

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0032.7
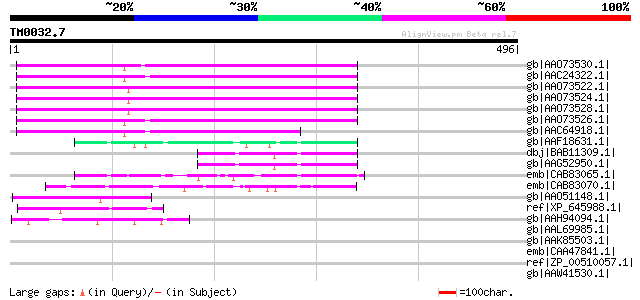
(496 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO73530.1| envelope-like protein [Glycine max] 270 1e-70
gb|AAC24322.1| envelope-like [Glycine max] gi|7488670|pir||T0889... 264 4e-69
gb|AAO73522.1| envelope-like protein [Glycine max] 263 1e-68
gb|AAO73524.1| envelope-like protein [Glycine max] 261 5e-68
gb|AAO73528.1| envelope-like protein [Glycine max] 260 8e-68
gb|AAO73526.1| envelope-like protein [Glycine max] 252 2e-65
gb|AAC64918.1| envelope-like [Glycine max] 202 3e-50
gb|AAF18631.1| F5J5.2 [Arabidopsis thaliana] 79 4e-13
dbj|BAB11309.1| unnamed protein product [Arabidopsis thaliana] 74 1e-11
gb|AAG52950.1| putative envelope protein [Arabidopsis thaliana] 74 1e-11
emb|CAB83065.1| putative protein [Arabidopsis thaliana] gi|11283... 60 2e-07
emb|CAB83070.1| putative protein [Arabidopsis thaliana] gi|11357... 58 6e-07
gb|AAO51148.1| similar to Y55B1BR.3.p [Caenorhabditis elegans] [... 51 1e-04
ref|XP_645988.1| hypothetical protein DDB0190281 [Dictyostelium ... 49 3e-04
gb|AAH94094.1| Unknown (protein for IMAGE:6643113) [Xenopus laevis] 48 8e-04
gb|AAL69985.1| putative envelope protein [Vicia faba] 47 0.001
gb|AAK85503.1| Hypothetical protein Y55B1BR.3 [Caenorhabditis el... 45 0.005
emb|CAA47841.1| S-layer protein [Clostridium thermocellum] gi|54... 45 0.007
ref|ZP_00510057.1| S-layer protein (SLH domain) [Clostridium the... 45 0.007
gb|AAW41530.1| protein kinase/endoribonuclease, putative [Crypto... 44 0.012
>gb|AAO73530.1| envelope-like protein [Glycine max]
Length = 685
Score = 270 bits (689), Expect = 1e-70
Identities = 144/340 (42%), Positives = 200/340 (58%), Gaps = 9/340 (2%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E + P ED T+EE+ E++P P+ PG + + SD+E P
Sbjct: 191 SSTPNAEVLSSPSKEDSTEEEDQATEETPAPRAPEPAPGDLIDLEEVESDEEPIANKLAP 250
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP------VDVEDSESD 120
+ R P+ R T+ K + T S + K +P + DS+ D
Sbjct: 251 GIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPTTSRRSKVAIPSKKRKEISSSDSDDD 310
Query: 121 AEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGP 180
E DV DI+ + K SGK++P NVP P+D +SFH ++V++WKFV + R+A RE+G
Sbjct: 311 VELDVPDIK---RAKTSGKKVPGNVPDAPLDNISFHSIDNVERWKFVYQRRLALERELGR 367
Query: 181 DVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVEC 240
D L+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +C
Sbjct: 368 DALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVFVRGKC 427
Query: 241 VVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILN 300
V FSPAVIN+ LGR D VS +AKE+TA V+ WP K LS G LSVKYAIL+
Sbjct: 428 VRFSPAVINKYLGRPTDGVVDITVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAILH 487
Query: 301 RIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
RI A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 488 RIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 527
>gb|AAC24322.1| envelope-like [Glycine max] gi|7488670|pir||T08898 envelope-like -
soybean (fragment)
Length = 648
Score = 264 bits (675), Expect = 4e-69
Identities = 144/340 (42%), Positives = 198/340 (57%), Gaps = 9/340 (2%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E + E+ T+EEE E++P P+ PG + + SD+E P
Sbjct: 161 SSTPNAEVLSSSSKEESTEEEEQATEETPAPRAPEPAPGDLIDLEEVESDEEPIANKLAP 220
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP------VDVEDSESD 120
+ R P+T R T+ K + T S K +P DS+ D
Sbjct: 221 GIAERLQSRKGKTPITRSGRIKTMAQKKSTPITPTTSRWSKVAIPSKKRKEFSSSDSDDD 280
Query: 121 AEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGP 180
E DV DI+ ++K SGK++P NVP P+D +SFH +V++WKFV + R+A RE+G
Sbjct: 281 VELDVPDIKRAKK---SGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALERELGR 337
Query: 181 DVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVEC 240
D L+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +C
Sbjct: 338 DALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVFVRGKC 397
Query: 241 VVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILN 300
V FSPAVIN+ LGR D VS +AKE+TA V+ WP K LS G LSVKYAIL+
Sbjct: 398 VRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAILH 457
Query: 301 RIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
