+P VDR++C+K+AIVHDIAEAIVGDITP+DG+PK EK+R EQ+AL+ MC+VLG
Sbjct: 204 MALIAGDLPAVDRERCIKIAIVHDIAEAIVGDITPSDGIPKAEKSRREQKALNEMCEVLG 263
Query: 174 GGSRAKEVAELWAEYEANSSPEAKFVKDLDK 204
GG A E+ ELW EYE NSS EA VKD DK
Sbjct: 264 GGPIADEIKELWEEYENNSSIEANLVKDFDK 294
Score = 78.2 bits (191), Expect = 2e-13
Identities = 38/51 (74%), Positives = 44/51 (85%)
Query: 204 KVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKKTNG 254
KVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+E+ +RR++ G
Sbjct: 408 KVEMILQALEYEKEHGKVLDEFFLSTAGKFQTEIGKSWAAEVNARREQRCG 458
>gb|AAQ09597.1| NS5ATP2 [Homo sapiens]
Length = 204
Score = 174 bits (442), Expect = 2e-42
Identities = 101/206 (49%), Positives = 138/206 (66%), Gaps = 13/206 (6%)
Query: 64 SAASATPS---ASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAAD 120
S +SAT S A S + FL + +LK R GW+ ++VQ PES++DHMYRM++MA++ D
Sbjct: 3 SVSSATFSGHGARSLLQFLRLVGQLKRVPRTGWVYRNVQRPESVSDHMYRMAVMAMVIKD 62
Query: 121 IPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKE 180
+++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE
Sbjct: 63 -DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KE 120
Query: 181 VAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQSTAGKFQTEI 237
+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ STAGKF
Sbjct: 121 LYELWEEYETQSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDSTAGKFNHPE 179
Query: 238 GKAWASEIVSRRKKTN---GSSHPHS 260
SE+ + R TN +S PHS
Sbjct: 180 IVQLVSELEAER-STNIAAAASEPHS 204
>gb|AAH03357.2| C6orf74 protein [Homo sapiens]
Length = 218
Score = 174 bits (442), Expect = 2e-42
Identities = 101/206 (49%), Positives = 138/206 (66%), Gaps = 13/206 (6%)
Query: 64 SAASATPS---ASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAAD 120
S +SAT S A S + FL + +LK R GW+ ++VQ PES++DHMYRM++MA++ D
Sbjct: 17 SVSSATFSGHGARSLLQFLRLVGQLKRVPRTGWVYRNVQRPESVSDHMYRMAVMAMVIKD 76
Query: 121 IPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKE 180
+++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE
Sbjct: 77 -DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KE 134
Query: 181 VAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQSTAGKFQTEI 237
+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ STAGKF
Sbjct: 135 LYELWEEYETQSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDSTAGKFNHPE 193
Query: 238 GKAWASEIVSRRKKTN---GSSHPHS 260
SE+ + R TN +S PHS
Sbjct: 194 IVQLVSELEAER-STNIAAAASEPHS 218
>gb|AAH66332.1| C6orf74 protein [Homo sapiens]
Length = 204
Score = 174 bits (442), Expect = 2e-42
Identities = 101/206 (49%), Positives = 138/206 (66%), Gaps = 13/206 (6%)
Query: 64 SAASATPS---ASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAAD 120
S +SAT S A S + FL + +LK R GW+ ++VQ PES++DHMYRM++MA++ D
Sbjct: 3 SVSSATFSGHGARSLLQFLRLVGQLKRVPRTGWVYRNVQRPESVSDHMYRMAVMAMVIKD 62
Query: 121 IPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKE 180
+++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE
Sbjct: 63 -DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KE 120
Query: 181 VAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQSTAGKFQTEI 237
+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ STAGKF
Sbjct: 121 LYELWEEYETRSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDSTAGKFNHPE 179
Query: 238 GKAWASEIVSRRKKTN---GSSHPHS 260
