

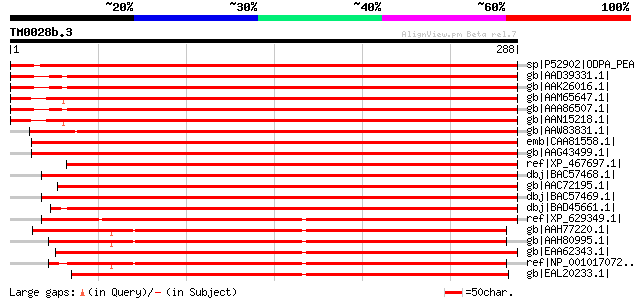
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.3
(288 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component alpha sub... 492 e-138
gb|AAD39331.1| pyruvate dehydrogenase E1 alpha subunit [Arabidop... 477 e-133
gb|AAK26016.1| putative pyruvate dehydrogenase e1 alpha subunit ... 477 e-133
gb|AAM65647.1| pyruvate dehydrogenase E1 alpha subunit [Arabidop... 475 e-133
gb|AAA86507.1| pyruvate dehydrogenase E1 alpha subunit gi|211753... 474 e-132
gb|AAN15218.1| pyruvate dehydrogenase E1a-like subunit IAR4 [Ara... 471 e-132
gb|AAW83831.1| E1 alpha subunit of pyruvate dehydrogenase [Petun... 470 e-131
emb|CAA81558.1| E1 alpha subunit of pyruvate dehydrogenase precu... 462 e-129
gb|AAG43499.1| pyruvate dehydrogenase [Lycopersicon esculentum] 459 e-128
ref|XP_467697.1| putative pyruvate dehydrogenase E1 alpha subuni... 447 e-124
dbj|BAC57468.1| pyruvate dehydrogenase E1alpha subunit [Beta vul... 446 e-124
gb|AAC72195.1| pyruvate dehydrogenase E1 alpha subunit [Zea mays] 443 e-123
dbj|BAC57469.1| pyruvate dehydrogenase E1 alpha subunit [Beta vu... 442 e-123
dbj|BAD45661.1| putative pyruvate dehydrogenase E1 alpha subunit... 427 e-118
ref|XP_629349.1| pyruvate dehydrogenase E1 subunit alpha [Dictyo... 295 8e-79
gb|AAH77220.1| Pdha1-A-prov protein [Xenopus laevis] 286 5e-76
gb|AAH80995.1| Pdha1-B-prov protein [Xenopus laevis] 284 2e-75
gb|EAA62343.1| hypothetical protein AN5162.2 [Aspergillus nidula... 283 5e-75
ref|NP_001017072.1| hypothetical protein LOC549826 [Xenopus trop... 282 7e-75
gb|EAL20233.1| hypothetical protein CNBF0450 [Cryptococcus neofo... 282 9e-75
>sp|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component alpha subunit, mitochondrial
precursor (PDHE1-A) gi|1263302|gb|AAA97411.1| pyruvate
dehydrogenase E1 alpha subunit gi|7431564|pir||T06531
pyruvate dehydrogenase (lipoamide) (EC 1.2.4.1) complex
E1 alpha chain - garden pea
Length = 397
Score = 492 bits (1267), Expect = e-138
Identities = 241/289 (83%), Positives = 261/289 (89%), Gaps = 4/289 (1%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNP-ITIETSVPFTAHNCDPPS 59
MALSRL+SSSSSS+ GS P +AAF+L R ISSD+ +TIETS+PFTAHNCDPPS
Sbjct: 1 MALSRLSSSSSSSN---GSNLFNPFSAAFTLNRPISSDTTATLTIETSLPFTAHNCDPPS 57
Query: 60 RSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKK 119
RSV TS +EL SFFR MA MRRMEIAADSLYKA LIRGFCHLYDGQEAVA+GMEAG TKK
Sbjct: 58 RSVTTSPSELLSFFRTMALMRRMEIAADSLYKANLIRGFCHLYDGQEAVAVGMEAGTTKK 117
Query: 120 DAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGA 179
D IITAYRDHCTFLGRGGTLL +++ELMGR DGCSKGKGGSMHFY+K++ FYGGHGIVGA
Sbjct: 118 DCIITAYRDHCTFLGRGGTLLRVYAELMGRRDGCSKGKGGSMHFYKKDSGFYGGHGIVGA 177
Query: 180 QVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGM 239
QVPLGCG+AF QKY KDE+VTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGM
Sbjct: 178 QVPLGCGLAFGQKYLKDESVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGM 237
Query: 240 GTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
GTA WR+AKSPAY+KRGDYVPGLKVDGMD LAVKQACKFAKEHALKNGP
Sbjct: 238 GTATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGP 286
>gb|AAD39331.1| pyruvate dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|24030439|gb|AAN41374.1| putative pyruvate
dehydrogenase e1 alpha subunit [Arabidopsis thaliana]
