

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
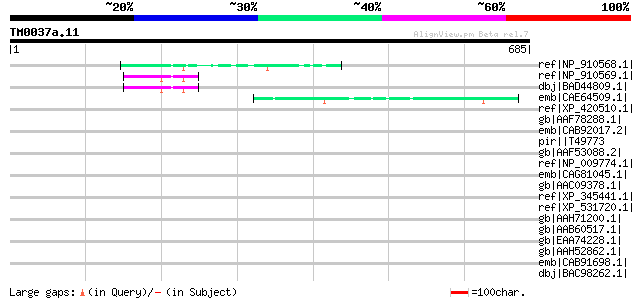
Query= TM0037a.11
(685 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_910568.1| Similar to Arabidopsis thaliana chromosome II B... 53 3e-05
ref|NP_910569.1| Similar to Zea mays putative AC9 transposase. (... 51 1e-04
dbj|BAD44809.1| hypothetical protein [Oryza sativa (japonica cul... 51 1e-04
emb|CAE64509.1| Hypothetical protein CBG09244 [Caenorhabditis br... 49 4e-04
ref|XP_420510.1| PREDICTED: similar to KSHV latent nuclear antig... 47 0.002
gb|AAF78288.1| merozoite surface protein 3g [Plasmodium vivax] 47 0.002
emb|CAB92017.2| related to actin-interacting protein AIP3 [Neuro... 47 0.003
pir||T49773 related to actin-interacting protein AIP3 [imported]... 47 0.003
gb|AAF53088.2| CG6392-PA [Drosophila melanogaster] gi|24583666|r... 46 0.005
ref|NP_009774.1| Highly charged, basic protein required for norm... 45 0.006
emb|CAG81045.1| unnamed protein product [Yarrowia lipolytica CLI... 45 0.006
gb|AAC09378.1| polymorphic antigen [Plasmodium falciparum] 45 0.008
ref|XP_345441.1| PREDICTED: similar to ABTAP [Rattus norvegicus]... 45 0.010
ref|XP_531720.1| PREDICTED: similar to Ran GTPase-activating pro... 45 0.010
gb|AAH71200.1| Rangap1 protein [Mus musculus] gi|42490971|gb|AAH... 44 0.013
gb|AAB60517.1| RNA1 homolog 44 0.013
gb|EAA74228.1| hypothetical protein FG10944.1 [Gibberella zeae P... 44 0.013
gb|AAH52862.1| Rangap1 protein [Mus musculus] 44 0.013
emb|CAB91698.1| related to BCS1 protein precursor [Neurospora cr... 44 0.013
dbj|BAC98262.1| mKIAA1835 protein [Mus musculus] 44 0.013
>ref|NP_910568.1| Similar to Arabidopsis thaliana chromosome II BAC F13B15 genomic
sequence, putative non-LTR retroelement reverse
transcriptase. (AC006300) [Oryza sativa (japonica
cultivar-group)]
Length = 1285
Score = 53.1 bits (126), Expect = 3e-05
Identities = 73/303 (24%), Positives = 106/303 (34%), Gaps = 61/303 (20%)
Query: 147 LDLNIKLPFSPFICHVLSFLNVAPCQLQPNAWGFLRCFEILCEHLSFTPTYPLFFLFYKS 206
L+ ++LP PF C +L L VAP Q+ PN W + F +L H + P+ +F F++
Sbjct: 305 LEAGMRLPLHPFACELLRHLGVAPSQITPNGWRVVAGFLLLSHHAAAPPSLAVFRRFFRL 364
Query: 207 VTTKPTSVKWVPLLARKNMSR----FTALKSHYKVWQKKYFKVMESPEIKNLFRDSDNNP 262
+K W ++ FT L K W+ +F V
Sbjct: 365 FLSKLNG--WYHFRGKRGAGADGVLFTGLPK-LKGWKGGFFFVSS--------------- 406
Query: 263 LFPFYWTKNPRRKISVSYDALDESEQGVADYLRTLPVISGHDLIEASKSGTLNQFFTTMG 322
P W V + S+ A+ PV+SG ++ A K L+ G
Sbjct: 407 --PSPW------PCQVRWGGPPPSKSSTAE-----PVLSGEEMELAGKLLHLH------G 447
Query: 323 KGKVDANPIKMKEYLA-------QSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTS 375
VD + LA AA + + K SG N + K + S
Sbjct: 448 GAAVDLRTYLCESNLAAVFSSNLAGAAPPQPPPPRPARAKGKFPFSGEFNAQKVKTEPES 507
Query: 376 GAAGGKPLHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEA 435
P L K R E +N PPP + P P G + SA G +