RI A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 458 RIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGKYIF 497
>gb|AAO73522.1| envelope-like protein [Glycine max]
Length = 680
Score = 263 bits (671), Expect = 1e-68
Identities = 142/337 (42%), Positives = 199/337 (58%), Gaps = 3/337 (0%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E P E T+E++ E++P P+ PG + + SD+E P
Sbjct: 183 SSAPNAEALPSPSEEGSTEEDDQAAEETPAPRAPEPAPGDLIDLEEVESDEEPIANRLAP 242
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV--EDSESDAEAD 124
+ R P+ R T+ K + S + K +P E S SD++ D
Sbjct: 243 GIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPATSRRSKVAIPSKKRKEISSSDSDKD 302
Query: 125 VQ-DIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVL 183
V+ D+ S+K K SGK++P NVP P+D +SFH +V+KWK+V + R+A RE+G D L
Sbjct: 303 VELDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDAL 362
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +CV F
Sbjct: 363 DCKEIMDLIKAAGLLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKF 422
Query: 244 SPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRID 303
SPAVIN+ LGR D +VS +AKE+TA V+ WP K LS G LSVKYAIL+RI
Sbjct: 423 SPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIG 482
Query: 304 AVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 483 AANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 519
>gb|AAO73524.1| envelope-like protein [Glycine max]
Length = 682
Score = 261 bits (666), Expect = 5e-68
Identities = 141/337 (41%), Positives = 198/337 (57%), Gaps = 3/337 (0%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E P E T+E++ E++P P+ PG + + SD+E P
Sbjct: 185 SSAPNAEALPSPGEEGSTEEDDQAAEETPAPRAPEPAPGDLIDLEEVESDEEPIANRLAP 244
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV--EDSESDAEAD 124
+ R P+ R T+ K + S + K +P E S SD++ D
Sbjct: 245 GIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPATSRRSKVAIPSKKRKEISSSDSDKD 304
Query: 125 VQ-DIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVL 183
V+ D+ S+K K SGK++P NVP P+D +SFH +V+KWK+V + R+A RE+G D L
Sbjct: 305 VELDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDAL 364
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
+CK+++ LI+ GL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +CV F
Sbjct: 365 DCKEIMDLIKAGGLLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKF 424
Query: 244 SPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRID 303
SPAVIN+ LGR D +VS +AKE+TA V+ WP K LS G LSVKYAIL+RI
Sbjct: 425 SPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIG 484
Query: 304 AVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 485 AANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 521
>gb|AAO73528.1| envelope-like protein [Glycine max]
Length = 680
Score = 260 bits (664), Expect = 8e-68
Identities = 140/337 (41%), Positives = 199/337 (58%), Gaps = 3/337 (0%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E P E T+E++ E++P P+ PG + + SD+E P
Sbjct: 183 SSAPNAEALPSPSEEGSTEEDDQAAEETPAPRAPEPAPGDLIDLEEVESDEEPIANRLAP 242
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV--EDSESDAEAD 124
+ R P+ R T+ K + S + K +P E S SD++ D
Sbjct: 243 GIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPATSRRSKVAIPSKKRKEISSSDSDKD 302
Query: 125 VQ-DIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVL 183
V+ D+ S+K K SGK++P NVP P+D +SFH +V+KWK+V + R+A RE+G D L
Sbjct: 303 VELDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDAL 362
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SD++++VFVR +CV F
Sbjct: 363 DCKEIMDLIKAAGLLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDDYQRVFVRGKCVRF 422
Query: 244 SPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRID 303
SPAVIN+ LGR D +VS +AKE+TA V+ WP K LS G LSVKYAIL+RI
Sbjct: 423 SPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIG 482
Query: 304 AVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 483 AANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 519
>gb|AAO73526.1| envelope-like protein [Glycine max]
Length = 656
Score = 252 bits (643), Expect = 2e-65
Identities = 139/340 (40%), Positives = 195/340 (56%), Gaps = 9/340 (2%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E + E T+EE+ E++P P+ PG + + SD+E K