gi|21593256|gb|AAM65205.1| pyruvate dehydrogenase e1
alpha subunit, putative [Arabidopsis thaliana]
gi|15218940|ref|NP_176198.1| pyruvate dehydrogenase E1
component alpha subunit, mitochondrial (PDHE1-A)
[Arabidopsis thaliana] gi|25284401|pir||B96623 pyruvate
dehydrogenase E1 alpha subunit [imported] - Arabidopsis
thaliana gi|27735220|sp|P52901|ODPA_ARATH Pyruvate
dehydrogenase E1 component alpha subunit, mitochondrial
precursor (PDHE1-A)
Length = 389
Score = 477 bits (1228), Expect = e-133
Identities = 236/288 (81%), Positives = 258/288 (88%), Gaps = 10/288 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSR 60
MALSRL+S S+ + +P +AAFS R IS+D+ PITIETS+PFTAH CDPPSR
Sbjct: 1 MALSRLSSRSNIIT--------RPFSAAFS--RLISTDTTPITIETSLPFTAHLCDPPSR 50
Query: 61 SVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKD 120
SVE+S+ EL FFR MA MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEA ITKKD
Sbjct: 51 SVESSSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKD 110
Query: 121 AIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQ 180
AIITAYRDHC FLGRGG+L E+FSELMGR GCSKGKGGSMHFY+KE++FYGGHGIVGAQ
Sbjct: 111 AIITAYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQ 170
Query: 181 VPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG 240
VPLGCG+AFAQKY+K+EAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMG
Sbjct: 171 VPLGCGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMG 230
Query: 241 TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
TAEWRAAKSP+YYKRGDYVPGLKVDGMD AVKQACKFAK+HAL+ GP
Sbjct: 231 TAEWRAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGP 278
>gb|AAK26016.1| putative pyruvate dehydrogenase e1 alpha subunit [Arabidopsis
thaliana]
Length = 389
Score = 477 bits (1228), Expect = e-133
Identities = 236/288 (81%), Positives = 258/288 (88%), Gaps = 10/288 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSR 60
MALSRL+S S+ + +P +AAFS R IS+D+ PITIETS+PFTAH CDPPSR
Sbjct: 1 MALSRLSSRSNIIT--------RPFSAAFS--RLISTDTTPITIETSLPFTAHLCDPPSR 50
Query: 61 SVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKD 120
SVE+S+ EL FFR MA MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEA ITKKD
Sbjct: 51 SVESSSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKD 110
Query: 121 AIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQ 180
AIITAYRDHC FLGRGG+L E+FSELMGR GCSKGKGGSMHFY+KE++FYGGHGIVGAQ
Sbjct: 111 AIITAYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQ 170
Query: 181 VPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG 240
VPLGCG+AFAQKY+K+EAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMG
Sbjct: 171 VPLGCGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMG 230
Query: 241 TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
TAEWRAAKSP+YYKRGDYVPGLKVDGMD AVKQACKFAK+HAL+ GP
Sbjct: 231 TAEWRAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGP 278
>gb|AAM65647.1| pyruvate dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|15293169|gb|AAK93695.1| putative pyruvate
dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|13430606|gb|AAK25925.1| putative pyruvate
dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|15221692|ref|NP_173828.1| pyruvate dehydrogenase E1
component alpha subunit, mitochondrial, putative
[Arabidopsis thaliana] gi|7431563|pir||T00648 pyruvate
