

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.4
(1177 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
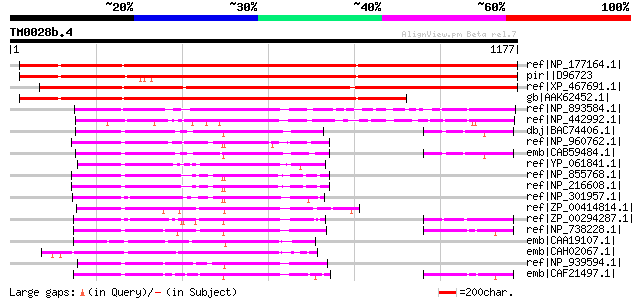
ref|NP_177164.1| DEAD/DEAH box helicase, putative [Arabidopsis t... 1550 0.0
pir||D96723 hypothetical protein F20P5.20 [imported] - Arabidops... 1454 0.0
ref|XP_467691.1| putative helicase [Oryza sativa (japonica culti... 1438 0.0
gb|AAK62452.1| Similar to Synechocystis antiviral protein [Arabi... 1152 0.0
ref|NP_893584.1| putative DNA helicase [Prochlorococcus marinus ... 513 e-143
ref|NP_442992.1| antiviral protein [Synechocystis sp. PCC 6803] ... 478 e-133
dbj|BAC74406.1| putative ATP-dependent RNA helicase [Streptomyce... 335 4e-90
ref|NP_960762.1| HelY [Mycobacterium avium subsp. paratuberculos... 335 7e-90
emb|CAB59484.1| putative helicase [Streptomyces coelicolor A3(2)... 330 2e-88
ref|YP_061841.1| ATP-dependent RNA helicase [Leifsonia xyli subs... 326 2e-87
ref|NP_855768.1| PROBABLE ATP-DEPENDENT DNA HELICASE HELY [Mycob... 323 3e-86
ref|NP_216608.1| PROBABLE ATP-DEPENDENT DNA HELICASE HELY [Mycob... 322 4e-86
ref|NP_301957.1| probable helicase, Ski2 subfamily [Mycobacteriu... 319 3e-85
ref|ZP_00414814.1| Helicase, C-terminal:Type III restriction enz... 317 1e-84
ref|ZP_00294287.1| COG4581: Superfamily II RNA helicase [Thermob... 316 3e-84
ref|NP_738228.1| putative helicase [Corynebacterium efficiens YS... 315 8e-84
emb|CAA19107.1| SPCC550.03c [Schizosaccharomyces pombe] gi|19075... 311 8e-83
emb|CAH02067.1| unnamed protein product [Kluyveromyces lactis NR... 306 3e-81
ref|NP_939594.1| Putative helicase [Corynebacterium diphtheriae ... 305 6e-81
emb|CAF21497.1| Superfamily II DNA and RNA helicase [Corynebacte... 302 4e-80
>ref|NP_177164.1| DEAD/DEAH box helicase, putative [Arabidopsis thaliana]
Length = 1171
Score = 1550 bits (4014), Expect = 0.0
Identities = 790/1160 (68%), Positives = 956/1160 (82%), Gaps = 14/1160 (1%)
Query: 24 YPSF--SLLPIPTTTLPLSLRSLSSTLKFRLSFSFNPPTSSSLPTFSDAGEDDVEDEDDD 81
+PS SL P + SL + L F+ + + P+ S L D E++ EDEDDD
Sbjct: 20 FPSLHRSLSHSPNFSFTKSLILNPNHLSFKSTLNSLSPSQSQLYEEEDDEEEEEEDEDDD 79
Query: 82 DEEEEEYE---EEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEKLCNEV 138
DE +EY+ +E DDDDD E+ ++P E ++ R EF+WQRVEKL + V
Sbjct: 80 DEAADEYDNISDEIRNSDDDDDDEETEFSVDLPTE-----SARERVEFRWQRVEKLRSLV 134
Query: 139 REFGAEIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATV 198
R+FG E+ID+DEL S+YDFRIDKFQRLA++AFLRGSSVVVSAPTSSGKTLIAEAAAV+TV
Sbjct: 135 RDFGVEMIDIDELISIYDFRIDKFQRLAIEAFLRGSSVVVSAPTSSGKTLIAEAAAVSTV 194
Query: 199 ARGRRIFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLY 258
A+GRR+FYTTPLKALSNQKFREFRETFGD VGLLTGDSA+NKDAQ++IMTTEILRNMLY
Sbjct: 195 AKGRRLFYTTPLKALSNQKFREFRETFGDDNVGLLTGDSAINKDAQIVIMTTEILRNMLY 254
Query: 259 QSVGNVSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPD 318
QSVG ++SS +GL +VD IVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPD
Sbjct: 255 QSVG-MASSGTGLFHVDAIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPD 313
Query: 319 ELAGWIGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAA 378
ELAGWIG+IHGKTELVTS++RPVPLTW+FS K+SLLPLLDEKG +NRKLSLNYLQL A+
Sbjct: 314 ELAGWIGEIHGKTELVTSTRRPVPLTWYFSTKHSLLPLLDEKGINVNRKLSLNYLQLSAS 373
Query: 379 GYK-PYKDDWSRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHI 437
+ DD RKR ++KRG SY++ ++ + LSKN IN IRRSQVPQI DTLWH+
Sbjct: 374 EARFRDDDDGYRKRRSKKRGGDTSYNNLVNVTDY-PLSKNEINKIRRSQVPQISDTLWHL 432
Query: 438 EPRDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVRETAVK 497
+ ++MLPAIWFIF+R+GCDAAVQY+E+ +LLD+CEKSEVELALK+FR+ YPDAVRE+A K
Sbjct: 433 QGKNMLPAIWFIFNRRGCDAAVQYVENFQLLDDCEKSEVELALKKFRVLYPDAVRESAEK 492
Query: 498 GLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRID 557
GLL+G+AAHHAGCLPLWK+FIEELFQ+GLVKVVFATETLAAGINMPARTAVISSLSK+
Sbjct: 493 GLLRGIAAHHAGCLPLWKSFIEELFQRGLVKVVFATETLAAGINMPARTAVISSLSKKAG 552
Query: 558 SGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTAS 617
+ R L NEL QMAGRAGRRGIDE G+ VL+QT EGAEECCK++FAG++PLVSQFTAS
Sbjct: 553 NERIELGPNELYQMAGRAGRRGIDEKGYTVLVQTAFEGAEECCKLVFAGVKPLVSQFTAS 612
Query: 618 YGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELK 677
YGMVLNL++G K +S+ ++ + GR+LE+A+KLVE+SFGNYVSSNV +AAK+EL
Sbjct: 613 YGMVLNLVAGSKVTRKSSGTEAGKVLQAGRSLEEAKKLVEKSFGNYVSSNVTVAAKQELA 672
Query: 678 KNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQMEAKRI 737
+ + +IE+L SEI+DEAID+KSRK LS + YKEI L+E+LR EKR R RR+ME +R
Sbjct: 673 EIDNKIEILSSEISDEAIDKKSRKLLSARDYKEITVLKEELREEKRKRAEQRRRMELERF 732
Query: 738 SALKPLLDDPESGHLPFLCLQYRDSEGVLHSIPAVFLGKVDSLNASKLKDMISSVDSFAL 797
ALKPLL E G+LPF+CL+++DSEG S+PAV+LG +DS SKL+ M+S +SFAL
Sbjct: 733 LALKPLLKGMEEGNLPFICLEFKDSEGREQSVPAVYLGHIDSFQGSKLQKMMSLDESFAL 792
Query: 798 NVADAERSLPDSVLNEDLEPSYHVALGSDNSWYLFTEKWIKTVYGTGFPDTPLAQGDARP 857
N+ + E + D +++PSY+VALGSDNSWYLFTEKW++TVY TGFP+ LA GDA P
Sbjct: 793 NLIEDELA-ADEPGKPNVKPSYYVALGSDNSWYLFTEKWVRTVYRTGFPNIALALGDALP 851
Query: 858 REIMSTLLDKEDMKWDKLAHSEHGGLWFMEGSLETWSWSLNVPVLSSFSENDELLLKSQA 917
REIM LLDK DM+WDKLA SE G LW +EGSLETWSWSLNVPVLSS S+ DE+L S+
Sbjct: 852 REIMKNLLDKADMQWDKLAESELGSLWRLEGSLETWSWSLNVPVLSSLSDEDEVLHMSEE 911
Query: 918 YRDAIEQYKDQRNKVSRLKKRISRTEGYKEYNKIIDGVKFIEEKIKRLKTRSKRLTNRIE 977
Y +A ++YK+QR+K+SRLKK++SR+EG++EY KI++ EK+KRLK RS+RL NR+E
Sbjct: 912 YDNAAQKYKEQRSKISRLKKKMSRSEGFREYKKILENANLTVEKMKRLKARSRRLINRLE 971
Query: 978 QIEPSGWKEFMQVSNVIHETRALDINTHVIFPLGETAAAIRGENELWLAMVLRSKILVGL 1037
QIEPSGWK+FM++SNVIHE+RALDINTH+IFPLGETAAAIRGENELWLAMVLR+K LV L
Sbjct: 972 QIEPSGWKDFMRISNVIHESRALDINTHLIFPLGETAAAIRGENELWLAMVLRNKALVDL 1031
Query: 1038 KPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCIGLLGEQRSALLAIQEKHGVTI 1097
KPPQLA VCA LVSEGIKVRPW++N++IYEPS TVV+ + L +QRS+L+ +QEKH V I
Sbjct: 1032 KPPQLAGVCASLVSEGIKVRPWRDNNYIYEPSDTVVDMVNFLEDQRSSLIKLQEKHEVMI 1091
Query: 1098 SCCLDTQFCGMVEAWASGLTWREIMMDCAMDDGDLARLLRRTIDLLAQIPKLADIDPLLQ 1157
CCLD QF GMVEAWASGL+W+E+MM+CAMD+GDLARLLRRTIDLLAQIPKL DIDP+LQ
Sbjct: 1092 PCCLDVQFSGMVEAWASGLSWKEMMMECAMDEGDLARLLRRTIDLLAQIPKLPDIDPVLQ 1151
Query: 1158 RNARAASDVMDRPPISELAG 1177
R+A AA+D+MDRPPISELAG
Sbjct: 1152 RSAAAAADIMDRPPISELAG 1171
>pir||D96723 hypothetical protein F20P5.20 [imported] - Arabidopsis thaliana
gi|2194131|gb|AAB61106.1| Similar to Synechocystis
antiviral protein (gb|D90917). [Arabidopsis thaliana]
Length = 1198
Score = 1454 bits (3764), Expect = 0.0
Identities = 759/1189 (63%), Positives = 932/1189 (77%), Gaps = 45/1189 (3%)
Query: 24 YPSF--SLLPIPTTTLPLSLRSLSSTLKFRLSFSFNPPTSSSLPTFSDAGEDDVEDEDDD 81
+PS SL P + SL + L F+ + + P+ S L D E++ EDEDDD
Sbjct: 20 FPSLHRSLSHSPNFSFTKSLILNPNHLSFKSTLNSLSPSQSQLYEEEDDEEEEEEDEDDD 79
Query: 82 DEEEEEYE---EEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEKLCNEV 138
DE +EY+ +E DDDDD E+ ++P E ++ R EF+WQRVEKL + V
Sbjct: 80 DEAADEYDNISDEIRNSDDDDDDEETEFSVDLPTE-----SARERVEFRWQRVEKLRSLV 134
Query: 139 REFGAEIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATV 198
R+FG E+ID+DEL S+YDFRIDKFQRLA++AFLRGSSVVVSAPTSSGKTLIAEAAAV+TV
Sbjct: 135 RDFGVEMIDIDELISIYDFRIDKFQRLAIEAFLRGSSVVVSAPTSSGKTLIAEAAAVSTV 194
Query: 199 ARGRRIFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLY 258
A+GRR+FYTTPLKALSNQKFREFRETFGD VGLLTGDSA+NKDAQ++IMTTEILRNMLY
Sbjct: 195 AKGRRLFYTTPLKALSNQKFREFRETFGDDNVGLLTGDSAINKDAQIVIMTTEILRNMLY 254
Query: 259 QSVGNVSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCP-------KEVQLICLS 311
Q ++S V +VL+ + D GTVWEEIV + + + L+ +S
Sbjct: 255 QRSFFLTSI---FVLKSFLVLEWLLQELDFFMGTVWEEIVSFFSHCFSSFLQRLYLLSVS 311
Query: 312 --------ATVANPDELAGWIGQ--------------IHGKTELVTSSKRPVPLTWHFSM 349
+VANPDELAGWIG+ IHGKTELVTS++RPVPLTW+FS
Sbjct: 312 KLVSVFQIRSVANPDELAGWIGEQLIYCFADEVSQFLIHGKTELVTSTRRPVPLTWYFST 371
Query: 350 KNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYK-PYKDDWSRKRNARKRGYRMSYDSDESM 408
K+SLLPLLDEKG +NRKLSLNYLQL A+ + DD RKR ++KRG SY++ ++
Sbjct: 372 KHSLLPLLDEKGINVNRKLSLNYLQLSASEARFRDDDDGYRKRRSKKRGGDTSYNNLVNV 431
Query: 409 FEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFIFSRKGCDAAVQYIEDCKLL 468
+ LSKN IN IRRSQVPQI DTLWH++ ++MLPAIWFIF+R+GCDAAVQY+E+ +LL
Sbjct: 432 TDY-PLSKNEINKIRRSQVPQISDTLWHLQGKNMLPAIWFIFNRRGCDAAVQYVENFQLL 490
Query: 469 DECEKSEVELALKRFRIQYPDAVRETAVKGLLQGVAAHHAGCLPLWKAFIEELFQKGLVK 528
D+CEKSEVELALK+FR+ YPDAVRE+A KGLL+G+AAHHAGCLPLWK+FIEELFQ+GLVK
Sbjct: 491 DDCEKSEVELALKKFRVLYPDAVRESAEKGLLRGIAAHHAGCLPLWKSFIEELFQRGLVK 550
Query: 529 VVFATETLAAGINMPARTAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVL 588
VVFATETLAAGINMPARTAVISSLSK+ + R L NEL QMAGRAGRRGIDE G+ VL
Sbjct: 551 VVFATETLAAGINMPARTAVISSLSKKAGNERIELGPNELYQMAGRAGRRGIDEKGYTVL 610
Query: 589 IQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRT 648