Sbjct: 508 DMPWWSP-----LSGKKRGFE---AEENNPPPSPTPT-----PPAPHGGSWSWSASGMCS 554
Query: 436 PPP 438
PPP
Sbjct: 555 PPP 557
>ref|NP_910569.1| Similar to Zea mays putative AC9 transposase. (P03010) [Oryza
sativa (japonica cultivar-group)]
Length = 947
Score = 51.2 bits (121), Expect = 1e-04
Identities = 35/107 (32%), Positives = 49/107 (45%), Gaps = 10/107 (9%)
Query: 151 IKLPFSPFICHVLSFLNVAPCQLQPNAWGFLRCFEILCEHLSFTPTYP----LFFLFYKS 206
++LP PF VL+ +AP QL PNAW L F LC P P +F F+ +
Sbjct: 98 MRLPLQPFFAAVLAHFGLAPSQLTPNAWRLLAGFAALCRRAGVAPPPPPPLTVFRHFFTA 157
Query: 207 VTTKPTSVKWVPLLARKNMSR---FTALKS-HYKVWQKKYFKVMESP 249
+ P + W AR + S FT L + W K+ F ++ SP
Sbjct: 158 IGCPPGA--WYSFRARGSASTGSLFTRLSNVTMNPWWKEQFILVSSP 202
>dbj|BAD44809.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 569
Score = 51.2 bits (121), Expect = 1e-04
Identities = 35/107 (32%), Positives = 49/107 (45%), Gaps = 10/107 (9%)
Query: 151 IKLPFSPFICHVLSFLNVAPCQLQPNAWGFLRCFEILCEHLSFTPTYP----LFFLFYKS 206
++LP PF VL+ +AP QL PNAW L F LC P P +F F+ +
Sbjct: 98 MRLPLQPFFAAVLAHFGLAPSQLTPNAWRLLAGFAALCRRAGVAPPPPPPLTVFRHFFTA 157
Query: 207 VTTKPTSVKWVPLLARKNMSR---FTALKS-HYKVWQKKYFKVMESP 249
+ P + W AR + S FT L + W K+ F ++ SP
Sbjct: 158 IGCPPGA--WYSFRARGSASTGSLFTRLSNVTMNPWWKEQFILVSSP 202
>emb|CAE64509.1| Hypothetical protein CBG09244 [Caenorhabditis briggsae]
Length = 3376
Score = 49.3 bits (116), Expect = 4e-04
Identities = 79/357 (22%), Positives = 140/357 (39%), Gaps = 21/357 (5%)
Query: 323 KGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTSGAAGGKP 382
K + DA K + A+ AK + E ++K K E + + D K +K + A
Sbjct: 1137 KKEADAKTKKEADDKAKKDLEAKTKK-EADEKAKKEAEAKTKKEADDKARKEAEAKKEAE 1195
Query: 383 LHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTL----INRPSTPFGQAGPSSAIGGEAPPP 438
+ D + A KK D+ + D + + + + EA
Sbjct: 1196 AKKEADDKAKKEAVAKKVADDKTKKEADDKARNEAEAKKEAEDKAKKEDDAKTKKEADDK 1255
Query: 439 LLNLSDPHFNGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKEL 498
+D N + + + KD+ + A L A + + KE
Sbjct: 1256 AKKKADEKAN------KEAEAKTKKDVEAKTKKEADDKA-KKDLEAKTKKEADEKAKKE- 1307
Query: 499 IAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLA-ASDLLLQKTKSL 557
A+ + +K+A K A ++AK D+E NKK A+EK K E A D ++ ++
Sbjct: 1308 --AEAKIKKEAEAKKEADDKAKKDSEAKNKK--EADEKARKEAETKKEAEDKAKKEAETK 1363
Query: 558 KEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRI 617
KEA K + K + ++KK D+ ++SI +S + F ++ VE D S
Sbjct: 1364 KEAELGKSDETKPKKRVVKKKTDKSDSSISQISSADTSSEFTVQEKPAEKVEQLPDESTS 1423
Query: 618 GWLKEI---KDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQA 671
+K+ ++ +V + +S D P +PEE E ++ + +K D A
Sbjct: 1424 ATIKKAAQPQEPEVEKKELLSDDKKPVEGKPPKPEESDETTKKRVIKKKTQKSDSVA 1480
Score = 40.8 bits (94), Expect = 0.15
Identities = 53/252 (21%), Positives = 95/252 (37%), Gaps = 26/252 (10%)
Query: 323 KGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTSGAAGGKP 382