Sbjct: 169 SSTPNAEVLSSSSKEKSTEEEDQATEETPAPRAPEPAPGDLIDLEEVESDEEPIVKKLAL 228
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP------VDVEDSESD 120
+ R P+T R T+ K + T S K +P + DS+ D
Sbjct: 229 GIAERLQSRKGKTPITRSGRIKTIAQKKSTPITPTTSRWSKVAIPSKKRKEISSSDSDDD 288
Query: 121 AEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGP 180
E DV DI+ ++K SGK++P NVP P+D +SFH +V++WKFV + R+A RE+G
Sbjct: 289 VELDVPDIKRAKK---SGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALERELGR 345
Query: 181 DVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVEC 240
D L+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++ VFVR +
Sbjct: 346 DALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQTVFVRGKG 405
Query: 241 VVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILN 300
+ FSPAVIN+ LGR D VS +AKE+TA V+ WP K LS G LSVKYAIL+
Sbjct: 406 IRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAILH 465
Query: 301 RIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
RI NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 466 RIGTANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 505
>gb|AAC64918.1| envelope-like [Glycine max]
Length = 444
Score = 202 bits (513), Expect = 3e-50
Identities = 113/284 (39%), Positives = 160/284 (55%), Gaps = 9/284 (3%)
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E + E+ T+EEE E++P P+ PG + + SD+E P
Sbjct: 161 SSTPNAEVLSSSSKEESTEEEEQATEETPAPRAPEPAPGDLIDLEEVESDEEPIANKLAP 220
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP------VDVEDSESD 120
+ R P+T R T+ K + T S K +P DS+ D
Sbjct: 221 GIAERLQSRKGKTPITRSGRIKTMAQKKSTPITPTTSRWSKVAIPSKKRKEFSSSDSDDD 280
Query: 121 AEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGP 180
E DV DI+ ++K SGK++P NVP P+D +SFH +V++WKFV + R+A RE+G
Sbjct: 281 VELDVPDIKRAKK---SGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALERELGR 337
Query: 181 DVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVEC 240
D L+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +C
Sbjct: 338 DALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVFVRGKC 397
Query: 241 VVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSK 284
V FSPAVIN+ LGR D VS +AKE+TA V+ WP K
Sbjct: 398 VRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKK 441
>gb|AAF18631.1| F5J5.2 [Arabidopsis thaliana]
Length = 463
Score = 78.6 bits (192), Expect = 4e-13
Identities = 80/308 (25%), Positives = 125/308 (39%), Gaps = 50/308 (16%)
Query: 64 STPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESD--A 121
+TP P PP P T + K K K V T +++K++ +
Sbjct: 57 ATPSDPPPPPVAPMFIPKTEPGICMSKKTKSK---VSTLTQRKRQCTAASAKFRSKAHLQ 113
Query: 122 EADVQDIRPS---------------EKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKF 166
+ DV +I P +KY + P P F EE + ++K
Sbjct: 114 DGDVTEIAPPLFLSCYRENRRLLAVRDRKYPELKFPSETPYSSC----FLTEEGLGRYKV 169
Query: 167 VCKIRMAKVREVGPDVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLP 226
+ +R + D L+ GL+ T+ + ++V EF NL P
Sbjct: 170 IGNRCFNDMRFLPLDGNNTASTQQLLFNGGLLPTVTEIDSYVHEVVMEFYANL------P 223
Query: 227 ESDE----FRKVFVRVECVVFSPAVINQAL----------GRSAIEFADKEVSMDDVAKE 272
+ +E VFVR FS A+INQ G S I+ + SMD+VA
Sbjct: 224 DGEEGDNLAYSVFVRRNMYEFSLAIINQMFQLPNPSYPLDGMSEIQVPE---SMDEVAIA 280
Query: 273 LTAGHVKKWPSKKLLSTGNLSVKYAILNRIDAVNWVPTQHTSGVSATLAKLIYMIGKSIA 332
L+ G W K+L++ LS + A+LN+I NW PT + S + L+YM+ K++
Sbjct: 281 LSNGKANSW---KMLTSRLLSPELALLNKICCHNWSPTVNRSVLKPERMTLLYMVAKALP 337
Query: 333 FDFGAFVF 340
F+FG +F
Sbjct: 338 FNFGKLIF 345
>dbj|BAB11309.1| unnamed protein product [Arabidopsis thaliana]
Length = 515
Score = 73.9 bits (180), Expect = 1e-11
Identities = 50/163 (30%), Positives = 83/163 (50%), Gaps = 12/163 (7%)
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
E +D++ +IE L+ T+ + ++++ EF N + E + KVFVR + F
Sbjct: 201 EMRDMMKVIEDNHLLDTVAEIDPFVKEVIWEFYANFHI---FNEETKKSKVFVRGKMYEF 257
Query: 244 SPAVINQALGRSAI------EFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYA 297
SP +IN+ AI EF +S +AK L+ G V W K L T + A
Sbjct: 258 SPQMINKTFRSPAIKWHAKAEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLA 314
Query: 298 ILNRIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
L I +NW+P+++ S V+ AK++Y++ + I F+FG V+
Sbjct: 315 GLYLICCINWIPSKNNSNVTVDRAKMLYLLNERIPFNFGKLVY 357
>gb|AAG52950.1| putative envelope protein [Arabidopsis thaliana]