+QT EGAEECCK++FAG++PLVSQFTASYGMVLNL++G K +S+ ++ + GR+
Sbjct: 611 VQTAFEGAEECCKLVFAGVKPLVSQFTASYGMVLNLVAGSKVTRKSSGTEAGKVLQAGRS 670
Query: 649 LEDARKLVEQSFGNYVSSNVMLAAKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQY 708
LE+A+KLVE+SFGNYVSSNV +AAK+EL + + +IE+L SEI+DEAID+KSRK LS + Y
Sbjct: 671 LEEAKKLVEKSFGNYVSSNVTVAAKQELAEIDNKIEILSSEISDEAIDKKSRKLLSARDY 730
Query: 709 KEIAELQEDLRAEKRVRTALRRQMEAKRISALKPLLDDPESGHLPFLCLQYRDSEGVLHS 768
KEI L+E+LR EKR R RR+ME +R ALKPLL E G+LPF+CL+++DSEG S
Sbjct: 731 KEITVLKEELREEKRKRAEQRRRMELERFLALKPLLKGMEEGNLPFICLEFKDSEGREQS 790
Query: 769 IPAVFLGKVDSLNASKLKDMISSVDSFALNVADAERSLPDSVLNEDLEPSYHVALGSDNS 828
+PAV+LG +DS SKL+ M+S +SFALN+ + E + D +++PSY+VALGSDNS
Sbjct: 791 VPAVYLGHIDSFQGSKLQKMMSLDESFALNLIEDELA-ADEPGKPNVKPSYYVALGSDNS 849
Query: 829 WYLFTEKWIKTVYGTGFPDTPLAQGDARPREIMSTLLDKEDMKWDKLAHSEHGGLWFMEG 888
WYLFTEKW++TVY TGFP+ LA GDA PREIM LLDK DM+WDKLA SE G LW +EG
Sbjct: 850 WYLFTEKWVRTVYRTGFPNIALALGDALPREIMKNLLDKADMQWDKLAESELGSLWRLEG 909
Query: 889 SLETWSWSLNVPVLSSFSENDELLLKSQAYRDAIEQYKDQRNKVSRLKKRISRTEGYKEY 948
SLETWSWSLNVPVLSS S+ DE+L S+ Y +A ++YK+QR+K+SRLKK++SR+EG++EY
Sbjct: 910 SLETWSWSLNVPVLSSLSDEDEVLHMSEEYDNAAQKYKEQRSKISRLKKKMSRSEGFREY 969
Query: 949 NKIIDGVKFIEEKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALDINTHVIF 1008
KI++ EK+KRLK RS+RL NR+EQIEPSGWK+FM++SNVIHE+RALDINTH+IF
Sbjct: 970 KKILENANLTVEKMKRLKARSRRLINRLEQIEPSGWKDFMRISNVIHESRALDINTHLIF 1029
Query: 1009 PLGETAAAIRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEP 1068
PLGETAAAIRGENELWLAMVLR+K LV LKPPQLA VCA LVSEGIKVRPW++N++IYEP
Sbjct: 1030 PLGETAAAIRGENELWLAMVLRNKALVDLKPPQLAGVCASLVSEGIKVRPWRDNNYIYEP 1089
Query: 1069 SATVVNCIGLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCAMD 1128
S TVV+ + L +QRS+L+ +QEKH V I CCLD QF GMVEAWASGL+W+E+MM+CAMD
Sbjct: 1090 SDTVVDMVNFLEDQRSSLIKLQEKHEVMIPCCLDVQFSGMVEAWASGLSWKEMMMECAMD 1149
Query: 1129 DGDLARLLRRTIDLLAQIPKLADIDPLLQRNARAASDVMDRPPISELAG 1177
+GDLARLLRRTIDLLAQIPKL DIDP+LQR+A AA+D+MDRPPISELAG
Sbjct: 1150 EGDLARLLRRTIDLLAQIPKLPDIDPVLQRSAAAAADIMDRPPISELAG 1198
>ref|XP_467691.1| putative helicase [Oryza sativa (japonica cultivar-group)]
gi|46390556|dbj|BAD16042.1| putative helicase [Oryza
sativa (japonica cultivar-group)]
Length = 1179
Score = 1438 bits (3723), Expect = 0.0
Identities = 748/1114 (67%), Positives = 886/1114 (79%), Gaps = 20/1114 (1%)
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKW 128
DA D+ D+DDD+E +EE EE E+++ DD + ++E EG A R EE+K
Sbjct: 81 DADVDEEYDDDDDEELDEESGGEEEEEEEGDDGVEELEEEEGGREGTAA-RRRRSEEYKS 139
Query: 129 QRVEKLCNEVREFGAEIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTL 188
QRV KL EVREFG +IID +ELA +YDF IDKFQRLA+QAFLRGSSVVVSAPTSSGKTL
Sbjct: 140 QRVGKLVAEVREFGEDIIDYNELAGIYDFPIDKFQRLAIQAFLRGSSVVVSAPTSSGKTL 199
Query: 189 IAEAAAVATVARGRRIFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIM 248
IAEAAAVATVARGRR+FYTTPLKALSNQKFR+FR TFGD VGLLTGDSA+NKDAQ+LIM
Sbjct: 200 IAEAAAVATVARGRRLFYTTPLKALSNQKFRDFRNTFGDHNVGLLTGDSAINKDAQILIM 259
Query: 249 TTEILRNMLYQSVGNVSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLI 308
TTEILRNMLYQSVG +++S L VDVIVLDEVHYLSDISRGTVWEE VIYCPKEVQLI
Sbjct: 260 TTEILRNMLYQSVG-MAASEGRLFQVDVIVLDEVHYLSDISRGTVWEETVIYCPKEVQLI 318
Query: 309 CLSATVANPDELAGWIGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKL 368
CLSATVANPDELAGWIGQIHGKTELVTS KRPVPLTWHFS K +L+PLLD KG +MNRKL
Sbjct: 319 CLSATVANPDELAGWIGQIHGKTELVTSHKRPVPLTWHFSKKFALVPLLDGKGKKMNRKL 378
Query: 369 SLNYLQ-LQAAGYKPYKDDWSRKRNARK--RGYRMSYDSDESMFEQRSLSKNNINAIRRS 425
+++ Q L + + Y RK K +G R D + +Q LSK+ + +RRS
Sbjct: 379 RMSHFQNLSSPKSEFYYVKGKRKLRTTKNEQGNRSPLD----ISKQVQLSKHELTNMRRS 434
Query: 426 QVPQIIDTLWHIEPRDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRI 485
QVP I DTL + DMLPAIWFIFSR+GCDAAV+Y+EDC+LL +CE SEVEL LKRFR+
Sbjct: 435 QVPLIRDTLSQLWENDMLPAIWFIFSRRGCDAAVEYLEDCRLLHDCEASEVELELKRFRM 494
Query: 486 QYPDAVRETAVKGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPAR 545
QYPDA+RE+AVKGLL+GVAAHHAGCLPLWK+FIEELFQ+GLVKVVFATETLAAGINMPAR
Sbjct: 495 QYPDAIRESAVKGLLRGVAAHHAGCLPLWKSFIEELFQRGLVKVVFATETLAAGINMPAR 554
Query: 546 TAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFA 605
T+VISSLSKRID+GR+ L+ NEL QMAGRAGRRGID GH VL+QT EG EECC V+FA
Sbjct: 555 TSVISSLSKRIDAGRQLLTPNELFQMAGRAGRRGIDTVGHSVLVQTTYEGPEECCDVIFA 614
Query: 606 GLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVS 665
GLEPLVSQFTASYGMVLNLL+G K H ESDD++ GRTLE+ARKLVEQSFGNYV
Sbjct: 615 GLEPLVSQFTASYGMVLNLLAGSKVTHNQKESDDIKVKRSGRTLEEARKLVEQSFGNYVG 674
Query: 666 SNVMLAAKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVR 725
SNVM+AAKEEL++ + EI+ L SEITDE+IDRK R+ LS++ Y EI+ LQ+ L+ EK++R
Sbjct: 675 SNVMVAAKEELERIQSEIQYLSSEITDESIDRKCREELSEEDYAEISLLQKKLKEEKQMR 734
Query: 726 TALRRQMEAKRISALKPLLDDPESGHLPFLCLQYRDSEGVLHSIPAVFLGKVDSLNASKL 785
L+++ME +R+ A K L++ ESGHLPF+CLQY+D + V H+IPAVF+G + S K+
Sbjct: 735 NELKKRMELERMVAWKTRLEEFESGHLPFMCLQYKDKDSVQHTIPAVFIGSLSSFADQKI 794
Query: 786 KDMISSVDSFALNVADAERSLPDSVLNEDLE--PSYHVALGSDNSWYLFTEKWIKTVYGT 843
++ + D+ + V NE + PSY+VAL SDNSWYLFTEKWIK VY T
Sbjct: 795 VSLVEN---------DSPVAGKQKVDNEGQQYYPSYYVALSSDNSWYLFTEKWIKAVYKT 845
Query: 844 GFPDTPLAQGDARPREIMSTLLDKEDMKWDKLAHSEHGGLWFMEGSLETWSWSLNVPVLS 903
G P P A+G PRE + LL +EDM WDK+A SE+G L M+GSL+TWSWSLNVPVL+
Sbjct: 846 GLPAVPSAEGGPLPRETLKQLLLREDMMWDKIAKSEYGSLLCMDGSLDTWSWSLNVPVLN 905
Query: 904 SFSENDELLLKSQAYRDAIEQYKDQRNKVSRLKKRISRTEGYKEYNKIIDGVKFIEEKIK 963
S SE+DE+ SQ ++DA+E +K QR KVS+LKK I T+G+KE+ KIID F +EKI+
Sbjct: 906 SLSEDDEVERFSQEHQDAVECHKQQRKKVSQLKKTIRSTKGFKEFQKIIDMRNFTKEKIE 965
Query: 964 RLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALDINTHVIFPLGETAAAIRGENEL 1023
RL+ RS+RLT RI QIEP+GWKEF+Q+S VI E RALDINT VI+PLGETAAAIRGENEL
Sbjct: 966 RLEARSRRLTRRIRQIEPTGWKEFLQISKVIQEARALDINTQVIYPLGETAAAIRGENEL 1025
Query: 1024 WLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCIGLLGEQR 1083
WLAMVLR+K+L+ LKP QLAA+C LVSEGIK+RPWKN+S++YEPS+ V I L EQR
Sbjct: 1026 WLAMVLRNKVLLDLKPSQLAAICGSLVSEGIKLRPWKNSSYVYEPSSVVTGVINYLEEQR 1085
Query: 1084 SALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCAMDDGDLARLLRRTIDLL 1143
++L+ +QEKH V I C +D QF GMVEAWASGLTWREIMMD AMDDGDLARLLRRTIDLL
Sbjct: 1086 NSLVDLQEKHSVKIPCEIDAQFAGMVEAWASGLTWREIMMDSAMDDGDLARLLRRTIDLL 1145
Query: 1144 AQIPKLADIDPLLQRNARAASDVMDRPPISELAG 1177
AQIPKL DIDP+LQ+NA+ A ++MDR PISELAG
Sbjct: 1146 AQIPKLPDIDPVLQKNAQIACNIMDRVPISELAG 1179
>gb|AAK62452.1| Similar to Synechocystis antiviral protein [Arabidopsis thaliana]
Length = 916
Score = 1152 bits (2981), Expect = 0.0
Identities = 600/904 (66%), Positives = 726/904 (79%), Gaps = 14/904 (1%)
Query: 24 YPSF--SLLPIPTTTLPLSLRSLSSTLKFRLSFSFNPPTSSSLPTFSDAGEDDVEDEDDD 81
+PS SL P + SL + L F+ + + P+ S L D E++ EDEDDD
Sbjct: 20 FPSLHRSLSHSPNFSFTKSLILNPNHLSFKSTLNSLSPSQSQLYEEEDDEEEEEEDEDDD 79
Query: 82 DEEEEEYE---EEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEKLCNEV 138
DE +EY+ +E DDDDD E+ ++P E ++ R EF+WQRVEKL + V
Sbjct: 80 DEAADEYDNISDEIRNSDDDDDDEETEFSVDLPTE-----SARERVEFRWQRVEKLRSLV 134
Query: 139 REFGAEIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATV 198
R+FG E+ID+DEL S+YDFRIDKFQRLA++AFLRGSSVVVSAPTSSGKTLIAEAAAV+TV
Sbjct: 135 RDFGVEMIDIDELISIYDFRIDKFQRLAIEAFLRGSSVVVSAPTSSGKTLIAEAAAVSTV 194
Query: 199 ARGRRIFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLY 258
A+GRR+FYTTPLKALSNQKFREFRETFGD VGLLTGDS +NKDAQ++IMTTEILRNMLY
Sbjct: 195 AKGRRLFYTTPLKALSNQKFREFRETFGDDNVGLLTGDSDINKDAQIVIMTTEILRNMLY 254
Query: 259 QSVGNVSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPD 318
QSVG ++SS +GL +VD IVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPD
Sbjct: 255 QSVG-MASSGTGLFHVDAIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPD 313
Query: 319 ELAGWIGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAA 378
ELAGWIG+IHGKTELVTS++RPVPLTW+FS K+SLLPLLDEKG +NRKLSLNYLQL A+
Sbjct: 314 ELAGWIGEIHGKTELVTSTRRPVPLTWYFSTKHSLLPLLDEKGINVNRKLSLNYLQLSAS 373
Query: 379 GYK-PYKDDWSRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHI 437
+ DD RKR ++KRG SY++ ++ + LSKN IN IRRSQVPQI DTLWH+
Sbjct: 374 EARFRDDDDGYRKRRSKKRGGDTSYNNLVNVTDY-PLSKNEINKIRRSQVPQISDTLWHL 432
Query: 438 EPRDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVRETAVK 497
+ ++MLPAIWFIF+R+GCDAAVQY+E+ +LLD+CEKSEVELALK+FR+ YPDAVRE+A K
Sbjct: 433 QGKNMLPAIWFIFNRRGCDAAVQYVENFQLLDDCEKSEVELALKKFRVLYPDAVRESAEK 492
Query: 498 GLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRID 557
GLL+G+AAHHAGCLPLWK+FIEELFQ+GLVKVVFATETLAAGINMPARTAVISSLSK+
Sbjct: 493 GLLRGIAAHHAGCLPLWKSFIEELFQRGLVKVVFATETLAAGINMPARTAVISSLSKKAG 552
Query: 558 SGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTAS 617
+ R L NEL QMAGRAGRRGIDE G+ VL+QT EGAEECCK++FAG++PLVSQFTAS