K + +A K + A++ A AKK A + +K + T + + K A K
Sbjct: 905 KKEAEAKTKKEADDKARNEAEAKKEAEDKAEKGADAKTKKEADDKAKKEAVAKKEADDKT 964
Query: 383 LHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNL 442
++ + ++ KK+ D V ++D + + + + I EA N
Sbjct: 965 KKEAEAEKEAEAKTKKEADDKV---KKDLEAKTKKEADEKAKKEAEAKIKKEADDKARNE 1021
Query: 443 SDPHFNGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKELIAAK 502
++ + + D + K+ + ++ +M K+ K
Sbjct: 1022 AEAKKEAEDKAKKEADAKTKKEADDKAKKDLEAMT------------------KKEADEK 1063
Query: 503 NRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAIN 562
+ E +A K A E+AK +AE K K E K K +D A DL + K+ KEA
Sbjct: 1064 AKKEAEAKTKKEADEKAKKEAEAKTK--KDVEAKTKKEADDKAKKDL---EAKTKKEADE 1118
Query: 563 DKHTAVQAKYQK 574
+AK +K
Sbjct: 1119 KAKKEAEAKIKK 1130
>ref|XP_420510.1| PREDICTED: similar to KSHV latent nuclear antigen interacting
protein 1 [Gallus gallus]
Length = 345
Score = 47.4 bits (111), Expect = 0.002
Identities = 47/217 (21%), Positives = 89/217 (40%), Gaps = 32/217 (14%)
Query: 381 KPLHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTLINRP--STPFGQAGPSSAIGGEAPPP 438
+PLH + +D+ + + VP PQ+ ++ + TP G S G P
Sbjct: 58 QPLHSTAVDACGEEHSENESSGYVPAPQRTNAERSEKMLLETPEGDVHEFSQSGSAREPL 117
Query: 439 LLNLSDPHFNGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKEL 498
+ NL+ P+ E + + D S P ++ L + + ++ + EL
Sbjct: 118 MENLNAPNTTRPEVKKKRPSKKSSSDSSVNSPSSV-------QLWCPNKLKRSSRDITEL 170
Query: 499 IAAKNRFEKKAADYK----------------TAYERAKTDAETANKKLKSAEEKCAKLTE 542
FEK AA+Y+ +A+E TD T ++LK+ ++K AK+
Sbjct: 171 DVVLAEFEKIAANYRQSIESKACRKAVSAFCSAFEDQVTDLITEVQELKNTKKKNAKVVA 230
Query: 543 DLAASDLLLQKTKSLKEAINDKHTAVQAKYQKLEKKY 579
D ++K + + +K + + + KL+K+Y
Sbjct: 231 D-------IKKKRQRLMQVREKLSRTEPQLIKLQKEY 260
>gb|AAF78288.1| merozoite surface protein 3g [Plasmodium vivax]
Length = 969
Score = 47.4 bits (111), Expect = 0.002
Identities = 79/357 (22%), Positives = 127/357 (35%), Gaps = 31/357 (8%)
Query: 342 AAAKKRAAETEQKK-KNEGTSGSDNVRDPKRQKTSGAAGGKPLHQSTLDSKSRPAEKKKG 400
A K + AE E KK K+E + + K+Q K + +K A+ +
Sbjct: 566 ACEKAKTAEQEAKKAKDEAVKAAKEAEEAKKQAEKAEKITKTATEEANKAKEEEAKASEA 625
Query: 401 HDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNLSDPHFNGLEFMTRTFDNQ 460
D + + F ++ P + L+ + +N
Sbjct: 626 KQEAETKAGDVDEEVYAVNVEFESVKAAAKAAAHHKVPEI---------LDKEKKNAENA 676
Query: 461 IHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKE---LIAAKNRFEKKAADYKTAYE 517
K + + + A T AG AQ E IAA EK + + ++ E
Sbjct: 677 AKKASAKATEAKTTAETATKKATEAKTAAGNAQKASENAKAIAADVLAEKASTEAQSLKE 736
Query: 518 RAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKYQ-KLE 576
AK A K + EEK + D AA+D Q + S +A K A QAK + LE
Sbjct: 737 EAKKLAADIKKSNVTNEEKAKR---DKAANDAAHQASLSASKAKEAKTAATQAKGEVALE 793
Query: 577 KKYDRLNASILGRASLQYAQGFLAAK----------EQINVVEPG-FDLSRIGWLKEIKD 625
KK + ++ A+ A + EQ++V G +L+ + ++
Sbjct: 794 KKKEESAKAVEAAKEAMKARDKAAFELLKLKKQDVLEQVDVSPSGNNNLNDVDEQVSLEV 853