dehydrogenase (lipoamide) (EC 1.2.4.1) E1 alpha chain -
Arabidopsis thaliana gi|2829869|gb|AAC00577.1| pyruvate
dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
Length = 393
Score = 475 bits (1223), Expect = e-133
Identities = 231/290 (79%), Positives = 259/290 (88%), Gaps = 10/290 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAF--SLRRSISSDSNPITIETSVPFTAHNCDPP 58
MALSRL+S S++ FLKP A S+RR +S+DS+PITIET+VPFT+H C+ P
Sbjct: 1 MALSRLSSRSNT--------FLKPAITALPSSIRRHVSTDSSPITIETAVPFTSHLCESP 52
Query: 59 SRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITK 118
SRSVETS+ E+ +FFR MA MRRMEIAADSLYKAKLIRGFCHLYDGQEA+A+GMEA ITK
Sbjct: 53 SRSVETSSEEILAFFRDMARMRRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITK 112
Query: 119 KDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 178
KDAIIT+YRDHCTF+GRGG L++ FSELMGR GCS GKGGSMHFY+K+ +FYGGHGIVG
Sbjct: 113 KDAIITSYRDHCTFIGRGGKLVDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVG 172
Query: 179 AQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 238
AQ+PLGCG+AFAQKY+KDEAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYG
Sbjct: 173 AQIPLGCGLAFAQKYNKDEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYG 232
Query: 239 MGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
MGTA WR+AKSPAY+KRGDYVPGLKVDGMD LAVKQACKFAKEHALKNGP
Sbjct: 233 MGTATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGP 282
>gb|AAA86507.1| pyruvate dehydrogenase E1 alpha subunit gi|2117533|pir||JC4358
pyruvate dehydrogenase (lipoamide) (EC 1.2.4.1) alpha
chain precursor - Arabidopsis thaliana
Length = 389
Score = 474 bits (1220), Expect = e-132
Identities = 234/288 (81%), Positives = 257/288 (88%), Gaps = 10/288 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSR 60
MALSRL+S S+ + +P +AAFS R IS+D+ PITIETS+PFTAH CDPPSR
Sbjct: 1 MALSRLSSRSNIIT--------RPFSAAFS--RLISTDTTPITIETSLPFTAHLCDPPSR 50
Query: 61 SVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKD 120
SVE+S+ EL FFR MA MRRMEIAADSLYKA +IRGFCHLYDGQEAVAIGMEA ITKKD
Sbjct: 51 SVESSSQELLDFFRTMALMRRMEIAADSLYKANVIRGFCHLYDGQEAVAIGMEAAITKKD 110
Query: 121 AIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQ 180
AIITAYRDHC FLGRGG+L E+FSELMGR GCSKGKGGSMHFY+KE++FYGGHGIVGAQ
Sbjct: 111 AIITAYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQ 170
Query: 181 VPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG 240
VPLGCG+AFAQKY+K+EAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMG
Sbjct: 171 VPLGCGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMG 230
Query: 241 TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
TAEWRAAKSP+YYKRGDYVPGLKVDGMD AVKQACKFAK+HAL+ GP
Sbjct: 231 TAEWRAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGP 278
>gb|AAN15218.1| pyruvate dehydrogenase E1a-like subunit IAR4 [Arabidopsis thaliana]
Length = 393
Score = 471 bits (1213), Expect = e-132
Identities = 230/290 (79%), Positives = 258/290 (88%), Gaps = 10/290 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAF--SLRRSISSDSNPITIETSVPFTAHNCDPP 58
MALSRL+S S++ FLKP A S+RR +S+DS+PITIET+VPFT+H C+ P
Sbjct: 1 MALSRLSSRSNT--------FLKPAITALPSSIRRHVSTDSSPITIETAVPFTSHLCESP 52
Query: 59 SRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITK 118
SRSVETS+ E+ +FFR MA MRRMEIAADSLYKAKLIRGFCHLYDGQEA+A+GMEA ITK