Sbjct: 553 NERIELGPNELYQMAGRAGRRGIDEKGYTVLVQTAFEGAEECCKLVFAGVKPLVSQFTAS 612
Query: 618 YGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELK 677
YGMVLNL++G K +S+ ++ + GR+LE+A+KLVE+SFGNYVSSNV +AAK+EL
Sbjct: 613 YGMVLNLVAGSKVTRKSSGTEAGKVLQAGRSLEEAKKLVEKSFGNYVSSNVTVAAKQELA 672
Query: 678 KNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQMEAKRI 737
+ + +IE+L SEI+DEAID+KSRK LS + YKEI L+E+LR EKR R RR+ME +R
Sbjct: 673 EIDNKIEILSSEISDEAIDKKSRKLLSARDYKEITVLKEELREEKRKRAEQRRRMELERF 732
Query: 738 SALKPLLDDPESGHLPFLCLQYRDSEGVLHSIPAVFLGKVDSLNASKLKDMISSVDSFAL 797
ALKPLL E G+LPF+CL+++DSEG S+PAV+LG +DS SKL+ M+S +SFAL
Sbjct: 733 LALKPLLKGMEEGNLPFICLEFKDSEGREQSVPAVYLGHIDSFQGSKLQKMMSLDESFAL 792
Query: 798 NVADAERSLPDSVLNEDLEPSYHVALGSDNSWYLFTEKWIKTVYGTGFPDTPLAQGDARP 857
N+ + E + D +++PSY+VALGSDNSWYLFTEKW++TVY TGFP+ LA GDA P
Sbjct: 793 NLIEDELA-ADEPGKPNVKPSYYVALGSDNSWYLFTEKWVRTVYRTGFPNIALALGDALP 851
Query: 858 REIMSTLLDKEDMKWDKLAHSEHGGLWFMEGSLETWSWSLNVPVLSSFSENDELLLKSQA 917
REIM LLDK DM+WDKLA SE G LW +EGSLETWSWSLNVPVLSS S+ DE+L S+
Sbjct: 852 REIMKNLLDKADMQWDKLAESELGSLWRLEGSLETWSWSLNVPVLSSLSDEDEVLHMSEE 911
Query: 918 YRDA 921
Y +A
Sbjct: 912 YDNA 915
>ref|NP_893584.1| putative DNA helicase [Prochlorococcus marinus subsp. pastoris str.
CCMP1986] gi|33634241|emb|CAE19926.1| putative DNA
helicase [Prochlorococcus marinus subsp. pastoris str.
CCMP1986]
Length = 908
Score = 513 bits (1321), Expect = e-143
Identities = 346/1032 (33%), Positives = 528/1032 (50%), Gaps = 157/1032 (15%)
Query: 151 LASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPL 210
L + F +D FQ A++A G+SVV++APT SGKTLI E A ++ R+FYTTPL
Sbjct: 4 LEEYFPFPLDPFQIEAIKAINSGNSVVLTAPTGSGKTLIGEFAIYRGLSHESRVFYTTPL 63
Query: 211 KALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSSRSG 270
KALSNQKFR+F FG+ VGLLTGD ++N+DA +L+MTTEI RNMLY G
Sbjct: 64 KALSNQKFRDFINQFGEKKVGLLTGDISINRDAPILVMTTEIFRNMLY---GEFEEFDDP 120
Query: 271 LVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGK 330
LVN++ ++LDE HY++D RGTVWEE +I+CP Q+I LSAT++N D+L WI ++HG
Sbjct: 121 LVNLESVILDECHYMNDPQRGTVWEETIIHCPSRAQIIALSATISNADQLQNWIEKVHGP 180
Query: 331 TELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKPYKDDWSRK 390
T LV S+KRPVPL + F L PLL+ KG G P W
Sbjct: 181 TVLVNSNKRPVPLDFIFCSAKGLHPLLNNKGN----------------GIHPNCKIWRAP 224
Query: 391 RNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFIF 450
+ +K+G + I + + P I + + R+MLPAI+FIF
Sbjct: 225 KGQKKKG--------------------KVGRIMQPKAPPIAFVVSKLAERNMLPAIYFIF 264
Query: 451 SRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVRETA-VKGLLQGVAAHHAG 509
SR+GCD AV+YI+D L+ E + + L + + +++ + L +G+A+HHAG
Sbjct: 265 SRRGCDKAVEYIKDLSLVSYSEANLISQRLDVYLKNNEEGIKDKFHCEALKRGIASHHAG 324
Query: 510 CLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRRPLSSNELL 569
LP WK +EELFQ+GL+KVVFATETLAAGINMPART VIS+LSKR D G R L S+E L
Sbjct: 325 LLPAWKELVEELFQQGLIKVVFATETLAAGINMPARTTVISTLSKRTDEGHRLLFSSEFL 384
Query: 570 QMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVLNLLSGVK 629
QM+GRAGRRG D G+VV +QT EGA+E + + PLVSQFT SYGMVLNLL
Sbjct: 385 QMSGRAGRRGKDLQGYVVTLQTRFEGAKEASSLAISEPNPLVSQFTPSYGMVLNLLQNY- 443
Query: 630 AIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYV---SSNVMLAAKEELKKNEKEIELL 686
+L+ +R+L+++SFG+++ S+ A L ++ KE++ +
Sbjct: 444 ------------------SLDKSRELIKRSFGSFLYLGESSEETAILYNLGRDLKELKKI 485
Query: 687 MSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQMEAKRISALKPLLDD 746
S I S + + +L+ L+ E+R+ L +Q K + L
Sbjct: 486 TSNI-------------SWQDFDSYEKLKSRLKEERRLLRILEKQAAEKLSEEITSALTF 532
Query: 747 PESGHLPFLCLQYRDSEGVLHSIPAVFLGKVDSLNASKLKDMIS-SVDSFALNVADAERS 805
+ G L + + + +PA+ K+ + K+K ++ ++D+ + +
Sbjct: 533 IKDGSL----ISIKAPQINRKVVPALICKKI--YESKKIKSLLCLTIDNLFILI------ 580
Query: 806 LPDSVLNEDLEPSYHVALGSDNSWYLFTEKWIKTVYGTGFPDTPLAQGDARPREIMSTLL 865
+PSY V + D + +G + +GD + + +
Sbjct: 581 ----------KPSYIVNIFPDLEEIEILRLEEPKMNFSG----EVVRGDNESQTFVDRIF 626
Query: 866 DKEDMKWDKLAHSEHGGLWFMEGSLETWSWSLNVPVLSS---FSENDELLLKSQAYRDAI 922
D + K+D L T + L+ VL+ + +E + A++
Sbjct: 627 DISE-KYD----------------LRTPQYDLSTEVLAQKKLITNLEETITNQPAHK--- 666
Query: 923 EQYKDQRNKVSRLKKRISRTEGYKEYNKIIDGVKFIEEKIKRLKTRSKRLTNRIEQIEPS 982
+ D + K+ R +KRI E +I+ +EEK E
Sbjct: 667 --FGDSK-KLKRYRKRIIEIE-----QEIVMKNNLMEEK------------------ENH 700
Query: 983 GWKEFMQVSNVIHETRALDINTHVIFPLGETAAAIRGENELWLAMVLRSKILVGLKPPQL 1042
W +F + +++ L N + +G++ AIR ENELW+ +VL S L L PP L
Sbjct: 701 NWIKFTDLIKILNHFGCL--NDLELTEVGQSVGAIRSENELWVGLVLLSGYLDDLAPPDL 758
Query: 1043 AAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCIGLLGEQRSALLAIQEKHGVTISCCLD 1102
AA+ + + + W N ++PS V++ L R + + Q K + L+
Sbjct: 759 AAIIQAICVDVRRPNLWCN----FKPSIKVIDVFNELESLRKLVASKQNKFNINTPIFLE 814
Query: 1103 TQFCGMVEAWASGLTWREIMMDCAMDDGDLARLLRRTIDLLAQIPKLADIDPLLQRNARA 1162
T+ G++ WA G W+E++ + ++D+GD+ R+LRR++D+L+QI + L+ A+
Sbjct: 815 TELTGIISEWARGKKWKELIFNTSLDEGDVVRILRRSMDVLSQIQYCVGVSNKLKNKAKL 874
Query: 1163 ASDVMDRPPISE 1174
A ++R P+SE
Sbjct: 875 ALKAINRFPVSE 886
>ref|NP_442992.1| antiviral protein [Synechocystis sp. PCC 6803]
gi|1653894|dbj|BAA18804.1| antiviral protein
[Synechocystis sp. PCC 6803] gi|7446068|pir||S76892
hypothetical protein - Synechocystis sp. (strain PCC
6803)
Length = 1006
Score = 478 bits (1229), Expect = e-133
Identities = 360/1121 (32%), Positives = 532/1121 (47%), Gaps = 224/1121 (19%)
Query: 152 ASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPLK 211
A ++ F +D FQ+ A+ A + SVVV APT SGKT++ E A +ARG+R+FYTTPLK
Sbjct: 10 ADLFAFPLDDFQQEAIAALDQDQSVVVCAPTGSGKTVVGEYAIYRAIARGKRVFYTTPLK 69
Query: 212 ALSNQKFREFRETF-------GDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQS-VGN 263
ALSNQK R+F+E VGL+TGD+ +N DA V++MTTEI RNMLY++ +G
Sbjct: 70 ALSNQKIRDFQEKLEKLGLENAAQLVGLITGDTVINADAPVVVMTTEIFRNMLYETPIGE 129
Query: 264 VSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGW 323
V +S L +V+ +V DEVHY+SD RGTVWEE +IYCP +QL+ LSAT+ NP++L W
Sbjct: 130 VGTS---LEDVETVVFDEVHYISDRGRGTVWEESIIYCPSTIQLVGLSATIGNPEQLTEW 186
Query: 324 IGQIHGKTEL-----------------VTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNR 366
I Q+ L V S RPVPLT+H+S + L PLL+EK T +N
Sbjct: 187 INQVRTGVSLKVLRQGEQRNDSPPCVLVNSDFRPVPLTFHYSTRKGLFPLLNEKKTGVNT 246
Query: 367 KLSLNYLQLQAAGYKPYKDDWSRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAI---- 422
+L L AG K R+ + R R D + + L + N+ +
Sbjct: 247 RL------LPKAGNK-------NTRSGKNRRMRRE-DCPYPLLVMQQLQERNLLPVIYVI 292
Query: 423 --RRS--QVPQIIDTLWHIEPRDM----LPAIWFIFSRKGC-----------------DA 457
RR Q Q ++ L +EP + L + F F + A
Sbjct: 293 FSRRGCEQAAQSLEDLSLVEPAEQESIQLQLLDFFFGKNPLLRQKLERDLANGLPDLVQA 352
Query: 458 AVQYIEDCKLLDECEKSEVELALK---RFRI-----QYPDAVRETAVKGLLQGVAAHHAG 509
V Y++ + + ++ LA+ F++ Q R ++ LL+G+A HHAG
Sbjct: 353 VVAYLDHPQDSEAAQRLLTALAIDPEGMFKLWGWIAQSSPMTRFEQIEPLLRGIAVHHAG 412
Query: 510 CLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRRPLSSNELL 569
LP K +E+LF++GL+KVVFAT TL+AGINMPART VIS+LSKR + G LS +E L
Sbjct: 413 ILPDMKTLVEKLFEQGLIKVVFATATLSAGINMPARTTVISALSKRTNEGHAMLSPSEFL 472
Query: 570 QMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVLNLLSGVK 629
Q+AGRAGRRG+D GHV+ +QTP EGA E + A EPL S FT SYGMVLNLL
Sbjct: 473 QIAGRAGRRGMDTEGHVITVQTPFEGANEASYLALAEAEPLKSWFTPSYGMVLNLLQ--- 529
Query: 630 AIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKKNEKEIELLMSE 689
LE+ + L+E+SF Y++ + K + + K++ L +
Sbjct: 530 ----------------KHDLEEVKSLLERSFAEYLAQFALEPTKVAIAETVKKLSQLDIK 573
Query: 690 ITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQMEAKRISALKPLLDDPES 749
+ + +K + + + LR E+R+ L Q E +R LK L
Sbjct: 574 LA----------GIGEKDLRSYEKFRGRLREEQRLLKILEEQAEKERKQQLKDQLKTLAP 623
Query: 750 GHLPFLCLQYRDSEGVLHSIPAVFLGKVDSLNASKLKDMISSVDSFALNVADAERSLPDS 809
G L +L G+ ++ +L +
Sbjct: 624 GQLLYL------------------KGRHVKIHQPRL-----------------------A 642
Query: 810 VLNEDLEPSYHV----ALGSDNSWYLFTEKWIKTVYGTGFPDTPLAQGDARPREIMSTLL 865
V+ L P++++ LGSDN WY T + +Y + AQ L
Sbjct: 643 VIVAPLAPAHNLPQWCCLGSDNRWYHVT---VGDIYAVPIGNLSPAQWQHLTPPPAELLA 699
Query: 866 DKEDMKWDKLAHSEHGGLWFMEGSLETWSWSLNVPVLSSFSENDELLLKSQAYRDAIEQY 925
+ +K D+ ET + + + + E L++ QA D +E+
Sbjct: 700 PGKSVKGDE----------------ETLAIAAGLDPQTYPLEPSPELVEQQARVDHVEKL 743
Query: 926 KDQRNKVSRLKKRISRTEGYKEYNKIIDGVKFIEEKIKRLKTRSKRLTNRIEQIEPSGWK 985
S+ K K+++ +E K L R + +R++ + W+
Sbjct: 744 LAVHPLASQ-----------KHPGKLLEQFHQRQELRKTLSKRQQEY-DRLQSRQSYYWQ 791
Query: 986 EFMQVSNVIHETRALDINTHVIFPLGETAAAIRGENELWLAMVLRSKILVGLKPPQLAAV 1045
EF+ + +++ E AL+ T + LGE AA +RGENELWL + L S L+P QLAA
Sbjct: 792 EFLDLIDILQEMEALEEYTPTL--LGEVAATLRGENELWLGLALISGKFNDLEPEQLAAA 849
Query: 1046 CAGLVSEGIKVRPWKNNSFIYEPSATVV-------NCIGL-------------------- 1078