Query: 626 GQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQGNNANNE 682
G+ + D D PQ +E E D ED E +E Q++ T A E
Sbjct: 854 GEQENESD---DAPPQETEEVTEEGDEEDEEEMEEDEIQDESGHTEETPTEQAAEQE 907
>emb|CAB92017.2| related to actin-interacting protein AIP3 [Neurospora crassa]
Length = 1127
Score = 46.6 bits (109), Expect = 0.003
Identities = 37/135 (27%), Positives = 61/135 (44%), Gaps = 10/135 (7%)
Query: 558 KEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRI 617
KE + + A+Q +Q + +R A L + LQ KE + VE G L +
Sbjct: 942 KEGVLGEVRALQPNHQSRLEAIER--AEKLRQKELQTRMVNPLTKELASFVEEG-KLKKS 998
Query: 618 GWLKEIKDGQVVGDDDISLD-------LLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQ 670
G ++E++ + DD I + L+ + DD+ E EED E+ E+ E+ E+E+ +
Sbjct: 999 GGVEEVERARKAKDDQIRREVWERMNGLIAENDDDGEDEEDDEEEEEEEEEEEGEEEEEE 1058
Query: 671 AGTSQGNNANNENLA 685
G Q E A
Sbjct: 1059 EGEEQEEGEEGEEKA 1073
>pir||T49773 related to actin-interacting protein AIP3 [imported] - Neurospora
crassa
Length = 972
Score = 46.6 bits (109), Expect = 0.003
Identities = 37/135 (27%), Positives = 61/135 (44%), Gaps = 10/135 (7%)
Query: 558 KEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRI 617
KE + + A+Q +Q + +R A L + LQ KE + VE G L +
Sbjct: 787 KEGVLGEVRALQPNHQSRLEAIER--AEKLRQKELQTRMVNPLTKELASFVEEG-KLKKS 843
Query: 618 GWLKEIKDGQVVGDDDISLD-------LLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQ 670
G ++E++ + DD I + L+ + DD+ E EED E+ E+ E+ E+E+ +
Sbjct: 844 GGVEEVERARKAKDDQIRREVWERMNGLIAENDDDGEDEEDDEEEEEEEEEEEGEEEEEE 903
Query: 671 AGTSQGNNANNENLA 685
G Q E A
Sbjct: 904 EGEEQEEGEEGEEKA 918
>gb|AAF53088.2| CG6392-PA [Drosophila melanogaster] gi|24583666|ref|NP_524993.2|
CG6392-PA [Drosophila melanogaster]
Length = 2013
Score = 45.8 bits (107), Expect = 0.005
Identities = 25/101 (24%), Positives = 54/101 (52%), Gaps = 3/101 (2%)
Query: 497 ELIAAKNRFEKKAADYKTAYE---RAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQK 553
E+++ +++ E + + A + TD E +K+L +++ +L E A D Q
Sbjct: 704 EIVSVRDKLESVESAFNLASSEIIQKATDCERLSKELSTSQNAFGQLQERYDALDQQWQA 763
Query: 554 TKSLKEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQY 594
++ +++KH VQ KYQKL+++Y++L + +S ++
Sbjct: 764 QQAGITTLHEKHEHVQEKYQKLQEEYEQLESRARSASSAEF 804
>ref|NP_009774.1| Highly charged, basic protein required for normal cell-cycle
regulation of histone gene transcription; mutants
display strong synthetic defects with subunits of FACT,
a complex that allows RNA Pol II to elongate through
nucleosomes; Hpc2p [Saccharomyces cerevisiae]
gi|536602|emb|CAA85179.1| HPC2 [Saccharomyces
cerevisiae] gi|417142|sp|Q01448|HPC2_YEAST Histone
promoter control 2 protein gi|171700|gb|AAA34684.1| HPC2
protein
Length = 623
Score = 45.4 bits (106), Expect = 0.006
Identities = 75/327 (22%), Positives = 133/327 (39%), Gaps = 26/327 (7%)