Sbjct: 53 SRSVETSSEEILAFFRDMARMRRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITK 112
Query: 119 KDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 178
KDAIIT+YRDHCTF+GRGG L++ FSELMGR GCS GKGGSMHFY+K+ +FYGGHGIVG
Sbjct: 113 KDAIITSYRDHCTFIGRGGKLVDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVG 172
Query: 179 AQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 238
AQ+PLGCG+AFAQKY+KDEAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNH G
Sbjct: 173 AQIPLGCGLAFAQKYNKDEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHDG 232
Query: 239 MGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
MGTA WR+AKSPAY+KRGDYVPGLKVDGMD LAVKQACKFAKEHALKNGP
Sbjct: 233 MGTATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGP 282
>gb|AAW83831.1| E1 alpha subunit of pyruvate dehydrogenase [Petunia x hybrida]
Length = 390
Score = 470 bits (1209), Expect = e-131
Identities = 228/277 (82%), Positives = 249/277 (89%), Gaps = 1/277 (0%)
Query: 12 SSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVETSTAELHS 71
S++ T SKFLKPLT A S R +S+ +N +TIETS+PFT HN DPPSR+VET+ EL +
Sbjct: 4 STTRTTVSKFLKPLTTAVSTTRHLST-TNTLTIETSLPFTGHNIDPPSRTVETNPNELLT 62
Query: 72 FFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCT 131
FF+ MA MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGME+ ITKKD IITAYRDHC
Sbjct: 63 FFKDMAEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMESAITKKDCIITAYRDHCI 122
Query: 132 FLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQ 191
FL RGGTL E FSELMGR DGCSKGKGGSMHFY+K++ FYGGHGIVGAQVPLG G+AFAQ
Sbjct: 123 FLSRGGTLFECFSELMGRKDGCSKGKGGSMHFYKKDSGFYGGHGIVGAQVPLGIGLAFAQ 182
Query: 192 KYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPA 251
KYSK++ VTFA+YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWRAAKSP+
Sbjct: 183 KYSKEDHVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPS 242
Query: 252 YYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
YYKRGDYVPGLKVDGMD LAVKQACKFAKEHALKNGP
Sbjct: 243 YYKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGP 279
>emb|CAA81558.1| E1 alpha subunit of pyruvate dehydrogenase precursor [Solanum
tuberosum] gi|1709453|sp|P52903|ODPA_SOLTU Pyruvate
dehydrogenase E1 component alpha subunit, mitochondrial
precursor (PDHE1-A) gi|7431569|pir||T07372 pyruvate
dehydrogenase (lipoamide) (EC 1.2.4.1) E1 alpha chain -
potato
Length = 391
Score = 462 bits (1188), Expect = e-129
Identities = 223/277 (80%), Positives = 246/277 (88%), Gaps = 1/277 (0%)
Query: 13 SSSTVGSKFLKPLTAAFSLRRSISSDSNP-ITIETSVPFTAHNCDPPSRSVETSTAELHS 71
S+S + +KPL+AA R +SSDS IT+ETS+PFT+HN DPPSRSVETS EL +
Sbjct: 4 STSRAINHIMKPLSAAVCATRRLSSDSTATITVETSLPFTSHNIDPPSRSVETSPKELMT 63
Query: 72 FFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCT 131
FF+ M MRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEA ITKKD IITAYRDHC
Sbjct: 64 FFKDMTEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDCIITAYRDHCI 123
Query: 132 FLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQ 191
FLGRGGTL+E F+ELMGR DGCS+GKGGSMHFY+KE+ FYGGHGIVGAQVPLG G+AFAQ
Sbjct: 124 FLGRGGTLVEAFAELMGRRDGCSRGKGGSMHFYKKESGFYGGHGIVGAQVPLGIGLAFAQ 183
Query: 192 KYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPA 251
KY K++ VTFA+YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWRAAKSPA
Sbjct: 184 KYKKEDYVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPA 243
Query: 252 YYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
YYKRGDYVPGL+VDGMDV AVKQAC FAK+HALKNGP
Sbjct: 244 YYKRGDYVPGLRVDGMDVFAVKQACTFAKQHALKNGP 280
>gb|AAG43499.1| pyruvate dehydrogenase [Lycopersicon esculentum]
Length = 391
Score = 459 bits (1182), Expect = e-128
Identities = 222/277 (80%), Positives = 245/277 (88%), Gaps = 1/277 (0%)
Query: 13 SSSTVGSKFLKPLTAAFSLRRSISSDSNP-ITIETSVPFTAHNCDPPSRSVETSTAELHS 71
S+S + +KPL+ A R +SSDS IT+ETS+PFT+HN DPPSRSVETS EL +
Sbjct: 4 STSRAINHIMKPLSRAVCATRRLSSDSTATITVETSLPFTSHNVDPPSRSVETSPMELMT 63
Query: 72 FFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCT 131
FF+ M MRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEA ITKKD IITAYRDHC
Sbjct: 64 FFKDMTEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDCIITAYRDHCI 123
Query: 132 FLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQ 191
FLGRGGTL+E F+ELMGR DGCS+GKGGSMHFY+KE+ FYGGHGIVGAQVPLG G+AFAQ
Sbjct: 124 FLGRGGTLVESFAELMGRRDGCSRGKGGSMHFYKKESGFYGGHGIVGAQVPLGIGLAFAQ 183
Query: 192 KYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPA 251
KY K++ VTFA+YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWRAAKSPA
Sbjct: 184 KYKKEDYVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPA 243
Query: 252 YYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
YYKRGDYVPGL+VDGMDV AVKQAC FAK+HALKNGP
Sbjct: 244 YYKRGDYVPGLRVDGMDVFAVKQACAFAKQHALKNGP 280
>ref|XP_467697.1| putative pyruvate dehydrogenase E1 alpha subunit [Oryza sativa
(japonica cultivar-group)] gi|51964350|ref|XP_506960.1|
PREDICTED P0684F11.25 gene product [Oryza sativa
(japonica cultivar-group)] gi|46390562|dbj|BAD16048.1|
putative pyruvate dehydrogenase E1 alpha subunit [Oryza
sativa (japonica cultivar-group)]
Length = 390
Score = 447 bits (1149), Expect = e-124
Identities = 212/256 (82%), Positives = 231/256 (89%)
Query: 33 RSISSDSNPITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMATMRRMEIAADSLYKA 92
RSIS + P+TIETSVPFT+H DPPSR V T+ AEL +FFR M+ MRRMEIAADSLYKA
Sbjct: 24 RSISDSTAPLTIETSVPFTSHIVDPPSRDVTTTPAELLTFFRDMSVMRRMEIAADSLYKA 83
Query: 93 KLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDG 152
KLIRGFCHLYDGQEAVA+GMEA IT+ D+IITAYRDHCT+L RGG L+ F+ELMGR G
Sbjct: 84 KLIRGFCHLYDGQEAVAVGMEAAITRSDSIITAYRDHCTYLARGGDLVSAFAELMGRQAG 143
Query: 153 CSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQG 212
CS+GKGGSMHFY+K+ NFYGGHGIVGAQVPLGCG+AFAQKY K+E TFALYGDGAANQG
Sbjct: 144 CSRGKGGSMHFYKKDANFYGGHGIVGAQVPLGCGLAFAQKYRKEETATFALYGDGAANQG 203
Query: 213 QLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAV 272
QLFEALNI+ALW LPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAV
Sbjct: 204 QLFEALNISALWKLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAV 263
Query: 273 KQACKFAKEHALKNGP 288
KQACKFAKEHA+ NGP
Sbjct: 264 KQACKFAKEHAIANGP 279
>dbj|BAC57468.1| pyruvate dehydrogenase E1alpha subunit [Beta vulgaris]
Length = 395
Score = 446 bits (1147), Expect = e-124
Identities = 215/271 (79%), Positives = 242/271 (88%), Gaps = 1/271 (0%)
Query: 19 SKFLKPLTAAFSLRRSISSDSNP-ITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMA 77
S L PLT+ + R +SSDS +TIETSVPF +H +PPSRSV+T+ AEL ++FR MA
Sbjct: 14 SSVLLPLTSTHTHSRRLSSDSTKTLTIETSVPFKSHIVEPPSRSVDTTPAELMTYFRDMA 73