+ L++E + W + ++PS V+ + GL
Sbjct: 850 ASALITETPRSDAWTD----FKPSPVVLAALRPSNDLFGLLFCLKHPQPPLALWQAMTVA 905
Query: 1079 -------LGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCAMDDGD 1131
L + R L+ Q + + I L+ F G+VE WA G+ W + ++D+GD
Sbjct: 906 DHLQRLNLWDLRRKLIKAQNQRAIAIPLWLEVDFMGLVEQWALGMEWENLCRQTSLDEGD 965
Query: 1132 LARLLRRTIDLLAQIPKLADIDPLLQRNARAASDVMDRPPI 1172
L RL RRT+DLL QIP++ + P L+RNAR A M R P+
Sbjct: 966 LVRLFRRTVDLLWQIPQVPHLSPRLKRNARIAVQQMKRFPL 1006
>dbj|BAC74406.1| putative ATP-dependent RNA helicase [Streptomyces avermitilis
MA-4680] gi|29833237|ref|NP_827871.1| putative
ATP-dependent RNA helicase [Streptomyces avermitilis
MA-4680]
Length = 937
Score = 335 bits (860), Expect = 4e-90
Identities = 213/583 (36%), Positives = 302/583 (51%), Gaps = 52/583 (8%)
Query: 154 VYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPLKAL 213
+YDF +D FQ A QA G V+V+APT SGKT++ E A + +G++ FYTTP+KAL
Sbjct: 31 MYDFGLDPFQIEACQALEAGKGVLVAAPTGSGKTIVGEFAVHLALQQGKKCFYTTPIKAL 90
Query: 214 SNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSSRSGLVN 273
SNQK+ + +G VGLLTGD++VN DA V++MTTE+LRNMLY + L+
Sbjct: 91 SNQKYADLARRYGADKVGLLTGDNSVNSDAPVVVMTTEVLRNMLY-------AGSQTLLG 143
Query: 274 VDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGKTEL 333
+ +V+DEVHYLSD RG VWEE++I+ P+ V L+ LSATV+N +E W+ + G T++
Sbjct: 144 LGYVVMDEVHYLSDRFRGAVWEEVIIHLPESVTLVSLSATVSNAEEFGDWLDTVRGDTQV 203
Query: 334 VTSSKRPVPLTWHFSMKNSLLPLLDE-KGTRMNRKLSLNYLQLQAAGYKPYKDDWSRKRN 392
+ S RPVPL H + L +E +G + L L A Y+D
Sbjct: 204 IVSEHRPVPLFQHVLAGRRMYDLFEEGEGHKKAVNPDLTRLARMEASRPSYQD------- 256
Query: 393 ARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFIFSR 452
RKRG M E QRS R P + + ++ +LPAI FIFSR
Sbjct: 257 -RKRGRAMREADRERERRQRS----------RVWTPGRPEVIERLDAEGLLPAITFIFSR 305
Query: 453 KGCDAAVQ--YIEDCKLLDECEKSEVELALKRFRIQYPDAVRETA-----VKGLLQGVAA 505
C+AAVQ +L DE + +V ++ P ++GL +G+AA
Sbjct: 306 AACEAAVQQCLYAGLRLNDEAARDKVRALVEERTASIPTEDLHVLGYYEWLEGLERGIAA 365
Query: 506 HHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRRPLSS 565
HHAG LP +K +EELF +GLVK VFATETLA GINMPAR+ V+ L K ++
Sbjct: 366 HHAGMLPTFKEVVEELFVRGLVKAVFATETLALGINMPARSVVLEKLVKWNGEQHADITP 425
Query: 566 NELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVLNLL 625
E Q+ GRAGRRGID GH V++ E + PL S F SY M +NL+
Sbjct: 426 GEYTQLTGRAGRRGIDVEGHAVVLWQRGFSPEHLAGLAGTRTYPLRSSFKPSYNMAVNLV 485
Query: 626 SGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKKNEKEIEL 685
HRS R+L+E SF + + ++ ++++NE+ +E
Sbjct: 486 DQF-GRHRS------------------RELLETSFAQFQADKSVVGISRQVQRNEEGLEG 526
Query: 686 LMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTAL 728
+T D + L ++ EL + A++R A+
Sbjct: 527 YKESMTCHLGDFEEYARLRRELKDRETELAKQGAAQRRAEAAV 569
Score = 52.4 bits (124), Expect = 9e-05
Identities = 58/221 (26%), Positives = 95/221 (42%), Gaps = 19/221 (8%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHET---RALDINTHVIFPLGETAAA 1016
E+ RL + +L RIE + + F ++ ++ E R ++ H G+ A
Sbjct: 716 ERYYRLMRDTSQLERRIEGRTNTIARTFDRIVALLTELDYLRGDEVTAH-----GKRLAR 770
Query: 1017 IRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCI 1076
+ GE +L + LR+ + GL P +LAA + LV E R + PS +
Sbjct: 771 LYGELDLLASECLRAGVWEGLDPAELAACVSALVYES---RVGDDAMAPKLPSGKAKAAL 827
Query: 1077 GLLGEQRSALLAIQEKHGVTISCCL-----DTQFCGMVEAWASGLTWREIMMDCAMDDGD 1131
G + L A++E +T + + D F WASG E++ + M GD
Sbjct: 828 GEMVRIWGRLDALEEDFRITQTEGVGQREPDLGFAWAAYMWASGKGLDEVLREAEMPAGD 887
Query: 1132 LARLLRRTIDLLAQIPKLADI---DPLLQRNARAASDVMDR 1169
R ++ ID+L QI A + + +NAR A D + R
Sbjct: 888 FVRWCKQVIDVLGQISAAAPVRGEGSTVAKNARKAVDELLR 928
>ref|NP_960762.1| HelY [Mycobacterium avium subsp. paratuberculosis str. k10]
gi|41396280|gb|AAS04145.1| HelY [Mycobacterium avium
subsp. paratuberculosis str. k10]
Length = 821
Score = 335 bits (858), Expect = 7e-90
Identities = 225/620 (36%), Positives = 317/620 (50%), Gaps = 75/620 (12%)
Query: 144 EIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRR 203
E++++ +S F +D FQR A A RG V+V APT +GKT++ E A +A G +
Sbjct: 13 ELVELTRFSSELPFALDGFQRRACAALERGHGVLVCAPTGAGKTVVGEFAVHLALAAGGK 72
Query: 204 IFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGN 263
FYTTPLKALSNQK + +G +GLLTGD +VN DA V++MTTE+LRNMLY
Sbjct: 73 CFYTTPLKALSNQKHTDLTARYGRDRIGLLTGDMSVNADAPVVVMTTEVLRNMLY----- 127
Query: 264 VSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGW 323
+ L + +V+DEVH+L+D RG VWEE++++ P EV+++ LSATV+N +E GW
Sbjct: 128 --ADSPALQGLSYVVMDEVHFLADRMRGPVWEEVILHLPDEVRVVSLSATVSNAEEFGGW 185
Query: 324 IGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKP- 382
I + G T +V RPVPL H + L L D + + AG +P
Sbjct: 186 IQTVRGDTTVVVDEHRPVPLWQHVLVGKRLFDLFDYRNAEAPGQ--------PGAGREPR 237
Query: 383 YKDDWSRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDM 442
D R R+ R+S D R R + P D + ++ +
Sbjct: 238 VNPDLLRHIAHRREADRLS-DWQPRRGAGRGRPPARAGRPRFYRTPGRPDVIATLDAEGL 296
Query: 443 LPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVR---------E 493
LPAI F+FSR GCDAAVQ +C +S ++L + R+Q + + +
Sbjct: 297 LPAITFVFSRAGCDAAVQ---------QCLRSPLQLTTQEERVQIAEVIEHRCGDLADAD 347
Query: 494 TAV-------KGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPART 546
AV +GLL+G+AAHHAG LP ++ +EELF GLVK VFATETLA GINMPART
Sbjct: 348 LAVLGYYEWREGLLRGLAAHHAGMLPAFRHTVEELFTAGLVKAVFATETLALGINMPART 407
Query: 547 AVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAG 606
V+ L K L+ E Q+ GRAGRRGID GH V++ P E E V AG
Sbjct: 408 VVLERLVKFNGEQHVALTPGEYTQLTGRAGRRGIDVEGHAVVLWNPTEETTEPSAV--AG 465
Query: 607 LE-----PLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFG 661
L PL S F SY M +NL+ Q G E A +L+EQSF
Sbjct: 466 LASTRTFPLRSSFAPSYNMTINLV----------------QQMGP---EQAHRLLEQSFA 506
Query: 662 NYVSSNVMLAAKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAE 721
Y + ++ +++ + ++ + +E+ KA + + A + E RA+
Sbjct: 507 QYQADRSVVGLVRGIERGQAMLDEIAAEL-------GGPKAPILEYARMRARISEMERAQ 559
Query: 722 KRVRTALRRQMEAKRISALK 741
R RRQ + ++AL+
Sbjct: 560 TRASRLHRRQAASDALAALR 579
>emb|CAB59484.1| putative helicase [Streptomyces coelicolor A3(2)]
gi|21220127|ref|NP_625906.1| putative helicase
[Streptomyces coelicolor A3(2)]
Length = 950
Score = 330 bits (846), Expect = 2e-88
Identities = 219/604 (36%), Positives = 309/604 (50%), Gaps = 73/604 (12%)
Query: 154 VYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPLKAL 213
+YDF +D FQ A QA G V+V+APT SGKT++ E A + +GR+ FYTTP+KAL
Sbjct: 46 MYDFGLDPFQIEACQALEAGKGVLVAAPTGSGKTIVGEFAVHLALQQGRKCFYTTPIKAL 105
Query: 214 SNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSSRSGLVN 273
SNQK+ + +G VGLLTGD++VN +A V++MTTE+LRNMLY + L+
Sbjct: 106 SNQKYADLCRRYGTDKVGLLTGDNSVNSEAPVVVMTTEVLRNMLY-------AGSQTLLG 158
Query: 274 VDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGKTEL 333
+ +V+DEVHYLSD RG VWEE++I+ P+ V L+ LSATV+N +E W+ + G T++
Sbjct: 159 LGHVVMDEVHYLSDRFRGAVWEEVIIHLPESVTLVSLSATVSNAEEFGDWLDTVRGDTQV 218
Query: 334 VTSSKRPVPLTWHFSMKNSLLPLLDE-KGTRMNRKLSLNYLQLQAAGYKPYKDDWSRKRN 392
+ S RPVPL H + L +E +G + L + A Y+D
Sbjct: 219 IVSEHRPVPLFQHVLAGRRMYDLFEEAEGHKKAVNPDLTRMARLEASRPSYQD------- 271
Query: 393 ARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFIFSR 452
R+RG M E QRS R P + + ++ +LPAI FIFSR
Sbjct: 272 -RRRGRAMKEADRERERRQRS----------RVWTPSRPEVIERLDSEGLLPAITFIFSR 320
Query: 453 KGCDAAVQ--YIEDCKLLDECEKSEVELALKRFRIQYPDAVRETA--------VKGLLQG 502
GC+AAVQ +L DE + V ++ P RE ++GL +G
Sbjct: 321 AGCEAAVQQCLYAGLRLNDEGARERVRALVEERTSSIP---REDLHVLGYYEWLEGLERG 377
Query: 503 VAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRRP 562
+AAHHAG LP +K +EELF +GLVK VFATETLA GINMPAR+ V+ L K
Sbjct: 378 IAAHHAGMLPTFKEVVEELFVRGLVKAVFATETLALGINMPARSVVLEKLVKWNGEQHAD 437
Query: 563 LSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVL 622
++ E Q+ GRAGRRGID GH V++ E + PL S F SY M +
Sbjct: 438 ITPGEFTQLTGRAGRRGIDVEGHAVVLWQRAMNPEHLAGLAGTRTYPLRSSFKPSYNMAV 497
Query: 623 NLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKKNEKE 682
NL+ HRS R+L+E SF + + ++ ++++NE+
Sbjct: 498 NLVDQF-GRHRS------------------RELLETSFAQFQADKSVVGISRQVQRNEEG 538
Query: 683 IELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEK----RVRTALRRQMEAKRIS 738
+E + +T D + E A L+ +L+ + R RR A +
Sbjct: 539 LEGYKASMTCHLGD-----------FDEYARLRRELKDREQELARQGANQRRAEAAVALE 587
Query: 739 ALKP 742
LKP
Sbjct: 588 KLKP 591
Score = 49.3 bits (116), Expect = 8e-04
Identities = 57/219 (26%), Positives = 93/219 (42%), Gaps = 17/219 (7%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHET---RALDINTHVIFPLGETAAA 1016
E+ RL + +L RIE + + F ++ ++ E R ++ H G+ A
Sbjct: 731 ERYHRLLRDTSQLERRIEGRTNTIARTFDRIVALLTELDYLRGDEVTEH-----GKRLAR 785
Query: 1017 IRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCI 1076
+ GE +L + LR + GL P +LAA + LV E R + + PS +