Query: 258 SDNNPLFPFYWTKNPRRKISVSYDALDESEQGVADYLRTLPVISGHDLIEASKSGTLNQF 317
SD+ LF + K RKI + L ++ V + +ISG S SG +
Sbjct: 45 SDSEDLFNKFSNKKTNRKIPNIAEELAKNRNYVKGASPSPIIISGSS--STSPSGPSSSS 102
Query: 318 FTTMG--KGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTS 375
MG + + N +++ ++ K +TE+K++N DN P+R +S
Sbjct: 103 TNPMGIPTNRFNKNTVELYQHSPSPVMTTNK--TDTEEKRQNN--RNMDNKNTPERGSSS 158
Query: 376 GAAGGKPLHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTLINR-PSTPFGQAGPSSAIGGE 434
AA K L S+L + S + K H N ++S+ N PS ++I
Sbjct: 159 FAA--KQLKISSLLTISSNEDSKTLHINDTNGNKNSNAASNNIPSAYAELHTEGNSIESL 216
Query: 435 APPPLLNLSDPHFNGLE----FMTRTFDNQIHKDIS-GQGPPN--IASMAIHHALSAAST 487
PP S P L T+ D I KD G P IA + L ST
Sbjct: 217 IKPP----SSPRNKSLTPKVILPTQNMDGTIAKDPHLGDNTPGILIAKTSSPVNLDVEST 272
Query: 488 VAGMAQCVKELIAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAAS 547
+ + K + K K A+ K + +R+ + ++ + S+++ ++ + +++
Sbjct: 273 AQSLGKFNKSTNSLKAALTKAPAE-KVSLKRSISSVTNSDSNISSSKKPTSEKAKKSSSA 331
Query: 548 DLLLQK---TKSLKEAINDKHTAVQAK 571
+L K TK+ K+A ++ + + K
Sbjct: 332 SAILPKPTTTKTSKKAASNSSDSTRKK 358
>emb|CAG81045.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50550769|ref|XP_502857.1| hypothetical protein
[Yarrowia lipolytica]
Length = 1098
Score = 45.4 bits (106), Expect = 0.006
Identities = 51/197 (25%), Positives = 87/197 (43%), Gaps = 24/197 (12%)
Query: 496 KELIAAKNRFEKKA--ADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQK 553
K + A+N+ + A A K +R K +AE KK K+ + E+ +++ K
Sbjct: 33 KNAVKAENKAKNDAREAKKKEHEQRKKEEAEAMKKKAKNFNKSKFGKKEEEKVTEVKPSK 92
Query: 554 TKSL-------------KEAINDKHTAVQAKYQKLEKKYDRLNASILG----RASLQYAQ 596
TK +++ +DK T + K + E K D L A IL L+ +
Sbjct: 93 TKGSDKTEKSKGSRTKSQDSSSDKSTKTKTKTK--EFKPDSLLAEILELGGTEEDLKLVE 150
Query: 597 GFLAAKEQINVVEP--GFDLSRIGWLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGED 654
++ + VV P G + LK +KD + GD ++ + D E E EED ++
Sbjct: 151 DLSDGEDDV-VVNPSNGSKIDSGDLLKFMKDIGLDGDVPQLVEDVKDEDVEEEQEEDDDE 209
Query: 655 GNEQHRNEDQEKEDPQA 671
G E +ED E++D ++
Sbjct: 210 GMEDDEDEDTEEDDEES 226
>gb|AAC09378.1| polymorphic antigen [Plasmodium falciparum]
Length = 380
Score = 45.1 bits (105), Expect = 0.008
Identities = 59/260 (22%), Positives = 106/260 (40%), Gaps = 26/260 (10%)
Query: 448 NGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQ-CVKELIAAK--NR 504
N +EF F KDI AS A A + A+ KE + K ++
Sbjct: 73 NSMEF-GEGFSADDQKDIEAYKKAKQASQDAEQAAKDAENASKEAEEAAKEAVNLKESDK 131
Query: 505 FEKKAADYKTAYERAKTDAETANKKLKSAEE--KCAKLTEDLAASDLLLQKTKSLKEAIN 562
KA + TA +AK ETA K AE K ++ E + +L +KTK E
Sbjct: 132 SYTKAKEACTAASKAKKAVETALKAKDDAETALKTSETPEKPSRINLFSRKTKEYAEKAK 191
Query: 563 DKHTAVQAKYQKLEK---------KYDRLNASILGRASLQYAQG-------FLAAKEQIN 606
+ + + YQK + YD + G ++ + ++++K++ N
Sbjct: 192 NAYEKAKNAYQKANQAVLKAKEASSYDYILGWEFGGGVPEHKKEENMLSHLYVSSKDKEN 251
Query: 607 VVEPGFDL--SRIGWLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQ 664