SE+ + R TN +S PHS
Sbjct: 180 IVQLVSELEAER-STNIAAAASEPHS 204
>ref|XP_419755.1| PREDICTED: similar to chromosome 6 open reading frame 74; CGI-130
protein [Gallus gallus]
Length = 196
Score = 170 bits (431), Expect = 3e-41
Identities = 89/197 (45%), Positives = 128/197 (64%), Gaps = 5/197 (2%)
Query: 67 SATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDR 126
+A P + FL + +LK R GW+ ++V+ PES++DHMYRM++MAL+ D +++
Sbjct: 2 AAGPGGGGMLPFLRLLGQLKRVPRTGWVYRNVEKPESVSDHMYRMAVMALVTED-KSLNK 60
Query: 127 DKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWA 186
D+C+++A+VHD+AE IVGDI P D + KEEK+R E+ A+ + ++L + KE+ ELW
Sbjct: 61 DRCIRLALVHDMAECIVGDIAPADNISKEEKHRREEAAMRQLTQLLSEDLK-KEIYELWE 119
Query: 187 EYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKF-QTEIGKAWAS 243
EYE + EAKFVK LD+ EMILQALEYE + L +F+ STAGKF EI + +
Sbjct: 120 EYENQCTAEAKFVKQLDQCEMILQALEYEELENTPGRLQDFYDSTAGKFIHPEILQLVSL 179
Query: 244 EIVSRRKKTNGSSHPHS 260
R KK + HPHS
Sbjct: 180 INTERNKKIAATCHPHS 196
>ref|NP_001019595.1| hypothetical protein LOC554129 [Danio rerio]
gi|63100943|gb|AAH95766.1| Hypothetical protein
LOC554129 [Danio rerio]
Length = 197
Score = 169 bits (427), Expect = 9e-41
Identities = 84/160 (52%), Positives = 115/160 (71%), Gaps = 4/160 (2%)
Query: 76 IDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIV 135
+ F+ + +LK R GW+ ++++ PES++DHMYRMS+MAL DI V++++C+K+A+V
Sbjct: 2 LQFMKLVGQLKRVPRTGWVYRNIKQPESVSDHMYRMSMMALTIQDI-SVNKERCMKLALV 60
Query: 136 HDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPE 195
HD+AE IVGDI P D V K EK+R E++A+ H+ +L G R KE+ LW EYE SSPE
Sbjct: 61 HDLAECIVGDIAPADNVSKAEKHRREKDAMVHITGLLDDGLR-KEIYNLWEEYETQSSPE 119
Query: 196 AKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKF 233
AK VK+LD +EMI+QA EYE +GK L EFF ST GKF
Sbjct: 120 AKLVKELDNLEMIIQAHEYEELEGKPGRLQEFFVSTEGKF 159
>ref|XP_125542.4| PREDICTED: hypothetical protein LOC69692 [Mus musculus]
Length = 203
Score = 169 bits (427), Expect = 9e-41
Identities = 90/200 (45%), Positives = 132/200 (66%), Gaps = 6/200 (3%)
Query: 65 AASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGV 124
A +++ S + + FL + +LK R GW+ ++V+ PES++DHMYRM++MA++ D +
Sbjct: 6 ALASSGSGAGLLRFLRLVGQLKRVPRTGWVYRNVEKPESVSDHMYRMAVMAMVTRD-DRL 64
Query: 125 DRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAEL 184
++D+C+++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE+ EL
Sbjct: 65 NKDRCIRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KELYEL 123
Query: 185 WAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKFQTEIGKAWA 242
W EYE SS EAKFVK LD+ EMILQA EYE + K L +F+ STAGKF
Sbjct: 124 WEEYETQSSEEAKFVKQLDQCEMILQASEYEDLENKPGRLQDFYDSTAGKFSHPEIVQLV 183
Query: 243 SEIVSRRKKT--NGSSHPHS 260
SE+ + R + S+ P S
Sbjct: 184 SELETERNASMATASAEPGS 203
>gb|EAL32858.1| GA10728-PA [Drosophila pseudoobscura]
Length = 380
Score = 167 bits (424), Expect = 2e-40
Identities = 100/275 (36%), Positives = 153/275 (55%), Gaps = 40/275 (14%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T T+ +SSL + +P + H P + TAA S+S ++S TT P
Sbjct: 104 TTTKFAMSSLAAEPLPQA---------------HKPDEI----TAAASTSDVAASGTTPP 144
Query: 63 PSAASATPSASSA--------------IDFLSICHRLKTTKRAGWIRKDVQDPESIADHM 108
++ A+S+ + F+ + LK TKR GW+ +DV D ESI+ HM