Sbjct: 786 LYGELDLLASECLREGVWEGLSPAELAACVSALVFES---RAADDATAPKVPSGRAKAAL 842
Query: 1077 GLLGEQRSALLAIQEKHGVTISCCL-----DTQFCGMVEAWASGLTWREIMMDCAMDDGD 1131
G L A++E ++ + + D F WASG E++ + M GD
Sbjct: 843 GETVRIWGRLDALEEDFRISQTEGVGQREPDLGFAWAAYMWASGKGLDEVLREVEMPAGD 902
Query: 1132 LARLLRRTIDLLAQIPKLA-DIDPLLQRNARAASDVMDR 1169
R ++ ID+L QI A + +NAR A D + R
Sbjct: 903 FVRWCKQVIDVLGQISAAAPGAGSTVPKNARKAVDELLR 941
>ref|YP_061841.1| ATP-dependent RNA helicase [Leifsonia xyli subsp. xyli str. CTCB07]
gi|50951035|gb|AAT88736.1| ATP-dependent RNA helicase
[Leifsonia xyli subsp. xyli str. CTCB07]
Length = 811
Score = 326 bits (836), Expect = 2e-87
Identities = 210/591 (35%), Positives = 323/591 (54%), Gaps = 63/591 (10%)
Query: 157 FRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGR-RIFYTTPLKALSN 215
F +D FQR A G SV+V+APT +GKT++AE A + + ++FYTTP+KALSN
Sbjct: 34 FDLDPFQREACTCLENGRSVLVAAPTGAGKTIVAEFAVFLAMRQANAKVFYTTPMKALSN 93
Query: 216 QKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSSRSGLVNVD 275
QKF+EF++T+G VGLLTGD+ +N A++++MTTE+LRNMLY + L ++
Sbjct: 94 QKFQEFQDTYGPESVGLLTGDTNINSHARIVVMTTEVLRNMLY-------ADSDLLGDLA 146
Query: 276 VIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGKTELVT 335
+V+DEVHYL+D RG VWEE++I+ P V+++ LSATV+N +E W+ + G T++V
Sbjct: 147 YVVMDEVHYLADRFRGAVWEEVIIHLPPAVRMVSLSATVSNAEEFGDWLQAVRGDTDVVV 206
Query: 336 SSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKPYKDDWSRKRNARK 395
S +RPVPL H M++ L+ L D G +++ +Q+ +G R ++R+
Sbjct: 207 SEERPVPLEQHILMRSKLIDLFDSSGLAAANRVNPELVQMARSG--------GRVLSSRQ 258
Query: 396 RGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFIFSRKGC 455
R D + R ++ + R+++ +++D ++LPAI+F+FSR GC
Sbjct: 259 R-------RDIGRYHSRGGRPDSFR-MNRAEIVRLLDE------HNLLPAIFFLFSRNGC 304
Query: 456 DAAVQYI--EDCKLLDECEKSEVE-LALKRFRIQYPDAVRETA----VKGLLQGVAAHHA 508
DAAV+ +L ++ E+ ++ + +R R + + ++GL GVAAHHA
Sbjct: 305 DAAVRQTLRAGVRLTEQRERDDIRSIVEERCRTLMDEDLAVLGYWEWLEGLEHGVAAHHA 364
Query: 509 GCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRRPLSSNEL 568
G LP +K +EELF++ LVKVVFATETLA GINMPART V+ L K R P++ E
Sbjct: 365 GMLPAFKEVVEELFRRKLVKVVFATETLALGINMPARTVVLEKLEKFNGESRVPITPGEY 424
Query: 569 LQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVLNLLSGV 628
Q+ GRAGRRGID G+ V+ + + PL S F +Y M +NL+
Sbjct: 425 TQLTGRAGRRGIDVEGNSVIQWEDGLDPQSVASLASRRSYPLNSSFRPTYNMAVNLIDQF 484
Query: 629 KAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSN--VMLAAK-----EELKKNEK 681
RQ T R+++E SF + + V LA K E L EK
Sbjct: 485 G-----------RQRT--------REILESSFAQFQADRAVVDLARKVRQQEESLAGYEK 525
Query: 682 EIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQM 732
+ + + + + R+ L +K + + + D +R T LR++M
Sbjct: 526 AMTCHLGDFREYSGVRRELTDLERKGQRLDSASRADRDRRQRQLTELRKRM 576
>ref|NP_855768.1| PROBABLE ATP-DEPENDENT DNA HELICASE HELY [Mycobacterium bovis
AF2122/97] gi|31618867|emb|CAD96972.1| PROBABLE
ATP-DEPENDENT DNA HELICASE HELY [Mycobacterium bovis
AF2122/97]
Length = 906
Score = 323 bits (827), Expect = 3e-86
Identities = 214/618 (34%), Positives = 310/618 (49%), Gaps = 88/618 (14%)
Query: 144 EIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRR 203
E+ ++D + F +D FQ+ A A RG V+V APT +GKT++ E A +A G +
Sbjct: 3 ELAELDRFTAELPFSLDDFQQRACSALERGHGVLVCAPTGAGKTVVGEFAVHLALAAGSK 62
Query: 204 IFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGN 263
FYTTPLKALSNQK + +G +GLLTGD +VN +A V++MTTE+LRNMLY
Sbjct: 63 CFYTTPLKALSNQKHTDLTARYGRDQIGLLTGDLSVNGNAPVVVMTTEVLRNMLY----- 117
Query: 264 VSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGW 323
+ L + +V+DEVH+L+D RG VWEE+++ P +V+++ LSATV+N +E GW
Sbjct: 118 --ADSPALQGLSYVVMDEVHFLADRMRGPVWEEVILQLPDDVRVVSLSATVSNAEEFGGW 175
Query: 324 IGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKPY 383
I + G T +V RPVPL H + + L D + + +N L+ ++
Sbjct: 176 IQMVRGDTTVVVDEHRPVPLWQHVLVGKRMFDLFDYRIGEAEGQPQVNRELLRHIAHRRE 235
Query: 384 KD---DWS-RKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEP 439
D DW R+R + + G+ R P++I L +
Sbjct: 236 ADRMADWQPRRRGSGRPGF-----------------------YRPPGRPEVIAKL---DA 269
Query: 440 RDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAV-------- 491
+LPAI F+FSR GCDAAV +C +S + L + R + + +
Sbjct: 270 EGLLPAITFVFSRAGCDAAVT---------QCLRSPLRLTSEEERARIAEVIDHRCGDLA 320
Query: 492 -RETAV-------KGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMP 543
+ AV +GLL+G+AAHHAG LP ++ +EELF GLVK VFATETLA GINMP
Sbjct: 321 DSDLAVLGYYEWREGLLRGLAAHHAGMLPAFRHTVEELFTAGLVKAVFATETLALGINMP 380
Query: 544 ARTAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVL 603
ART V+ L K PL+ E Q+ GRAGRRGID GH V+I P E +
Sbjct: 381 ARTVVLERLVKFNGEQHMPLTPGEYTQLTGRAGRRGIDVEGHAVVIWHPEIEPSEVAGLA 440
Query: 604 FAGLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNY 663
PL S F SY M +NL +HR + A +L+EQSF Y
Sbjct: 441 STRTFPLRSSFAPSYNMTINL------VHRMGP-------------QQAHRLLEQSFAQY 481
Query: 664 VSSNVMLAAKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKR 723
+ ++ +++ + ++ EI E + + ++EL+ RA+ R
Sbjct: 482 QADRSVVGLVRGIERGNR----ILGEIAAELGGSDAPILEYARLRARVSELE---RAQAR 534
Query: 724 VRTALRRQMEAKRISALK 741
RRQ ++AL+
Sbjct: 535 ASRLQRRQAATDALAALR 552
Score = 39.7 bits (91), Expect = 0.61
Identities = 50/217 (23%), Positives = 90/217 (41%), Gaps = 11/217 (5%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALD--INTHVIFPLGETAAAI 1017
E+ R++ + +L ++ S + F + ++ E +D V+ G A I
Sbjct: 681 ERYLRIERDNAQLERKVAAATNSLARTFDRFVGLLTEREFIDGPATDPVVTDDGRLLARI 740
Query: 1018 RGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYE-PSATVVNCI 1076
E++L +A LR+ GLKP +LA V + +V E + + F + P+ + +
Sbjct: 741 YSESDLLVAECLRTGAWEGLKPAELAGVVSAVVYE-TRGGDGQGAPFGADVPTPRLRQAL 799
Query: 1077 GLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAW------ASGLTWREIM-MDCAMDD 1129
+ L A ++ H +T S D F ++ W A+ L ++ +
Sbjct: 800 TQTSRLSTTLRADEQAHRITPSREPDDGFVRVIYRWSRTGDLAAALAAADVNGSGSPLLA 859
Query: 1130 GDLARLLRRTIDLLAQIPKLADIDPLLQRNARAASDV 1166
GD R R+ +DLL Q+ A L RA D+
Sbjct: 860 GDFVRWCRQVLDLLDQVRNAAPNPELRATAKRAIGDI 896
>ref|NP_216608.1| PROBABLE ATP-DEPENDENT DNA HELICASE HELY [Mycobacterium
tuberculosis H37Rv] gi|1370259|emb|CAA98204.1| PROBABLE
ATP-DEPENDENT DNA HELICASE HELY [Mycobacterium
tuberculosis H37Rv] gi|13881831|gb|AAK46434.1|
ATP-dependent RNA helicase, DEAD/DEAH box family
[Mycobacterium tuberculosis CDC1551]
gi|1731349|sp|Q10701|HELY_MYCTU Probable helicase helY
gi|15841583|ref|NP_336620.1| ATP-dependent RNA helicase,
DEAD/DEAH box family [Mycobacterium tuberculosis
CDC1551]
Length = 906
Score = 322 bits (826), Expect = 4e-86
Identities = 214/618 (34%), Positives = 310/618 (49%), Gaps = 88/618 (14%)
Query: 144 EIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRR 203
E+ ++D + F +D FQ+ A A RG V+V APT +GKT++ E A +A G +
Sbjct: 3 ELAELDRFTAELPFSLDDFQQRACSALERGHGVLVCAPTGAGKTVVGEFAVHLALAAGSK 62
Query: 204 IFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGN 263
FYTTPLKALSNQK + +G +GLLTGD +VN +A V++MTTE+LRNMLY
Sbjct: 63 CFYTTPLKALSNQKHTDLTARYGRDQIGLLTGDLSVNGNAPVVVMTTEVLRNMLY----- 117
Query: 264 VSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGW 323
+ L + +V+DEVH+L+D RG VWEE+++ P +V+++ LSATV+N +E GW
Sbjct: 118 --ADSPALQGLSYVVMDEVHFLADRMRGPVWEEVILQLPDDVRVVSLSATVSNAEEFGGW 175
Query: 324 IGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKPY 383
I + G T +V RPVPL H + + L D + + +N L+ ++
Sbjct: 176 IQTVRGDTTVVVDEHRPVPLWQHVLVGKRMFDLFDYRIGEAEGQPQVNRELLRHIAHRRE 235
Query: 384 KD---DWS-RKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEP 439
D DW R+R + + G+ R P++I L +
Sbjct: 236 ADRMADWQPRRRGSGRPGF-----------------------YRPPGRPEVIAKL---DA 269
Query: 440 RDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAV-------- 491
+LPAI F+FSR GCDAAV +C +S + L + R + + +
Sbjct: 270 EGLLPAITFVFSRAGCDAAVT---------QCLRSPLRLTSEEERARIAEVIDHRCGDLA 320
Query: 492 -RETAV-------KGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMP 543
+ AV +GLL+G+AAHHAG LP ++ +EELF GLVK VFATETLA GINMP
Sbjct: 321 DSDLAVLGYYEWREGLLRGLAAHHAGMLPAFRHTVEELFTAGLVKAVFATETLALGINMP 380
Query: 544 ARTAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVL 603
ART V+ L K PL+ E Q+ GRAGRRGID GH V+I P E +
Sbjct: 381 ARTVVLERLVKFNGEQHMPLTPGEYTQLTGRAGRRGIDVEGHAVVIWHPEIEPSEVAGLA 440
Query: 604 FAGLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNY 663
PL S F SY M +NL +HR + A +L+EQSF Y
Sbjct: 441 STRTFPLRSSFAPSYNMTINL------VHRMGP-------------QQAHRLLEQSFAQY 481
Query: 664 VSSNVMLAAKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKR 723
+ ++ +++ + ++ EI E + + ++EL+ RA+ R
Sbjct: 482 QADRSVVGLVRGIERGNR----ILGEIAAELGGSDAPILEYARLRARVSELE---RAQAR 534
Query: 724 VRTALRRQMEAKRISALK 741
RRQ ++AL+
Sbjct: 535 ASRLQRRQAATDALAALR 552