+ + D+ + +E ++ ++ ++ + D+E E EE+ E+ NE + ++Q
Sbjct: 252 ISKENDDVLDEKEEEAEETEEEELEEKNEEETESEISEDEEEEEEEEKEEENE--KKKEQ 309
Query: 665 EKEDPQAGTSQGNNANNENL 684
EKE Q + +NL
Sbjct: 310 EKEQSNENNDQKKDMEAQNL 329
>ref|XP_345441.1| PREDICTED: similar to ABTAP [Rattus norvegicus]
gi|42491219|dbj|BAD10946.1| ABTAP [Rattus rattus]
Length = 842
Score = 44.7 bits (104), Expect = 0.010
Identities = 40/169 (23%), Positives = 71/169 (41%), Gaps = 27/169 (15%)
Query: 513 KTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKY 572
K +++ K D+E + K +EE + +DL ++ + K D T+ K
Sbjct: 142 KNSHKSPKIDSEVSPK---DSEESLQSRKKKRDTTDLSVEASPKGKLRTKDPSTSAMVK- 197
Query: 573 QKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVV-------EPGFDLSRIGWLKEIKD 625
++++ G + + Q + AK+ + E S +G +E +
Sbjct: 198 ----------SSTVSGSKAKREKQAVIMAKDNAGRMLHEEAPEEDSDSASELGRDEE-SE 246
Query: 626 GQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTS 674
G++ DD S D DDE+E EE+ EDG E+ E++E E S
Sbjct: 247 GEITSDDRASAD-----DDENEDEEEEEDGEEEEEEEEEEDESDDESDS 290
>ref|XP_531720.1| PREDICTED: similar to Ran GTPase-activating protein 1 [Canis
familiaris]
Length = 991
Score = 44.7 bits (104), Expect = 0.010
Identities = 39/147 (26%), Positives = 74/147 (49%), Gaps = 27/147 (18%)
Query: 539 KLTE-DLAASDLLLQKTKSLKEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQG 597
KL E +L+ ++ ++ EA+ DK A+ +KL+ LN ++LG
Sbjct: 674 KLKELNLSFCEIKRDAALAVAEAVADK-----AELEKLD-----LNGNMLGEEGC----- 718
Query: 598 FLAAKEQINVVEPGFDLSRIGWLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNE 657
EQ+ V GF+++R+ L + D + G+DD + + ++E E EE+ ED E
Sbjct: 719 -----EQLQEVLDGFNMARV--LASLSDDE--GEDDEEEEEEEEEEEEGEEEEEEED--E 767
Query: 658 QHRNEDQEKEDPQAGTSQGNNANNENL 684
+ E++E+E+PQ G + ++ + +
Sbjct: 768 EEEEEEEEEEEPQQGQGEESSTPSRKI 794
>gb|AAH71200.1| Rangap1 protein [Mus musculus] gi|42490971|gb|AAH66213.1| Rangap1
protein [Mus musculus]
Length = 589
Score = 44.3 bits (103), Expect = 0.013
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>gb|AAB60517.1| RNA1 homolog
Length = 589
Score = 44.3 bits (103), Expect = 0.013
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>gb|EAA74228.1| hypothetical protein FG10944.1 [Gibberella zeae PH-1]
gi|46138859|ref|XP_391120.1| hypothetical protein
FG10944.1 [Gibberella zeae PH-1]
Length = 1493
Score = 44.3 bits (103), Expect = 0.013
Identities = 55/204 (26%), Positives = 79/204 (37%), Gaps = 39/204 (19%)
Query: 492 AQCVKELIAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLL 551
A+ KEL AA EK D ERA E ++ AEE A ++L A L
Sbjct: 1063 AKSSKELAAASRDLEKLENDINNQGERA----EELQAQVAEAEEGLATKKQELKALKAEL 1118
Query: 552 -QKTKSLKEA----------INDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLA 600
+KT L E + + A+ Q+L D+L+ +L
Sbjct: 1119 DEKTDELNETRAMEIEMRNKLEENQKALAENQQRLRHWNDKLSKIVL------------- 1165