Sbjct: 145 NGLGASCQEAASSTGKKPCFSTGLAEILQFMELIGNLKHTKRTGWVLRDVNDCESISGHM 204
Query: 109 YRMSLMALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHM 168
YRMS++ + G+++ +C+++A+VHD+AE++VGDITP GV KEEK +E +A++ +
Sbjct: 205 YRMSMLTFLLDGSEGLNQIRCMELALVHDLAESLVGDITPFCGVSKEEKRAMEFKAMEDI 264
Query: 169 CKVLGGGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDL--DEFF 226
CK++ R K + EL+ EYE S E++FVKDLD+++M++QA EYE L EFF
Sbjct: 265 CKLI--EPRGKRIMELFEEYEHAESAESRFVKDLDRLDMVMQAFEYEKRDNCLLKHQEFF 322
Query: 227 QSTAGKFQTEIGKAWASEIVSRRK---KTNGSSHP 258
ST GKF K +EI +R K G++ P
Sbjct: 323 DSTEGKFNHPFVKKLVNEIYEQRNVLAKAKGATPP 357
>ref|XP_341742.1| PREDICTED: similar to chromosome 6 open reading frame 74 [Rattus
norvegicus]
Length = 199
Score = 167 bits (422), Expect = 4e-40
Identities = 85/171 (49%), Positives = 122/171 (70%), Gaps = 5/171 (2%)
Query: 66 ASATPSASSAI-DFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGV 124
ASAT + + + FL + +LK R GW+ ++V+ PES++DHMYRM++MA++ D +
Sbjct: 2 ASATSNCQAGLLRFLRLVGQLKRVPRTGWVYRNVEKPESVSDHMYRMAVMAMVTRD-DRL 60
Query: 125 DRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAEL 184
++D+C+++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE+ EL
Sbjct: 61 NKDRCIRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKEITQLLPEDLR-KELYEL 119
Query: 185 WAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKF 233
W EYE SS EA+FVK LD+ EMILQA EYE + K L +F+ STAGKF
Sbjct: 120 WEEYETQSSEEARFVKQLDQCEMILQASEYEDMENKPGRLQDFYDSTAGKF 170
>ref|XP_518725.1| PREDICTED: similar to C6orf74 protein [Pan troglodytes]
Length = 412
Score = 167 bits (422), Expect = 4e-40
Identities = 96/214 (44%), Positives = 132/214 (60%), Gaps = 19/214 (8%)
Query: 52 SPPSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRM 111
S P +SVT+ + ATP R GW+ ++VQ PES++DHMYRM
Sbjct: 213 SRPQNSVTSRNKTFLPATPQTG-----------FNRVPRTGWVYRNVQRPESVSDHMYRM 261
Query: 112 SLMALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKV 171
++MA++ D +++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++
Sbjct: 262 AVMAMVIKD-DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQL 320
Query: 172 LGGGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQS 228
L R KE+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ S
Sbjct: 321 LPEDLR-KELYELWEEYETQSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDS 378
Query: 229 TAGKFQTEIGKAWASEIVSRRKK--TNGSSHPHS 260
TAGKF SE+ + R +S PHS
Sbjct: 379 TAGKFNHPEIVQLVSELEAERNANIAAAASEPHS 412
>ref|XP_624893.1| PREDICTED: similar to GA10728-PA [Apis mellifera]
Length = 190
Score = 166 bits (421), Expect = 5e-40
Identities = 84/177 (47%), Positives = 126/177 (70%), Gaps = 7/177 (3%)
Query: 77 DFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVH 136
+F+ + RLK KR GW+ K+V DPE+IA HMYRM++ + + D +D+ K ++MA++H
Sbjct: 8 EFMELVGRLKHMKRTGWVLKNVSDPETIAGHMYRMAMFSFLV-DNENLDKVKIMQMALIH 66
Query: 137 DIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEA 196
D+AE IVGDITP+ G+P E K++LE EA++ +CK+L G R + E++ EYE SPEA
Sbjct: 67 DLAECIVGDITPSCGIPSEIKHKLEDEAMEDICKLL--GDRGPMILEIFREYEKQESPEA 124
Query: 197 KFVKDLDKVEMILQALEYEHEQ---GKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
K+VKDLD++++I+QA EYE GK L+EFF +T+GK + + ASEI++RR+
Sbjct: 125 KYVKDLDRLDLIMQAYEYEKRDNIPGK-LEEFFTTTSGKIRHPFIQKLASEIIARRQ 180
>gb|AAH78480.1| MGC85244 protein [Xenopus laevis]