Score = 39.7 bits (91), Expect = 0.61
Identities = 50/217 (23%), Positives = 90/217 (41%), Gaps = 11/217 (5%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALD--INTHVIFPLGETAAAI 1017
E+ R++ + +L ++ S + F + ++ E +D V+ G A I
Sbjct: 681 ERYLRIERDNAQLERKVAAATNSLARTFDRFVGLLTEREFIDGPATDPVVTDDGRLLARI 740
Query: 1018 RGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYE-PSATVVNCI 1076
E++L +A LR+ GLKP +LA V + +V E + + F + P+ + +
Sbjct: 741 YSESDLLVAECLRTGAWEGLKPAELAGVVSAVVYE-TRGGDGQGAPFGADVPTPRLRQAL 799
Query: 1077 GLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAW------ASGLTWREIM-MDCAMDD 1129
+ L A ++ H +T S D F ++ W A+ L ++ +
Sbjct: 800 TQTSRLSTTLRADEQAHRITPSREPDDGFVRVIYRWSRTGDLAAALAAADVNGSGSPLLA 859
Query: 1130 GDLARLLRRTIDLLAQIPKLADIDPLLQRNARAASDV 1166
GD R R+ +DLL Q+ A L RA D+
Sbjct: 860 GDFVRWCRQVLDLLDQVRNAAPNPELRATAKRAIGDI 896
>ref|NP_301957.1| probable helicase, Ski2 subfamily [Mycobacterium leprae TN]
gi|13093245|emb|CAC31714.1| probable helicase, Ski2
subfamily [Mycobacterium leprae]
gi|4200282|emb|CAA22943.1| putative helicase
[Mycobacterium leprae] gi|18203674|sp|Q9ZBD8|HELY_MYCLE
Probable helicase helY
Length = 920
Score = 319 bits (818), Expect = 3e-85
Identities = 223/615 (36%), Positives = 315/615 (50%), Gaps = 89/615 (14%)
Query: 147 DVDELA---SVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRR 203
D+ ELA + F +D FQ+ A A RG V+V APT +GKT++ E A +A G +
Sbjct: 3 DLAELARFTAELPFSLDDFQQRACAALERGHGVLVCAPTGAGKTVVGEFAVHLALAAGGK 62
Query: 204 IFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGN 263
FYTTPLKALSNQK+ + +G + +GLLTGD +VN D+ V++MTTE+LRNMLY
Sbjct: 63 CFYTTPLKALSNQKYTDLTARYGRNRIGLLTGDQSVNGDSPVVVMTTEVLRNMLYAD--- 119
Query: 264 VSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGW 323
S + GL +V V+DEVH+++D RG VWEE++++ P +V+++ LSATV+N +E GW
Sbjct: 120 -SFALQGLSHV---VMDEVHFIADRMRGPVWEEVILHLPDDVRMVSLSATVSNAEEFGGW 175
Query: 324 IGQIHGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQ-LQAAGYKP 382
+ + G T +V RPVPL H + L L D ++ N L+ +
Sbjct: 176 VQTVRGDTTVVVDEHRPVPLWQHVLVGKRLFDLFDYDSNVDQSPVNPNLLRHIAHCREAD 235
Query: 383 YKDDW--SRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPR 440
DW R+R R G R + RSL++ + AI ++
Sbjct: 236 RMSDWRNPRRRAGRGSGVRPRF--------YRSLARPEVIAI--------------LDAE 273
Query: 441 DMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAV--------- 491
+LPAI F+FSR GCDAAVQ +C +S + L + R Q + +
Sbjct: 274 GLLPAITFVFSRFGCDAAVQ---------QCLRSPLRLTSEEERAQIAEVIDHRCGDLAD 324
Query: 492 RETAV-------KGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPA 544
+ AV +GLL+G+AAHHAG LP ++ +EELF GLVK VFATETLA GINMPA
Sbjct: 325 ADLAVLGYYEWREGLLRGLAAHHAGMLPAFRHAVEELFTAGLVKAVFATETLALGINMPA 384
Query: 545 RTAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNE---GAEECCK 601
RT V+ L K PL+ E Q+ GRAGRRGID GH V+I P+E G
Sbjct: 385 RTVVLERLVKFNGKQHVPLTPGEYTQLTGRAGRRGIDVEGHAVVIWHPSEDTSGPSAVAG 444
Query: 602 VLFAGLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFG 661
+ A PL S F SY M +NL+ + E A L+EQSF
Sbjct: 445 LASARTFPLRSSFVPSYNMTINLVHWMSP-------------------ERAHALLEQSFA 485
Query: 662 NYVSSNVMLAAKEELKKNEKEIELLMSEITD------EAIDRKSRKALSQKQYKEIAELQ 715
Y + ++ +++ + + + SE+ E ++R A ++ + LQ
Sbjct: 486 QYQADRSVVGLVRGIERCTQVLGDISSELGGPDAPILEYARLRARIAEMERAQSFVFRLQ 545
Query: 716 EDLRAEKRVRTALRR 730
+A V ALRR
Sbjct: 546 RK-QAANDVLAALRR 559
Score = 44.3 bits (103), Expect = 0.025
Identities = 40/163 (24%), Positives = 67/163 (40%), Gaps = 7/163 (4%)
Query: 1011 GETAAAIRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSA 1070
G A I E++L +A LR+ GL+P +LAAV + ++ E P+
Sbjct: 748 GRLLARIYSESDLLVAECLRTGAWAGLRPAELAAVVSAVLYETRGDDGPGGPVDAEAPTP 807
Query: 1071 TVVNCIGLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMM------- 1123
+ + + L A +++H + +S D F G++ WA ++
Sbjct: 808 RLRQALQHTSRLSATLRADEQRHRIALSREPDDGFVGVIYCWARTGDLAAVLAAADASGN 867
Query: 1124 DCAMDDGDLARLLRRTIDLLAQIPKLADIDPLLQRNARAASDV 1166
+ GD R R+ +DLL Q+ A L RA +DV
Sbjct: 868 GAPLSAGDFVRWCRQVLDLLDQLRNAAPEPDLRATAKRAINDV 910
>ref|ZP_00414814.1| Helicase, C-terminal:Type III restriction enzyme, res
subunit:DEAD/DEAH box helicase, N-terminal [Arthrobacter
sp. FB24] gi|66866940|gb|EAL94334.1| Helicase,
C-terminal:Type III restriction enzyme, res
subunit:DEAD/DEAH box helicase, N-terminal [Arthrobacter
sp. FB24]
Length = 964
Score = 317 bits (813), Expect = 1e-84
Identities = 225/691 (32%), Positives = 336/691 (48%), Gaps = 71/691 (10%)
Query: 156 DFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPLKALSN 215
DF +D FQR A + +G V+V+APT +GKT++ E A + R + FYTTP+KALSN
Sbjct: 36 DFELDDFQRQACLSLQQGRGVLVAAPTGAGKTIVGEFAIYLALQRALKAFYTTPIKALSN 95
Query: 216 QKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSSRSGLVNVD 275
QK+ E + +G VGLLTGD+++N DA V++MTTE+LRNMLY + L ++
Sbjct: 96 QKYSELADKYGPENVGLLTGDTSINGDAPVVVMTTEVLRNMLY-------ADSDTLDDLG 148
Query: 276 VIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGKTELVT 335
+V+DEVHYL+D RG VWEE++I+ P EVQ+ LSATV+N +E W+ + G T+++
Sbjct: 149 FVVMDEVHYLADRFRGAVWEEVIIHLPSEVQVASLSATVSNAEEFGAWLDTVRGHTDVIV 208
Query: 336 SSKRPVPLTWHFSMKNSLLPL------LDEKGTRMNRKLSLNYLQLQAA---GYKPYKDD 386
S RPVPL H + ++ L DE + + AA G++ D
Sbjct: 209 SEHRPVPLWQHVMVGREIVDLFAGETTFDEIAPEGESDPAATAMTANAALERGFEVNPDL 268
Query: 387 WSRKR-----NARKR---GYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIE 438
+ R N+R R G R + R + +R++ PQ+I +L +
Sbjct: 269 LAMARTESQMNSRARFGHGGRSQRRQQHQRGDNRHGQGGQQSPVRKASRPQVIASL---D 325
Query: 439 PRDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVRETAV-- 496
+D+LP+I FIFSR GCDAAV L E+ + +A + + V
Sbjct: 326 RQDLLPSITFIFSRAGCDAAVAQCVSAGLWLTTEREQQVIARRVDEAAQDIPSDDLDVLG 385
Query: 497 -----KGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISS 551
GLL+G+AAHHAG LP +K +E+LF +GLVK VFATETLA G+NMPAR+ V+
Sbjct: 386 FWSWRDGLLRGLAAHHAGMLPTFKEVVEKLFVEGLVKAVFATETLALGVNMPARSVVLEK 445
Query: 552 LSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLV 611
L K +++ E Q+ GRAGRRGID GH V++ P + PL
Sbjct: 446 LDKFNGEAHVGITAGEYTQLTGRAGRRGIDVEGHAVVLWQPGTDPTAVAGLASRRTYPLN 505
Query: 612 SQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLA 671
S F +Y M +NLL+ GR AR+++E SF + + ++
Sbjct: 506 SSFRPTYNMSINLLAQF-----------------GR--PRAREILESSFAQFQADRSVVG 546
Query: 672 AKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQ 731
+++ E+ + +T D L + EL + R + R+
Sbjct: 547 LARQVRSREESLAGFQKSMTCHLGDFTEYSRLRR-------ELSDAENIASRSTSRARKS 599
Query: 732 MEAKRISALKP--LLDDPESGHLPFLCLQYRDSEGVLHSIPAVFL--GKVDSLNASKLKD 787
+ +S L P ++D P +G P + PAV ++ + L+
Sbjct: 600 IAEDSLSRLLPGDVVDVP-TGRAPGFAVVLGSDHNSREPRPAVLTLDNQLRRIGLQDLEG 658
Query: 788 MISSV------DSFALNVADAERSLPDSVLN 812
I+ V SF V + + L SV N
Sbjct: 659 PITPVTRVRIPKSFNAKVPKSRKDLASSVRN 689
Score = 47.8 bits (112), Expect = 0.002
Identities = 36/152 (23%), Positives = 71/152 (46%), Gaps = 11/152 (7%)
Query: 1011 GETAAAIRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIK----VRPWKNNSFIY 1066
G+ I GE +L ++ LR L ++AA+ + LV + + +RP
Sbjct: 800 GQKLRRIYGEKDLLISQSLRQGAFSDLDATEVAALASVLVYQAKREERGLRPRM------ 853
Query: 1067 EPSATVVNCIGLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCA 1126
PS ++ + ++ + S L +E++ + ++ + + WA G +E++
Sbjct: 854 -PSVSLETAVDIVVREWSVLEDAEEENKLPLTGEPELGLVWPIFKWAKGKHLQEVLNGTD 912
Query: 1127 MDDGDLARLLRRTIDLLAQIPKLADIDPLLQR 1158
+ GD R +++ IDLL Q+ K+ +DP L R
Sbjct: 913 LAAGDFVRWVKQVIDLLDQLAKIPGLDPRLSR 944
>ref|ZP_00294287.1| COG4581: Superfamily II RNA helicase [Thermobifida fusca]
Length = 947
Score = 316 bits (810), Expect = 3e-84
Identities = 218/620 (35%), Positives = 319/620 (51%), Gaps = 78/620 (12%)
Query: 148 VDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYT 207
++E ++Y F D FQ A +A G V+V+APT SGKT++ E A + GR+ FYT
Sbjct: 22 IEEFQNLYGFDFDDFQIRACKALETGHGVLVAAPTGSGKTVVGEFAVHLALRDGRKCFYT 81
Query: 208 TPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSS 267
TP+KALSNQK+ + +G VGLLTGD+++N +A V++MTTE+LRNMLY S++
Sbjct: 82 TPIKALSNQKYTDLVRRYGSDKVGLLTGDNSINGEAPVVVMTTEVLRNMLYAG----SAT 137
Query: 268 RSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQI 327
SGL V V+DEVHYL+D RG VWEE++I+ P+ VQ+ LSATV+N +E W+ Q+
Sbjct: 138 LSGLAYV---VMDEVHYLADRFRGAVWEEVIIHLPESVQIAALSATVSNAEEFGQWLQQV 194
Query: 328 HGKTELVTSSKRPVPLTWHFSMKNSLLPLLDE-KGTRMNRKLSLNYLQLQAAGYKPYKDD 386
G T ++ KRPVPL H + + L E +GT ++ N G + +
Sbjct: 195 RGDTSVIVDEKRPVPLWQHMMVGTRIHDLFVEPEGTDTGQEEEKN-----GRGSRKRR-- 247
Query: 387 WSRKRNARKR-------GYRMS-------YDSDESMFEQRSLSKNNINAIRRSQVP---- 428
R R+AR+R G R+ + +++ Q + + + A R P
Sbjct: 248 --RSRHARQRTVEIEVGGERLHVNPKLIRFAEEDARLTQLAYQRRHPQARARGGAPRPRS 305
Query: 429 --------QIIDTLWHIEPRDMLPAIWFIFSRKGCDAAVQYI--EDCKLLDECEKSEVEL 478
QI++ L + +LPAI FIFSR GCD AV+ L E +E+