Length = 461
Score = 73.9 bits (180), Expect = 1e-11
Identities = 50/163 (30%), Positives = 83/163 (50%), Gaps = 12/163 (7%)
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
E +D++ +IE L+ T+ + ++++ EF N + E + KVFVR + F
Sbjct: 201 EMRDMMKVIEDNHLLDTVAEIDPFVKEVIWEFYANFHI---FNEETKKSKVFVRGKMYEF 257
Query: 244 SPAVINQALGRSAI------EFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYA 297
SP +IN+ AI EF +S +AK L+ G V W K L T + A
Sbjct: 258 SPQMINKTFRSPAIKWHAKAEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLA 314
Query: 298 ILNRIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
L I +NW+P+++ S V+ AK++Y++ + I F+FG V+
Sbjct: 315 GLYLICCINWIPSKNNSNVTVDRAKMLYLLNERIPFNFGKLVY 357
>emb|CAB83065.1| putative protein [Arabidopsis thaliana] gi|11283487|pir||T47400
hypothetical protein F23N14.20 - Arabidopsis thaliana
Length = 428
Score = 59.7 bits (143), Expect = 2e-07
Identities = 73/293 (24%), Positives = 120/293 (40%), Gaps = 43/293 (14%)
Query: 64 STPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEA 123
+TP P PP P T + K K K V P+++K++ + S S A A
Sbjct: 57 ATPSDPPPPPVAPMFIPKTEPGICMSKKTKSK---VSAPTQRKRQCTAASAK-SRSKAPA 112
Query: 124 DVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVL 183
S K S Q +V R S L++ V E+ P +
Sbjct: 113 PALRRLSSRKSNRSASAAGQTEDLV---RDSSDLQDG-------------DVTEIAPPLF 156
Query: 184 -----ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLV----NLSVDVGLPESDEFRKV 234
E + ++ + ++ G K I N RC+ + FL N + L +
Sbjct: 157 LSRYRENRRLLAVRDRLGRYKVIGN--RCFNDM--RFLPLDGNNTASTQQLLFNAGLLPT 212
Query: 235 FVRVECVVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSV 294
F ++ + +++ + D SMD+VA L+ G W K+L++ LS
Sbjct: 213 FTEID------SYVHEVVMEFYANLPDVPESMDEVAIALSNGKANSW---KMLTSRLLSP 263
Query: 295 KYAILNRIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF*ADIEAC 347
+ A+LN+I NW PT + S + L+YM+ K++ F+FG +F +I AC
Sbjct: 264 ELALLNKICCHNWSPTVNHSVLKPERMTLLYMVAKALPFNFGKLIF-DEIWAC 315
>emb|CAB83070.1| putative protein [Arabidopsis thaliana] gi|11357591|pir||T47405
hypothetical protein F23N14.70 - Arabidopsis thaliana
Length = 539
Score = 58.2 bits (139), Expect = 6e-07
Identities = 76/317 (23%), Positives = 130/317 (40%), Gaps = 37/317 (11%)
Query: 36 VKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKGK 95
V PP P + S S++ P ++ P P+P PV +
Sbjct: 135 VSIPPPSAPPLVLSDSKDAE----PAGLTNPSAPPSPLAPKNITPVASPVADVPMPDP-- 188
Query: 96 QKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSF 155
+ T + VP + A A Q ++ + +R P+VP F
Sbjct: 189 -LISPTAETAEGASVPDAAVSYAARAAAHRQVFAERDELDRTLRR-----PLVPPHTKRF 242
Query: 156 HLEESVQKWKFVCK--IRMAKVREVGPDVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVR 213
+ +++K + K K + P+VL +E +G+ +T+ V + ++VR
Sbjct: 243 LSAAAAERYKHIAKRDFIFQKTLPLDPEVLTATKYF--LEHSGMAQTVVAVEQFVPEVVR 300
Query: 214 EFLVNLSVDVGLPESDEFRK-----VFVRVECVVFSPAVINQ--ALGRSAIE----FADK 262
EF NL PE E+R+ V+VR + FSPA+IN ++ SA++
Sbjct: 301 EFYANL------PEM-EYRECGLDLVYVRGKMYEFSPALINHMFSIDDSALDPEAPVTLS 353
Query: 263 EVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRIDAVNWVPTQHTSGVSATLAK 322
S DD+A +T G ++W +L +L +L+++ NW PT +TS + +
Sbjct: 354 TASRDDLALMMTGGTTRRW--LRLQPADHLDTM-KMLHKVCCGNWFPTTNTSTLRVDRLR 410
Query: 323 LIYMIGKSIAFDFGAFV 339
LI M +F+ G V
Sbjct: 411 LIDMGTHGKSFNLGKLV 427
>gb|AAO51148.1| similar to Y55B1BR.3.p [Caenorhabditis elegans] [Dictyostelium
discoideum] gi|66821633|ref|XP_644266.1| hypothetical
protein DDB0167567 [Dictyostelium discoideum]
gi|60472037|gb|EAL69990.1| hypothetical protein
DDB0167567 [Dictyostelium discoideum]
Length = 727
Score = 50.8 bits (120), Expect = 1e-04
Identities = 36/144 (25%), Positives = 62/144 (43%), Gaps = 8/144 (5%)
Query: 3 KTYMSKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGK 62
+T +S + K P E+ +EEE NDE+ V V + ++ +P K
Sbjct: 370 ETKLSSSKKKTPVKKTSKEEEEEEEEDNDEEEQSSKKKTPVKKTSKIVEDDDEEETSPSK 429
Query: 63 VSTPEVTPTPPKRSKSCPVTYKSRQ-------ATVKGKGKQKVVKTPSEKKKRKVPVDVE 115
TP TP KR P + + K K +K K+ + + +K P +
Sbjct: 430 KKTPTKNTTPSKRKSVEPKPKEKEEEDEEKESPKKKTKKAEKSPKSDTTRSGKKTPKRTK 489
Query: 116 DSESDA-EADVQDIRPSEKKKYSG 138
E+D+ +++V+D + S+ KK G
Sbjct: 490 QDENDSDDSEVEDEKSSKGKKKKG 513
Score = 36.6 bits (83), Expect = 1.9