Query: 78 TMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGG 137
MRRMEIA+DSLYKAKLIRGFCHLYDGQEAVA+GMEA ITKKD IITAYRDHC +L RGG
Sbjct: 74 LMRRMEIASDSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDNIITAYRDHCIYLARGG 133
Query: 138 TLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDE 197
+LL F+ELMGR DGCS+GKGGSMHFY+K++ FYGGHGIVGAQVPLG G+AFAQKY+K++
Sbjct: 134 SLLSAFAELMGRQDGCSRGKGGSMHFYKKDSGFYGGHGIVGAQVPLGVGLAFAQKYNKED 193
Query: 198 AVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGD 257
V+FALYGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWRAAKSP+YYKRGD
Sbjct: 194 CVSFALYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPSYYKRGD 253
Query: 258 YVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
YVPGLKVDGMDVLAVKQACKFAKE+ LKNGP
Sbjct: 254 YVPGLKVDGMDVLAVKQACKFAKEYVLKNGP 284
>gb|AAC72195.1| pyruvate dehydrogenase E1 alpha subunit [Zea mays]
Length = 392
Score = 443 bits (1139), Expect = e-123
Identities = 211/261 (80%), Positives = 231/261 (87%)
Query: 28 AFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMATMRRMEIAAD 87
AF R IS + +TIETSVPFT+H DPPSR V T+ AEL +FFR M+ MRRMEIAAD
Sbjct: 21 AFMAARPISDSTAALTIETSVPFTSHLVDPPSRDVTTTPAELVTFFRDMSLMRRMEIAAD 80
Query: 88 SLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELM 147
SLYKAKLIRGFCHLYDGQEAVA+GMEA IT+ D+IITAYRDHCT+L RGG L+ FSELM
Sbjct: 81 SLYKAKLIRGFCHLYDGQEAVAVGMEAAITRSDSIITAYRDHCTYLARGGDLVSAFSELM 140
Query: 148 GRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDG 207
GR GCS+GKGGSMHFY+K+ NFYGGHGIVGAQVPLGCG+AFAQKY K++ TFALYGDG
Sbjct: 141 GREAGCSRGKGGSMHFYKKDANFYGGHGIVGAQVPLGCGLAFAQKYKKEDTATFALYGDG 200
Query: 208 AANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGM 267
AANQGQLFEALNI+ALW LPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGM
Sbjct: 201 AANQGQLFEALNISALWKLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGM 260
Query: 268 DVLAVKQACKFAKEHALKNGP 288
DVLAVKQACKFAK+HA+ NGP
Sbjct: 261 DVLAVKQACKFAKDHAVANGP 281
>dbj|BAC57469.1| pyruvate dehydrogenase E1 alpha subunit [Beta vulgaris]
Length = 395
Score = 442 bits (1136), Expect = e-123
Identities = 213/271 (78%), Positives = 241/271 (88%), Gaps = 1/271 (0%)
Query: 19 SKFLKPLTAAFSLRRSISSDSNP-ITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMA 77
S L PLT+ + R +SSDS +TIETSVPF +H +PPSRSV+T+ AEL ++FR MA
Sbjct: 14 SSLLLPLTSTHTHSRRLSSDSTKTLTIETSVPFKSHIVEPPSRSVDTTPAELMTYFRDMA 73
Query: 78 TMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGG 137
MRRMEIA+DSLYKAKLIRGFCHLYDGQEAVA+GMEA ITKKD IITAYRDHC +L RGG
Sbjct: 74 LMRRMEIASDSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDNIITAYRDHCIYLARGG 133
Query: 138 TLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDE 197
+LL F+EL+GR DGCS+GKGGSMHFY+K++ FYGGHGIVGAQVPLG G+AFAQKY+K++
Sbjct: 134 SLLSAFAELVGRQDGCSRGKGGSMHFYKKDSGFYGGHGIVGAQVPLGVGLAFAQKYNKED 193
Query: 198 AVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGD 257
V+FALYGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWRAAKSP+YYKRGD
Sbjct: 194 CVSFALYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPSYYKRGD 253
Query: 258 YVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
YVPGLKVDGMDVLAVKQACK AKE+ LKNGP
Sbjct: 254 YVPGLKVDGMDVLAVKQACKSAKEYVLKNGP 284
>dbj|BAD45661.1| putative pyruvate dehydrogenase E1 alpha subunit [Oryza sativa