Sbjct: 306 RFAPPTRAQIVEQL---DREGLLPAITFIFSRAGCDEAVRQCVASGLVLTTPEEAAEIRE 362
Query: 479 ALKRFRIQYPDAVRETA-----VKGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFAT 533
+R + P A ++ L G+AAHHAG LP +K +E LF +GL++ VFAT
Sbjct: 363 YAERQCAEIPPADLNVLGYSEWLQALECGIAAHHAGMLPTFKEVVEVLFSRGLIRAVFAT 422
Query: 534 ETLAAGINMPARTAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPN 593
ETLA GINMPART VI L K L+ E Q+ GRAGRRGID GH V++ P
Sbjct: 423 ETLALGINMPARTVVIEKLDKWNGETHVALTPGEYTQLTGRAGRRGIDVEGHAVVVWQPG 482
Query: 594 EGAEECCKVLFAGLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDAR 653
E + PL S F SY M +NL++ V GR E R
Sbjct: 483 TDPEMVAGLAGTRTYPLNSSFQPSYNMAVNLVAQV-----------------GR--ERGR 523
Query: 654 KLVEQSFGNYVSSNVMLAAKEELKKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAE 713
++E SF + + ++ +L+ ++ +E D D A+ ++ +++
Sbjct: 524 AMLESSFAQFQADRSVVGLVRKLRAQQEALEGYAKAAYDPRGDFMEYAAMRRR----LSD 579
Query: 714 LQEDLRAEKRVRTALRRQME 733
L+ A++ RTA R++ E
Sbjct: 580 LES--AAQRNRRTARRKEAE 597
Score = 59.3 bits (142), Expect = 7e-07
Identities = 58/210 (27%), Positives = 88/210 (41%), Gaps = 6/210 (2%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALDINTHVIFPLGETAAAIRG 1019
E+ RLK ++ L RIE + F +V V+ E L+ +T + G + I
Sbjct: 735 ERYFRLKKETEELERRIESRSHVIARTFDRVCGVLQELDYLEGDT--VTEDGRLLSRIYS 792
Query: 1020 ENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCIGLL 1079
E +L A LR + L P +LAA + LV E + + F PS V + +
Sbjct: 793 ELDLLAAESLRRGVWDALGPEELAACVSALVYESRR----PDEVFARVPSGPVEEALNAM 848
Query: 1080 GEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCAMDDGDLARLLRRT 1139
L I+ +H V+ D F WA G I+ + M GD R ++
Sbjct: 849 MRLWGELSDIEHRHRVSFLREPDLGFVWPTYRWARGDQLDHILNEAGMPAGDFVRSTKQL 908
Query: 1140 IDLLAQIPKLADIDPLLQRNARAASDVMDR 1169
IDLL QI + ++ AR A+D + R
Sbjct: 909 IDLLGQIAQAVPEASGVRTTARQAADKLMR 938
>ref|NP_738228.1| putative helicase [Corynebacterium efficiens YS-314]
gi|23493458|dbj|BAC18428.1| putative helicase
[Corynebacterium efficiens YS-314]
Length = 940
Score = 315 bits (806), Expect = 8e-84
Identities = 200/607 (32%), Positives = 319/607 (51%), Gaps = 54/607 (8%)
Query: 148 VDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYT 207
+ E + F +D FQ +A V+V APT +GKT++ E A ++RG + FYT
Sbjct: 11 LSEFVADLGFDLDDFQIQGCRAVEGDHGVLVCAPTGAGKTIVGEFAVSLALSRGTKCFYT 70
Query: 208 TPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSS 267
TP+KALSNQK+ + G VGLLTGD ++N DA++++MTTE+LRNM+Y
Sbjct: 71 TPIKALSNQKYHDLVAKHGADAVGLLTGDVSINHDAEIVVMTTEVLRNMIY-------GQ 123
Query: 268 RSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQI 327
L + +V+DE+H+L+D SRG VWEE+++ + V++I LSATV+N +E W+ +
Sbjct: 124 SFALERLSHVVMDEIHFLADASRGAVWEEVILNLDESVKIIGLSATVSNSEEFGEWLTTV 183
Query: 328 HGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGT--RMNRKLSLNYLQLQAAGYKPYKD 385
G T+++ + RPVPL + ++ +LPL E GT R+N++L +L + +P +
Sbjct: 184 RGDTQVIVTDHRPVPLDQYMMVQRKVLPLF-EPGTDGRLNKQLEATIDRLNSQASEPGRS 242
Query: 386 DW-------SRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIE 438
++ +R + R RG + ++ + ++ + R +V +I+ +
Sbjct: 243 EYRSGEGFRTRAKGGRDRGRDDGRQNRDTRRSGQVREQDRHRPLGRPEVLRILQGM---- 298
Query: 439 PRDMLPAIWFIFSRKGCDAAVQYIEDCKLL--DECEKSEVELALKRFRIQYPDAVRETA- 495
+MLPAI FIFSR GCD A+ KL+ D+ E E+ + + P+ +
Sbjct: 299 --NMLPAITFIFSRAGCDGALYQCLRSKLILTDQDEAQEIARIIDAGVVGIPEEDLQVLN 356
Query: 496 ----VKGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISS 551
L++G AAHHAG LP ++ +EELF KGLV+ VFATETLA GINMPART V+
Sbjct: 357 FKQWRSALMRGFAAHHAGMLPAFRHIVEELFVKGLVRAVFATETLALGINMPARTVVLEK 416
Query: 552 LSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLV 611
L K G L+ + Q+ GRAGRRGID G+ V+ +P + PL+
Sbjct: 417 LVKFDGEGHVDLTPGQYTQLTGRAGRRGIDTLGNAVVQWSPAMDPRFVAGLASTRTYPLI 476
Query: 612 SQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLA 671
S F Y M +NLL + E + +L+E+SF + + ++
Sbjct: 477 STFQPGYNMSVNLLKTI-------------------GYEQSLRLLEKSFAQFQADGSVVG 517
Query: 672 AKEELKKNEKEIELLMSEITDEAIDRK---SRKALSQKQYKEIAELQEDLRAEKRVRTAL 728
E+++ E+++E L +++ E S + + + E EL+ +L E+ +T
Sbjct: 518 DVREIERAERKVEELRAQLDREITATNPAVSPDKTAVESFLEYMELRRELNEEE--KTHR 575
Query: 729 RRQMEAK 735
R ME +
Sbjct: 576 RDSMEQR 582
Score = 51.6 bits (122), Expect = 2e-04
Identities = 53/218 (24%), Positives = 95/218 (43%), Gaps = 13/218 (5%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALDI---NTHVIFPLGETAAA 1016
E++ R + +LT +++ + + F ++ +++ E +D + VI GE A
Sbjct: 721 ERMLRKQRDLNKLTGTVDRARETLGRTFERILSLLSEMDYVDYADPDNPVITEEGERLAK 780
Query: 1017 IRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCI 1076
I E++L +A L+ I GL P +LA V + E + + + E A +N
Sbjct: 781 IHNESDLLVAQCLKRGIWEGLDPAELAGVVSMCTFENRRETGGSPEA-VTEAMAEAMNHT 839
Query: 1077 GLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCA-----MDDGD 1131
+ S L+ + +H + ++ + F + WA+G M A + GD
Sbjct: 840 ERIW---SELVDDERRHRLPLTRQPEAGFAMAIHQWAAGAPLGYCMAAAAENGAELTPGD 896
Query: 1132 LARLLRRTIDLLAQIPKLADIDPLLQRNARAASDVMDR 1169
R R+ IDLL Q+ K + D + R AR D + R
Sbjct: 897 FVRWCRQVIDLLEQVAKTSYSDE-MARTARQGIDAIRR 933
>emb|CAA19107.1| SPCC550.03c [Schizosaccharomyces pombe]
gi|19075595|ref|NP_588095.1| hypothetical protein
SPCC550.03c [Schizosaccharomyces pombe 972h-]
gi|7492647|pir||T41378 probable helicase - fission yeast
(Schizosaccharomyces pombe)
Length = 1213
Score = 311 bits (797), Expect = 8e-83
Identities = 202/572 (35%), Positives = 306/572 (53%), Gaps = 46/572 (8%)
Query: 148 VDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYT 207
V E+A + F +D FQ+ A+ G SV V+A TS+GKT++AE A + YT
Sbjct: 270 VPEMALDFPFELDNFQKEAIYHLEMGDSVFVAAHTSAGKTVVAEYAIALAQKHMTKAIYT 329
Query: 208 TPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSS 267
+P+KALSNQKFR+F+ F D VG+LTGD VN + L+MTTEILR+MLY+ +
Sbjct: 330 SPIKALSNQKFRDFKHKFED--VGILTGDVQVNPEGSCLLMTTEILRSMLYRGADLIR-- 385
Query: 268 RSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQI 327
+V+ ++ DEVHY++D+ RG VWEE++I P V LI LSATV N E A W+G+
Sbjct: 386 -----DVEFVIFDEVHYVNDLERGVVWEEVIIMLPPHVTLILLSATVPNTKEFASWVGRT 440
Query: 328 HGKTELVTSS-KRPVPLTWHFSMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKPYKDD 386
K V S+ KRPVPL + +K ++ ++D+ G R L Y A KP K
Sbjct: 441 KKKNIYVISTLKRPVPLEHYLWVKQNMFKIVDQHG----RFLMDGYKSANDALKKPDKPV 496
Query: 387 WSR--KRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLP 444
++ K +AR RG +M R +K+ + R + + H+ +++LP
Sbjct: 497 IAKDNKNSARGRGAARGRGVQTNMMRGRGSAKS----VERRDANTWVHLIGHLHKQNLLP 552
Query: 445 AIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVRETAVKG-----L 499
I F+FS+K C+ V + + L + EKSEV + +++ + R G L
Sbjct: 553 VIVFVFSKKRCEEYVDTLTNRDLNNHQEKSEVHVVIEKAVARLKKEDRLLPQIGRMREML 612
Query: 500 LQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSG 559
+G+A HH G LP+ K +E LFQ+GLVKV+FATET A G+NMPA++ V S K
Sbjct: 613 SRGLAVHHGGLLPIIKEIVEILFQRGLVKVLFATETFAMGVNMPAKSVVFSGTQKHDGRN 672
Query: 560 RRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAG-LEPLVSQFTASY 618
R L E Q +GRAGRRG+D +G V+++ + + G L+SQF +Y
Sbjct: 673 FRDLLPGEYTQCSGRAGRRGLDVTGTVIILSRSELPDTASLRHMIMGPSSKLISQFRLTY 732
Query: 619 GMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKK 678
M+LNLL V+ + +ED ++++SF V+ ++ +E++K
Sbjct: 733 NMILNLLR-VETLR----------------IED---MIKRSFSENVNQTLVPQHEEKIKS 772
Query: 679 NEKEIELLMSEITDEAIDRKSRKALSQKQYKE 710
E+++ L E++D + LS + +KE
Sbjct: 773 FEEKLSALKKEMSDVDLKEIKSCLLSSESFKE 804
>emb|CAH02067.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50303465|ref|XP_451674.1| unnamed protein product
[Kluyveromyces lactis]
Length = 1001
Score = 306 bits (783), Expect = 3e-81
Identities = 218/660 (33%), Positives = 343/660 (51%), Gaps = 64/660 (9%)
Query: 74 DVEDEDDDDEEEEEYEEEEYEDDD-----DDDVAADEYDDEVPDEGE------AFDASAR 122
++ + ++ +E E+ E EY+ D+ + + E D +P + F+ +A
Sbjct: 226 NLNNSQENKKEVEKLRELEYQKMASTAVVDEALGSSEIDKLLPVDLNFARHIRTFNNTA- 284
Query: 123 REEFKWQRVEKLCNEVREFGAEIIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPT 182
E+ +W + L + + F V A + F +D FQ+ AV +G SV V+A T
Sbjct: 285 -EKVEWAHMVDLNSNMDNFNEL---VPNPARTWPFELDTFQKEAVYHLEQGDSVFVAAHT 340
Query: 183 SSGKTLIAEAAAVATVARGRRIFYTTPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKD 242
S+GKT++AE A + + YT+P+KALSNQKFR+F+E F D VGL+TGD +N +
Sbjct: 341 SAGKTVVAEYAIAMSKRNMTKTIYTSPIKALSNQKFRDFKEDFDDVDVGLITGDVQINPE 400
Query: 243 AQVLIMTTEILRNMLYQSVGNVSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCP 302
A LIMTTEILR+MLY+ + +V+ ++ DEVHY++D +RG VWEE++I P
Sbjct: 401 ADCLIMTTEILRSMLYRGADLIR-------DVEFVIFDEVHYVNDQTRGVVWEEVIIMLP 453
Query: 303 KEVQLICLSATVANPDELAGWIGQIHGKTELVTSS-KRPVPLTWHFSMKNSLLPLLDEKG 361
+ V+ I LSATV N E A W+G+ K V S+ KRPVPL + K ++P++++K
Sbjct: 454 QHVKFILLSATVPNTFEFANWVGRTKQKNIYVISTPKRPVPLEINIWAKKKVIPVINDKR 513