Identities = 36/135 (26%), Positives = 58/135 (42%), Gaps = 18/135 (13%)
Query: 21 EDVTDEEEINDEDSPVKSPPDVVPGVETSVS------QEISDQENPGKVSTPEVTPTPPK 74
E+ +EEE +++ K P ++ S +E + E P K + E PP
Sbjct: 229 EEEEEEEEEEEDEEEKKPKKKSTPKKDSKSSKKKEEEEEEEEDETPKKSTEKETKKKPPA 288
Query: 75 RSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKR-KVPV------DVEDSESDAEADVQD 127
+K K + K ++KV KT KK KVP +++ S S + ++
Sbjct: 289 ATKKSNKKLKDDEEEEK---EEKVEKTTKVKKSSFKVPTAVTPNKEIKSSTSKKTPNKKE 345
Query: 128 IRPSEKKKYSGKRIP 142
I E+KK S K+IP
Sbjct: 346 I--EEEKKTSTKKIP 358
>ref|XP_645988.1| hypothetical protein DDB0190281 [Dictyostelium discoideum]
gi|60474143|gb|EAL72080.1| hypothetical protein
DDB0190281 [Dictyostelium discoideum]
Length = 2221
Score = 49.3 bits (116), Expect = 3e-04
Identities = 33/145 (22%), Positives = 67/145 (45%), Gaps = 7/145 (4%)
Query: 8 KVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVET--SVSQEISDQENPGKVST 65
K N+ NV ++ DE+E +++ K P + E + Q+ ++E P + +
Sbjct: 2021 KSNNQNEENVQPTQEKQDEQEPEEQEEQEKQPEEEEEEEEKVKEIEQQEEEEEEPKEEQS 2080
Query: 66 PEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADV 125
E P+ + K Q + G+++ + P E+++ K P + E+ E + E D
Sbjct: 2081 EEEEEEQPEEEQPEEEQTKEEQPGEEQPGEEE--EEPEEEEEEKEPEEEEEEEKEMEEDE 2138
Query: 126 QDIRPSEKKKYSGKRIPQNVPVVPI 150
++ E++K + P+ P+VPI
Sbjct: 2139 EEKEDEEEEK---EDTPKKQPIVPI 2160
>gb|AAH94094.1| Unknown (protein for IMAGE:6643113) [Xenopus laevis]
Length = 722
Score = 47.8 bits (112), Expect = 8e-04
Identities = 52/197 (26%), Positives = 81/197 (40%), Gaps = 36/197 (18%)
Query: 2 RKTYMSKVQNKEPTNV---------PHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQ 52
RKT K +N +P V P + +DEEE +ED PVK+ ++
Sbjct: 251 RKTRAKKSKNVKPVKVEEDTKSESSPPSSEPSDEEEEQEEDEPVKTN-----------NK 299
Query: 53 EISDQENPGKVSTPEVTPTPPKRSKSCPVTYK--SRQATVKGKGKQKVVKTPSEKKKRKV 110
++EN + P+ K K ++ K + K+ K +KK+ KV
Sbjct: 300 RRVEKENEDRTELDVSVNKEPEEKKEIKENKKDLKKEKKEKEEKKEPAEKEARDKKEIKV 359
Query: 111 PVDVEDSESD---AEADVQDIRPSEKKKYSGKRIPQNVPV--------VPIDRVSFHLEE 159
V+++ + E V +++ ++K+ S K I PV VPI S EE
Sbjct: 360 KEPVKEAVKEKEIKEEKVNEVKCEKEKEKSKKSISAETPVHITASSEPVPI---SEEAEE 416
Query: 160 SVQKWKFVCKIRMAKVR 176
QK VCK RM V+
Sbjct: 417 LDQKTFSVCKERMRPVK 433
>gb|AAL69985.1| putative envelope protein [Vicia faba]
Length = 59
Score = 47.4 bits (111), Expect = 0.001
Identities = 19/39 (48%), Positives = 25/39 (63%)
Query: 302 IDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
I A NWVPT H S +S L +LIY +G FD+G ++F
Sbjct: 1 IGAANWVPTNHKSTISTGLGRLIYAVGTRTEFDYGRYIF 39
>gb|AAK85503.1| Hypothetical protein Y55B1BR.3 [Caenorhabditis elegans]
gi|17556242|ref|NP_497198.1| chromo domain containing
protein (77.9 kD) (3B49) [Caenorhabditis elegans]
Length = 679
Score = 45.1 bits (105), Expect = 0.005
Identities = 42/167 (25%), Positives = 72/167 (42%), Gaps = 11/167 (6%)
Query: 2 RKTYMSKVQNKEPTNVPHCEDVTDEEEIND--EDSPVKSPPDVVPGVETSVS-QEISDQE 58
+K K +K+P E+ +D EE + E+SP+K D P + S ++ E
Sbjct: 445 KKVKSPKKSSKKPAAKVESEEPSDNEEEEEEVEESPIKK--DKTPRKYSRKSAAKVESTE 502
Query: 59 NPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVV-----KTPSEKKKRKVPVD 113
+ G EV +P K+ K+ + K A + +++ V K S +K K
Sbjct: 503 SSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEESDNEEEDVEESPKKRTSPRKSSKKRAA 562
Query: 114 VEDSESDAEADVQDIRPSEKKKYSGKRIPQNVPVVPID-RVSFHLEE 159
E+SE ++ +V+++ S KK R P ++ SF EE
Sbjct: 563 KEESEESSDNEVEEVEDSPKKNDKTLRKSPRKPAAKVESEESFGNEE 609
Score = 44.7 bits (104), Expect = 0.007
Identities = 42/171 (24%), Positives = 74/171 (42%), Gaps = 15/171 (8%)
Query: 8 KVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPE 67
K + N ++ + +EE E+SP K + S ++E S++E+ E
Sbjct: 379 KKKKSSKINKRKAKESSSDEEEEVEESPKKKTKSPRKSSKKSAAKEESEEESSDNEEEEE 438
Query: 68 VTPTPPKRSKSCPVTYKSRQATVKG-------KGKQKVVKTPSEKKK------RKVPVDV 114
V +P K+ KS + K A V+ + +++V ++P +K K RK V
Sbjct: 439 VDYSPKKKVKSPKKSSKKPAAKVESEEPSDNEEEEEEVEESPIKKDKTPRKYSRKSAAKV 498
Query: 115 EDSESDAEADVQDIRPSEKK--KYSGKRIPQNVPVVPIDRVSFHLEESVQK 163
E +ES + +++ S KK K K ++ V D +EES +K
Sbjct: 499 ESTESSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEESDNEEEDVEESPKK 549
Score = 41.2 bits (95), Expect = 0.076
Identities = 35/134 (26%), Positives = 54/134 (40%), Gaps = 8/134 (5%)