Query: 601 AKEQINVVEPGFDLSRIGWLKEIKDGQVVGDDDISLDLLPQFDDE--SEPEEDGEDGNEQ 658
+ I+ + G K+ K Q D DI +D PQ D + PEE+GE
Sbjct: 1166 --QNIDDLTGGSTNGHSKPRKQPKPQQNDDDGDIDMDDAPQDDVDMTDAPEEEGE----- 1218
Query: 659 HRNEDQEKEDPQAGTSQGNNANNE 682
ED ++E+ + G Q N NE
Sbjct: 1219 --QEDADEEEEEEGEEQMNGQPNE 1240
>gb|AAH52862.1| Rangap1 protein [Mus musculus]
Length = 589
Score = 44.3 bits (103), Expect = 0.013
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>emb|CAB91698.1| related to BCS1 protein precursor [Neurospora crassa]
gi|32405254|ref|XP_323240.1| hypothetical protein (
related to BCS1 protein precursor [imported] -
Neurospora crassa emb|CAB91698.1| (AL356172) related to
BCS1 protein precursor [Neurospora crassa] )
gi|11359615|pir||T49717 related to BCS1 protein
precursor [imported] - Neurospora crassa
gi|28918653|gb|EAA28324.1| hypothetical protein (
related to BCS1 protein precursor [imported] -
Neurospora crassa emb|CAB91698.1| (AL356172) related to
BCS1 protein precursor [Neurospora crassa] )
Length = 779
Score = 44.3 bits (103), Expect = 0.013
Identities = 53/189 (28%), Positives = 74/189 (39%), Gaps = 24/189 (12%)
Query: 399 KGHDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNLSDPHFNGLEFMTRTFD 458
+G D P DS L PST AG ++A EA L ++ +E + D
Sbjct: 477 EGDDVDTPSDADSKDLFKTPSTSTSPAGAAAAAAAEARRELAR-NEATLKVVELADQFAD 535
Query: 459 NQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKELIAAKNRFEKKAADYKTAYE- 517
+ S P I + H +A + V G Q V + A K + EK D + A E
Sbjct: 536 KIPAHEFS---PAEIQGFLLKHKRNATAAVEGAEQWVVD--ARKEKMEK---DLREARER 587
Query: 518 RAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKYQKLEK 577
R K + A KK EEK K KT++ K + KH V +K K
Sbjct: 588 REKEEKAEAEKKGTKKEEKGGK------------DKTETKKSKKSSKHNKVTSK--KSSS 633
Query: 578 KYDRLNASI 586
K + A+I
Sbjct: 634 KKSTMKAAI 642
>dbj|BAC98262.1| mKIAA1835 protein [Mus musculus]
Length = 646
Score = 44.3 bits (103), Expect = 0.013
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 372 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 410
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 411 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 464
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,252,628,453
Number of Sequences: 2540612
Number of extensions: 58068200
Number of successful extensions: 274305
Number of sequences better than 10.0: 1217
Number of HSP's better than 10.0 without gapping: 287
Number of HSP's successfully gapped in prelim test: 961
Number of HSP's that attempted gapping in prelim test: 267701
Number of HSP's gapped (non-prelim): 4988
length of query: 685
length of database: 863,360,394
effective HSP length: 135
effective length of query: 550
effective length of database: 520,377,774
effective search space: 286207775700
effective search space used: 286207775700
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0037a.11