Query: 362 TRMNRKLSLNYLQLQAAGYKPYKDDWSRKRNARKRGYRMSYDSDESMFEQRSLSKNNINA 421
+ + + L A+ K+ +N + G + + + + +
Sbjct: 514 EFLPQNFREHKELLTASSIGSSKNS---PKNGKPSGNQKTITKGSKGVGAKGSNMSTFYK 570
Query: 422 IRRSQVPQIIDTLWHIEPRDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSE----VE 477
+ D L ++ D+LPA+ F+FS+K C+ IE LL E+S +E
Sbjct: 571 YDGASKTTWYDLLKNLRANDLLPAVVFVFSKKRCEEYADSIEAADLLTAKERSAIHIFIE 630
Query: 478 LALKRFRIQYPDAVRETAVKGLL-QGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETL 536
++ R R + + T ++ LL +G+A HH G LP+ K IE LF KG VK++FATET
Sbjct: 631 KSISRLRKDDRELPQITKIRSLLSRGIAVHHGGLLPIVKELIEILFAKGFVKLLFATETF 690
Query: 537 AAGINMPARTAVISSLSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGA 596
A G+N+P RT V S + K +R L E QMAGRAGRRG D++G V+++
Sbjct: 691 AMGLNLPTRTVVFSEIQKHDGEKKRYLLPGEFTQMAGRAGRRGKDKTGTVIVMSYSRPID 750
Query: 597 EECCKVLFAGLEP-LVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKL 655
E K + G+ L SQF +Y M+LNLL ++A+ ++
Sbjct: 751 EASFKDVSLGVPTRLQSQFRLTYNMILNLLR-IEALR-------------------VEEM 790
Query: 656 VEQSFGNYVSSNVMLAAKEELK-KNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAEL 714
++ SF S N +KE LK + EKEI+ L S++ ++ID + K+Y+ + L
Sbjct: 791 IKFSF----SEN----SKETLKPEQEKEIKELQSKV--DSIDISEFSEETVKEYESVHNL 840
>ref|NP_939594.1| Putative helicase [Corynebacterium diphtheriae NCTC 13129]
gi|38200088|emb|CAE49768.1| Putative helicase
[Corynebacterium diphtheriae]
Length = 930
Score = 305 bits (781), Expect = 6e-81
Identities = 203/604 (33%), Positives = 300/604 (49%), Gaps = 66/604 (10%)
Query: 157 FRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPLKALSNQ 216
F +D+FQ QA G V+V APT +GKT++ E A +++ + FYTTP+KALSNQ
Sbjct: 20 FPLDEFQIRGCQAVESGRGVLVCAPTGAGKTIVGEFAVSLAISQQTKCFYTTPIKALSNQ 79
Query: 217 KFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSSRSGLVNVDV 276
KF + + G+ VGLLTGD ++N+DA +++MT E+LRNM+Y + L +
Sbjct: 80 KFHDLVKAHGEDKVGLLTGDVSINRDADIVVMTAEVLRNMIY-------ADSEALDRLSH 132
Query: 277 IVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGKTELVTS 336
+V+DE+H+LSD SRG VWEE ++ V +I LSATV+N +E W+ + G TE++ S
Sbjct: 133 VVMDEIHFLSDSSRGPVWEEAILGLDASVNVIGLSATVSNSEEFGRWLNTVRGDTEIIVS 192
Query: 337 SKRPVPLTWHFSMKNSLLPLLDEKGT--RMNRKL--SLNYLQLQAAGYKPYKDDWSRKRN 392
RPVPL + + PL E GT +N +L + L+ Y +
Sbjct: 193 ENRPVPLNQWMLVARRMHPLF-EPGTTGEVNSRLVEHIERLEGTTTDGTEYGRAQLKAGG 251
Query: 393 ARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFIFSR 452
R R M D+ S LS + A+ R D L ++ ++MLPAI FIFSR
Sbjct: 252 FRNRSRAMGSDNRASRRSPHRLSDKH-RALSRP------DVLHILQEQNMLPAITFIFSR 304
Query: 453 KGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVRETAV---------------- 496
GCDAA L +C +S + L + + D +
Sbjct: 305 AGCDAA---------LHQCMRSSLVLTTQEESEEISDIITAGVAGIPEEDLRLLEYRRWR 355
Query: 497 KGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRI 556
+ L++G AAHHAG LP ++ +E+LF KGLV+ VFATETLA GINMPART V+ L K
Sbjct: 356 QALMRGFAAHHAGMLPAFRHVVEKLFTKGLVRAVFATETLALGINMPARTVVLEKLVKFN 415
Query: 557 DSGRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTA 616
G L+ + Q+ GRAGRRGID G+ V+ TP + + PLVS F
Sbjct: 416 GEGHADLTPGQYTQLTGRAGRRGIDVIGNAVVQWTPAMDPGQVAGLASTRTYPLVSTFAP 475
Query: 617 SYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEEL 676
Y M +NLL + E A +L+E+SF + + ++ + +
Sbjct: 476 GYNMSVNLLKTI-------------------GFEPAHRLLEKSFAQFQADGSVVDDIKAI 516
Query: 677 KKNEKEIELLMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTAL---RRQME 733
+ E+ ++ L ++ A + E ++ +L E+R L +R ME
Sbjct: 517 ESLEESVDQLTKQLHAAFDSHDQGGAGDVDSFLEYIRIRRELTEEERKHKRLSIEQRDME 576
Query: 734 AKRI 737
R+
Sbjct: 577 TTRV 580
Score = 36.6 bits (83), Expect = 5.2
Identities = 44/219 (20%), Positives = 93/219 (42%), Gaps = 12/219 (5%)
Query: 958 IEEKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALDIN--THVIFPLGETAA 1015
+ E++ R + + ++ + + + F ++ N++ E ++ + VI GE
Sbjct: 710 VGEQLVREQRMLEAAQRKVNEATDTLGRTFDRIINLLAEYDYVEFHDGVPVITEEGENLC 769
Query: 1016 AIRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNC 1075
I E++L +A L+ I L P +L+ V + E + + ++ E + + N
Sbjct: 770 QIHNESDLLVAQCLKRGIWDDLDPAELSGVVSMCTFENRRETRGEPDAATEEMAKAMNNT 829
Query: 1076 IGLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCA-----MDDG 1130
+ + E L + ++ + ++ + F + WA+G + A + G
Sbjct: 830 VRIWEE----LSTDERRYRLPVTRYPEGGFALAMHQWAAGAPLGYCIAAAAESGAELTPG 885
Query: 1131 DLARLLRRTIDLLAQIPKLADIDPLLQRNARAASDVMDR 1169
D R R+ +DLL Q+ A + +R AR A D + R
Sbjct: 886 DFVRQCRQVVDLLEQVRSTA-YNNDTRRAARKAIDAIRR 923
>emb|CAF21497.1| Superfamily II DNA and RNA helicase [Corynebacterium glutamicum
ATCC 13032] gi|21324257|dbj|BAB98882.1| Superfamily II
DNA and RNA helicases [Corynebacterium glutamicum ATCC
13032] gi|62390371|ref|YP_225773.1| Superfamily II DNA
and RNA helicase [Corynebacterium glutamicum ATCC 13032]
gi|19552703|ref|NP_600705.1| putative helicase
[Corynebacterium glutamicum ATCC 13032]
Length = 929
Score = 302 bits (774), Expect = 4e-80
Identities = 196/610 (32%), Positives = 315/610 (51%), Gaps = 55/610 (9%)
Query: 148 VDELASVYDFRIDKFQRLAVQAFLRGSSVVVSAPTSSGKTLIAEAAAVATVARGRRIFYT 207
+ E + F +D+FQ A V+V APT +GKT++ E A ++RG + FYT
Sbjct: 11 LSEFIADLGFDLDEFQIKGCHAVEEDHGVLVCAPTGAGKTIVGEFAVSLALSRGTKCFYT 70
Query: 208 TPLKALSNQKFREFRETFGDSYVGLLTGDSAVNKDAQVLIMTTEILRNMLYQSVGNVSSS 267
TP+KALSNQK+ + G VGLLTGD ++N DA +++MTTE+LRNM+Y +
Sbjct: 71 TPIKALSNQKYHDLVAKHGSDAVGLLTGDVSINHDADIVVMTTEVLRNMIY-------AG 123
Query: 268 RSGLVNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQI 327
L + +V+DE+H+L+D SRG VWEE+++ V +I LSATV+N +E W+ +
Sbjct: 124 SFALERLSHVVMDEIHFLADASRGAVWEEVILNLDDSVNIIGLSATVSNSEEFGEWLTTV 183
Query: 328 HGKTELVTSSKRPVPLTWHFSMKNSLLPLLDEKGT--RMNRKLSLNYLQLQAAGYKPYKD 385
G T ++ + RPVPL + ++ ++PL E GT R+N++L +L + +
Sbjct: 184 RGDTRVIVTDHRPVPLDQYMMVQRKVMPLF-EPGTDGRVNKELEATIDRLNSK-----QS 237
Query: 386 DWSRKRNARKRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPA 445
+ R G+R D+ + R+ + R P+++ L I +MLPA
Sbjct: 238 EQGRAAYRSGEGFRARSKGDKQ--DSRTGKPREQDRHRPLGRPEVLSILKGI---NMLPA 292
Query: 446 IWFIFSRKGCDAAVQYIEDCKLL--DECEKSEVELALKRFRIQYPDAVRETA-----VKG 498
I FIFSR GCD A+ KL+ D+ E E+ + + P+ +
Sbjct: 293 ITFIFSRAGCDGALYQCLRSKLVLTDQAESEEIARIVDAGVVGIPEEDLQVLNFKQWRAA 352
Query: 499 LLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDS 558
L++G AAHHAG LP ++ +EELF KGLV+ VFATETLA GINMPART V+ + K
Sbjct: 353 LMRGFAAHHAGMLPAFRHIVEELFVKGLVRAVFATETLALGINMPARTVVLEKMVKFDGE 412
Query: 559 GRRPLSSNELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASY 618
G L+ + Q+ GRAGRRGID G+ V+ +P + PL+S F Y
Sbjct: 413 GHVDLTPGQYTQLTGRAGRRGIDVLGNAVVQWSPALDPRWVAGLASTRTYPLISTFQPGY 472
Query: 619 GMVLNLLSGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKK 678
M +NLL + E + +L+E+SF + + ++ E+++
Sbjct: 473 NMSVNLLKTI-------------------GYEPSLRLLEKSFAQFQADGSVVGDVREIER 513
Query: 679 NEKEIELLMSEITDEAIDRKSRKALSQKQ-----YKEIAELQEDLRAEKRVRTALRRQME 733
E ++ L +++ E + A+ + Q + + EL+ +L E+++ ++ +E
Sbjct: 514 AEAKVAELRAQLNKEI--AATNPAVREDQDAVEVFIDYMELRRELNEEEKLNR--KQSIE 569
Query: 734 AKRISALKPL 743
+ + ++ L
Sbjct: 570 DRNVETVRVL 579
Score = 55.5 bits (132), Expect = 1e-05
Identities = 56/218 (25%), Positives = 96/218 (43%), Gaps = 13/218 (5%)
Query: 960 EKIKRLKTRSKRLTNRIEQIEPSGWKEFMQVSNVIHETRALDINTH---VIFPLGETAAA 1016
E++ R + +LT +++ + + F ++ +++ E +D + VI GE A
Sbjct: 710 ERMIRKERDLAKLTGNVDKARETLGRTFERILSLLSEMDYVDYSNPDNPVITDEGERLAK 769
Query: 1017 IRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCI 1076
I E +L +A L+ I L P +LA V + E + + + + E A +N +
Sbjct: 770 IHSEADLLVAQCLKRGIWDNLDPAELAGVVSMCTFENRRETGGEAQA-VTEAMADSMNSV 828
Query: 1077 GLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGLTWREIMMDCA-----MDDGD 1131
+ + L+ + +H + I+ + F + WASG M A + GD
Sbjct: 829 ERIWGE---LVEDERRHRLPITRQPEAGFATAIHQWASGAPLGYCMAAAAENGAELTPGD 885
Query: 1132 LARLLRRTIDLLAQIPKLADIDPLLQRNARAASDVMDR 1169
R R+ IDLL Q+ K A D RNAR A D + R
Sbjct: 886 FVRWCRQVIDLLEQVAKTAYFDE-TTRNARQAIDAIRR 922
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,941,359,192
Number of Sequences: 2540612
Number of extensions: 84456963
Number of successful extensions: 915203
Number of sequences better than 10.0: 7582
Number of HSP's better than 10.0 without gapping: 4879
Number of HSP's successfully gapped in prelim test: 2941
Number of HSP's that attempted gapping in prelim test: 672536
Number of HSP's gapped (non-prelim): 75259
length of query: 1177
length of database: 863,360,394
effective HSP length: 139
effective length of query: 1038
effective length of database: 510,215,326
effective search space: 529603508388
effective search space used: 529603508388
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 81 (35.8 bits)
Lotus: description of TM0028b.4