Query: 8 KVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPE 67
K K V E +EEE E+SP K + S + E SD E +P+
Sbjct: 489 KYSRKSAAKVESTESSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEESDNEEEDVEESPK 548
Query: 68 VTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSE------KKKRKVPVDVEDSESDA 121
+P K SK +S +++ ++V +P + K RK VE ES
Sbjct: 549 KRTSPRKSSKKRAAKEESEESS--DNEVEEVEDSPKKNDKTLRKSPRKPAAKVESEESFG 606
Query: 122 EADVQDIRPSEKKK 135
+ +++ S KKK
Sbjct: 607 NEEEEEVEESPKKK 620
Score = 40.8 bits (94), Expect = 0.099
Identities = 36/133 (27%), Positives = 53/133 (39%), Gaps = 4/133 (3%)
Query: 9 VQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEV 68
V NK P H + D EE DE+ KSP + S SD ++ +
Sbjct: 177 VMNKPPKKSKHQDTSDDSEESEDEEE--KSPRKRSSKKRRAASVSDSDSDSDSDFDKKKW 234
Query: 69 TPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDI 128
KRSK + + + K +K K+ KK K V DS SD E + +
Sbjct: 235 KKNKAKRSKRDDSSDDDSEMERRRKKSKKSKKSKKFKKSEKRKRAVNDSSSDDEDEEE-- 292
Query: 129 RPSEKKKYSGKRI 141
+P ++ K S K +
Sbjct: 293 KPEKRSKKSKKAV 305
Score = 38.5 bits (88), Expect = 0.49
Identities = 35/113 (30%), Positives = 52/113 (45%), Gaps = 12/113 (10%)
Query: 72 PPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEK--KKRKVPVDVEDSESDAEADVQDIR 129
P KRSK KS++A + + + + S+K KK K D E SD+E +V +++
Sbjct: 294 PEKRSK------KSKKAVIDSSSEDEEEEKSSKKRSKKSKKESDEEQQASDSEEEVVEVK 347
Query: 130 PSEKKKYSGKRIPQNVPV-VPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPD 181
+ K S K+ P+ V + S EE V K K KI K +E D
Sbjct: 348 KNSK---SPKKTPKKTAVKEESEESSGDEEEEVVKKKKSSKINKRKAKESSSD 397
>emb|CAA47841.1| S-layer protein [Clostridium thermocellum]
gi|548937|sp|Q06852|SLAP1_CLOTM Cell surface glycoprotein
1 precursor (Outer layer protein B) (S-layer protein 1)
Length = 1664
Score = 44.7 bits (104), Expect = 0.007
Identities = 34/130 (26%), Positives = 51/130 (39%), Gaps = 15/130 (11%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 981 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP----------- 1029
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
S + T + TPSE + +P D E + SD + PS++ S +
Sbjct: 1030 -SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1088
Query: 141 IPQNVPVVPI 150
P P PI
Sbjct: 1089 TPSETPEEPI 1098
Score = 41.6 bits (96), Expect = 0.058
Identities = 32/126 (25%), Positives = 50/126 (39%), Gaps = 15/126 (11%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 926 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP----------- 974
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
S + T + TPSE + +P D E + SD + PS++ S +
Sbjct: 975 -SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1033
Query: 141 IPQNVP 146
P + P
Sbjct: 1034 TPSDEP 1039
Score = 41.6 bits (96), Expect = 0.058
Identities = 32/126 (25%), Positives = 50/126 (39%), Gaps = 15/126 (11%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 1251 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP----------- 1299
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
S + T + TPSE + +P D E + SD + PS++ S +
Sbjct: 1300 -SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1358
Query: 141 IPQNVP 146
P + P
Sbjct: 1359 TPSDEP 1364
Score = 41.2 bits (95), Expect = 0.076
Identities = 36/149 (24%), Positives = 55/149 (36%), Gaps = 22/149 (14%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTP-----PKRSKS 78
+DE +DE +P ++P + +P S SD+ P TP PTP P +
Sbjct: 1079 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 1138
Query: 79 CPV--------------TYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDA 121
P+ S + T + TPSE + +P D E + SD
Sbjct: 1139 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE 1198
Query: 122 EADVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1199 PTPSDEPTPSDEPTPSDEPTPSETPEEPI 1227
Score = 41.2 bits (95), Expect = 0.076
Identities = 36/149 (24%), Positives = 55/149 (36%), Gaps = 22/149 (14%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTP-----PKRSKS 78
+DE +DE +P ++P + +P S SD+ P TP PTP P +
Sbjct: 797 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 856
Query: 79 CPV--------------TYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDA 121
P+ S + T + TPSE + +P D E + SD
Sbjct: 857 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE 916