Length = 201
Score = 164 bits (415), Expect = 2e-39
Identities = 80/173 (46%), Positives = 122/173 (70%), Gaps = 4/173 (2%)
Query: 64 SAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPG 123
+A S + + S + F+ + +LK R GWI + V+ PES++DHMYRM++MA++ D
Sbjct: 3 AAGSCSSAGKSLLQFMKLVGQLKRVPRTGWIYRQVEKPESVSDHMYRMAVMAMLTEDRK- 61
Query: 124 VDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAE 183
+++D+C+++A+VHD+AE IVGDI P D + KEEK+R E+ A++H+ ++L + EV +
Sbjct: 62 LNKDRCIRLALVHDMAECIVGDIAPADNIAKEEKHRKEKAAMEHLTQLLPDNLKT-EVYD 120
Query: 184 LWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKFQ 234
LW EYE + EAKFVK+LD+ EMILQALEYE + + L +F+ STAGKF+
Sbjct: 121 LWEEYEHQFTAEAKFVKELDQCEMILQALEYEELEKRPGRLQDFYNSTAGKFK 173
>emb|CAG61113.1| unnamed protein product [Candida glabrata CBS138]
gi|50291459|ref|XP_448162.1| unnamed protein product
[Candida glabrata]
Length = 209
Score = 162 bits (411), Expect = 7e-39
Identities = 80/201 (39%), Positives = 128/201 (62%), Gaps = 5/201 (2%)
Query: 54 PSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSL 113
P + + A + T +++ I L+I +LK KR GW+ V+DPESI+DHMYRM +
Sbjct: 7 PEDHIPDNVKKALAGTKNSNYVIALLNIVQQLKIQKRTGWVDHKVKDPESISDHMYRMGI 66
Query: 114 MALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMC-KVL 172
+++ + P ++RDKCV++++VHD+AE++VGDITP D + KEEK+R E + + ++C K++
Sbjct: 67 TSMLITN-PDINRDKCVRISLVHDLAESLVGDITPNDPMTKEEKHRREYDTIKYLCDKII 125
Query: 173 G--GGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTA 230
AKE+ E W YE SSPEA++VKD+DK EM++Q EYE LD+F+ S
Sbjct: 126 APFNSKAAKEILEDWLAYENVSSPEARYVKDIDKFEMLVQCFEYEQSNNIRLDQFW-SAK 184
Query: 231 GKFQTEIGKAWASEIVSRRKK 251
+T W ++++ R++
Sbjct: 185 DAIKTPEVSEWCNDLIKMREE 205
>dbj|BAB26609.1| unnamed protein product [Mus musculus]
Length = 182
Score = 162 bits (411), Expect = 7e-39
Identities = 86/181 (47%), Positives = 122/181 (66%), Gaps = 6/181 (3%)
Query: 84 RLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAEAIV 143
+LK R GW+ ++V+ PES++DHMYRM++MA++ D +++D+C+++A+VHD+AE IV
Sbjct: 4 QLKRVPRTGWVYRNVEKPESVSDHMYRMAVMAMVTRD-DRLNKDRCIRLALVHDMAECIV 62
Query: 144 GDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVKDLD 203
GDI P D +PKEEK+R E+EA+ + ++L R KE+ ELW EYE SS EAKFVK LD
Sbjct: 63 GDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KELYELWEEYETQSSEEAKFVKQLD 121
Query: 204 KVEMILQALEYEHEQGKD--LDEFFQSTAGKFQTEIGKAWASEIVSRRKKT--NGSSHPH 259
+ EMILQA EYE + K L +F+ STAGKF SE+ + R + S+ P
Sbjct: 122 QCEMILQASEYEDLENKPGRLQDFYDSTAGKFSHPEIVQLVSELETERNASMATASAEPG 181
Query: 260 S 260
S
Sbjct: 182 S 182
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 413,192,812
Number of Sequences: 2540612
Number of extensions: 16010399
Number of successful extensions: 112374
Number of sequences better than 10.0: 618
Number of HSP's better than 10.0 without gapping: 264
Number of HSP's successfully gapped in prelim test: 363
Number of HSP's that attempted gapping in prelim test: 104250
Number of HSP's gapped (non-prelim): 5027
length of query: 261
length of database: 863,360,394
effective HSP length: 125
effective length of query: 136
effective length of database: 545,783,894
effective search space: 74226609584
effective search space used: 74226609584
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0041.3