Query: 122 EADVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 917 PTPSDEPTPSDEPTPSDEPTPSETPEEPI 945
Score = 37.4 bits (85), Expect = 1.1
Identities = 30/126 (23%), Positives = 46/126 (35%), Gaps = 27/126 (21%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 1208 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE-------- 1259
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
TPSE + +P D E + SD + PS++ S +
Sbjct: 1260 ----------------PTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1303
Query: 141 IPQNVP 146
P + P
Sbjct: 1304 TPSDEP 1309
Score = 36.6 bits (83), Expect = 1.9
Identities = 37/147 (25%), Positives = 52/147 (35%), Gaps = 28/147 (19%)
Query: 14 PTNVPHCEDV-------TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
PT+ P E +DE +DE +P P E + SD+ P TP
Sbjct: 1056 PTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTP 1115
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEA 123
PTP S + T + TPSE + +P D E + SD
Sbjct: 1116 SDEPTP------------SDEPTPSDE------PTPSETPEEPIPTDTPSDEPTPSDEPT 1157
Query: 124 DVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1158 PSDEPTPSDEPTPSDEPTPSETPEEPI 1184
Score = 36.6 bits (83), Expect = 1.9
Identities = 37/147 (25%), Positives = 52/147 (35%), Gaps = 28/147 (19%)
Query: 14 PTNVPHCEDV-------TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
PT+ P E +DE +DE +P P E + SD+ P TP
Sbjct: 774 PTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTP 833
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEA 123
PTP S + T + TPSE + +P D E + SD
Sbjct: 834 SDEPTP------------SDEPTPSDE------PTPSETPEEPIPTDTPSDEPTPSDEPT 875
Query: 124 DVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 876 PSDEPTPSDEPTPSDEPTPSETPEEPI 902
>ref|ZP_00510057.1| S-layer protein (SLH domain) [Clostridium thermocellum ATCC 27405]
gi|67849694|gb|EAM45291.1| S-layer protein (SLH domain)
[Clostridium thermocellum ATCC 27405]
Length = 2313
Score = 44.7 bits (104), Expect = 0.007
Identities = 34/130 (26%), Positives = 51/130 (39%), Gaps = 15/130 (11%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 1587 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP----------- 1635
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
S + T + TPSE + +P D E + SD + PS++ S +
Sbjct: 1636 -SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1694
Query: 141 IPQNVPVVPI 150
P P PI
Sbjct: 1695 TPSETPEEPI 1704
Score = 41.6 bits (96), Expect = 0.058
Identities = 32/126 (25%), Positives = 50/126 (39%), Gaps = 15/126 (11%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 1900 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP----------- 1948
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
S + T + TPSE + +P D E + SD + PS++ S +
Sbjct: 1949 -SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 2007
Query: 141 IPQNVP 146
P + P
Sbjct: 2008 TPSDEP 2013
Score = 41.2 bits (95), Expect = 0.076
Identities = 36/149 (24%), Positives = 55/149 (36%), Gaps = 22/149 (14%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTP-----PKRSKS 78
+DE +DE +P ++P + +P S SD+ P TP PTP P +
Sbjct: 1403 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 1462
Query: 79 CPV--------------TYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDA 121
P+ S + T + TPSE + +P D E + SD
Sbjct: 1463 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE 1522
Query: 122 EADVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1523 PTPSDEPTPSDEPTPSDEPTPSETPEEPI 1551
Score = 41.2 bits (95), Expect = 0.076
Identities = 36/149 (24%), Positives = 55/149 (36%), Gaps = 22/149 (14%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTP-----PKRSKS 78
+DE +DE +P ++P + +P S SD+ P TP PTP P +
Sbjct: 1771 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 1830
Query: 79 CPV--------------TYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDA 121
P+ S + T + TPSE + +P D E + SD
Sbjct: 1831 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE 1890
Query: 122 EADVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1891 PTPSDEPTPSDEPTPSDEPTPSETPEEPI 1919
Score = 41.2 bits (95), Expect = 0.076
Identities = 36/149 (24%), Positives = 55/149 (36%), Gaps = 22/149 (14%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTP-----PKRSKS 78
+DE +DE +P ++P + +P S SD+ P TP PTP P +
Sbjct: 1685 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 1744
Query: 79 CPV--------------TYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDA 121
P+ S + T + TPSE + +P D E + SD
Sbjct: 1745 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE 1804
Query: 122 EADVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1805 PTPSDEPTPSDEPTPSDEPTPSETPEEPI 1833
Score = 37.4 bits (85), Expect = 1.1
Identities = 30/126 (23%), Positives = 46/126 (35%), Gaps = 27/126 (21%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 1489 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE-------- 1540
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
TPSE + +P D E + SD + PS++ S +
Sbjct: 1541 ----------------PTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1584
Query: 141 IPQNVP 146
P + P
Sbjct: 1585 TPSDEP 1590
Score = 37.4 bits (85), Expect = 1.1
Identities = 30/126 (23%), Positives = 46/126 (35%), Gaps = 27/126 (21%)
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
+DE +DE +P ++P + +P S SD+ P TP PTP
Sbjct: 1857 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE-------- 1908
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEADVQDIRPSEKKKYSGKR 140
TPSE + +P D E + SD + PS++ S +
Sbjct: 1909 ----------------PTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1952
Query: 141 IPQNVP 146
P + P
Sbjct: 1953 TPSDEP 1958
Score = 36.6 bits (83), Expect = 1.9
Identities = 37/147 (25%), Positives = 52/147 (35%), Gaps = 28/147 (19%)
Query: 14 PTNVPHCEDV-------TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
PT+ P E +DE +DE +P P E + SD+ P TP
Sbjct: 1662 PTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTP 1721
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEA 123
PTP S + T + TPSE + +P D E + SD
Sbjct: 1722 SDEPTP------------SDEPTPSDE------PTPSETPEEPIPTDTPSDEPTPSDEPT 1763
Query: 124 DVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1764 PSDEPTPSDEPTPSDEPTPSETPEEPI 1790
Score = 36.6 bits (83), Expect = 1.9
Identities = 37/147 (25%), Positives = 52/147 (35%), Gaps = 28/147 (19%)
Query: 14 PTNVPHCEDV-------TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
PT+ P E +DE +DE +P P E + SD+ P TP
Sbjct: 1380 PTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTP 1439
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV---EDSESDAEA 123
PTP S + T + TPSE + +P D E + SD
Sbjct: 1440 SDEPTP------------SDEPTPSDE------PTPSETPEEPIPTDTPSDEPTPSDEPT 1481
Query: 124 DVQDIRPSEKKKYSGKRIPQNVPVVPI 150
+ PS++ S + P P PI
Sbjct: 1482 PSDEPTPSDEPTPSDEPTPSETPEEPI 1508
>gb|AAW41530.1| protein kinase/endoribonuclease, putative [Cryptococcus neoformans
var. neoformans JEC21] gi|50259858|gb|EAL22526.1|
hypothetical protein CNBB4040 [Cryptococcus neoformans
var. neoformans B-3501A] gi|58262854|ref|XP_568837.1|
protein kinase/endoribonuclease, putative [Cryptococcus
neoformans var. neoformans JEC21]
Length = 1073
Score = 43.9 bits (102), Expect = 0.012
Identities = 39/142 (27%), Positives = 54/142 (37%), Gaps = 12/142 (8%)
Query: 7 SKVQNKEPTNVPHCEDVT---DEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKV 63
SKV K P ++V EEE +E V S VP V + P
Sbjct: 492 SKVDEKMPLLAAPVQEVRLEQKEEEQKEEGEEVLSVVKSVPVVRIQEPSPTPEDSPPRSE 551
Query: 64 STPEV--TPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDA 121
TPE T TPP + KS + V+GK K+ + D E+ + D
Sbjct: 552 GTPETDQTATPPPKKKST-------RRRVRGKKKKPDATAATAASLIPEEHDGENEDEDH 604
Query: 122 EADVQDIRPSEKKKYSGKRIPQ 143
E D +D+ P K K +P+
Sbjct: 605 EKDKEDVSPRPTPKGGNKPLPE 626
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.333 0.142 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 782,839,220
Number of Sequences: 2540612
Number of extensions: 32524729
Number of successful extensions: 198942
Number of sequences better than 10.0: 780
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 794
Number of HSP's that attempted gapping in prelim test: 195271
Number of HSP's gapped (non-prelim): 3107
length of query: 496
length of database: 863,360,394
effective HSP length: 132
effective length of query: 364
effective length of database: 527,999,610
effective search space: 192191858040
effective search space used: 192191858040
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0032.7