(japonica cultivar-group)]
Length = 398
Score = 427 bits (1097), Expect = e-118
Identities = 202/265 (76%), Positives = 227/265 (85%), Gaps = 3/265 (1%)
Query: 24 PLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMATMRRME 83
PLT + R +S + P+TIETSVP+ +H DPP R V T+ EL +FFR M+ MRR E
Sbjct: 26 PLTTSV---RGVSDSTEPLTIETSVPYKSHIVDPPPREVATTARELATFFRDMSAMRRAE 82
Query: 84 IAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIF 143
IAADSLYKAKLIRGFCHLYDGQEAVA+GMEA T+ DAIITAYRDHC +L RGG L +F
Sbjct: 83 IAADSLYKAKLIRGFCHLYDGQEAVAVGMEAATTRADAIITAYRDHCAYLARGGDLAALF 142
Query: 144 SELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFAL 203
+ELMGR GCS+GKGGSMH Y+K+ NFYGGHGIVGAQVPLGCG+AFAQ+Y K+ AVTF L
Sbjct: 143 AELMGRRGGCSRGKGGSMHLYKKDANFYGGHGIVGAQVPLGCGLAFAQRYRKEAAVTFDL 202
Query: 204 YGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLK 263
YGDGAANQGQLFEALN+AALW LP +LVCENNHYGMGTAEWRA+KSPAYYKRGDYVPGLK
Sbjct: 203 YGDGAANQGQLFEALNMAALWKLPVVLVCENNHYGMGTAEWRASKSPAYYKRGDYVPGLK 262
Query: 264 VDGMDVLAVKQACKFAKEHALKNGP 288
VDGMDVLAVKQACKFAK+HAL+NGP
Sbjct: 263 VDGMDVLAVKQACKFAKQHALENGP 287
>ref|XP_629349.1| pyruvate dehydrogenase E1 subunit alpha [Dictyostelium discoideum]
gi|60462647|gb|EAL60849.1| pyruvate dehydrogenase E1
alpha subunit [Dictyostelium discoideum]
Length = 377
Score = 295 bits (756), Expect = 8e-79
Identities = 158/271 (58%), Positives = 185/271 (67%), Gaps = 4/271 (1%)
Query: 19 SKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMAT 78
S FLK + A R+ +S S I T + CD PS S T+ EL SFF +M+
Sbjct: 3 SNFLKVNSKALGHIRTFASKSGEIKHNFKKADT-YLCDGPSDSTVTNKDELISFFTEMSR 61
Query: 79 MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGT 138
RR+E D LYK KLIRGFCHLY GQEAV G+E+ ITK D IITAYRDH L RG T
Sbjct: 62 FRRLETVCDGLYKKKLIRGFCHLYTGQEAVCAGLESAITKDDHIITAYRDHTYMLSRGAT 121
Query: 139 LLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEA 198
EIF+EL+ + GCSKGKGGSMH + K NFYGG+GIVGAQ PLG G+AFAQKY+K
Sbjct: 122 PEEIFAELLMKETGCSKGKGGSMHMFTK--NFYGGNGIVGAQCPLGAGIAFAQKYNKTGN 179
Query: 199 VTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDY 258
V A+YGDGAANQGQLFEA N+A+LW LP I +CENN YGMGT++ R+ +Y RG Y
Sbjct: 180 VCLAMYGDGAANQGQLFEAFNMASLWKLPVIFICENNKYGMGTSQKRSTAGHDFYTRGHY 239
Query: 259 VPGLKVDGMDVLAVKQACKFAKEHA-LKNGP 288
V GLKVDGMDV AVK+A K+A E NGP
Sbjct: 240 VAGLKVDGMDVFAVKEAGKYAAEWCRAGNGP 270
>gb|AAH77220.1| Pdha1-A-prov protein [Xenopus laevis]
Length = 400
Score = 286 bits (732), Expect = 5e-76
Identities = 150/277 (54%), Positives = 186/277 (66%), Gaps = 11/277 (3%)
Query: 14 SSTVGSKFLKPLT-AAFSLRRSISSDSNPITIETSVPFTAHNCD-------PPSRSVETS 65
S + K KP+T AA R + + N + F CD PP+++V T
Sbjct: 9 SRVLKGKAQKPVTGAANEAVRVMVASRNYADFASEATFDVKKCDVHRLEEGPPTQAVLTR 68
Query: 66 TAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITA 125
L ++R M T+RRME+ +D LYK K+IRGFCHLYDGQEA +G+E+GI D +ITA
Sbjct: 69 EQGLQ-YYRTMQTIRRMELKSDQLYKQKIIRGFCHLYDGQEACCVGLESGINPTDHLITA 127
Query: 126 YRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGC 185
YR H RG ++ EI +EL GR GC+KGKGGSMH Y K NFYGG+GIVGAQVPLG
Sbjct: 128 YRAHGYTYTRGVSVKEILAELTGRKGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGA 185
Query: 186 GVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWR 245
GVA A K+ + + +LYGDGAANQGQ+FE N+AALW LP I +CENN YGMGT+ R
Sbjct: 186 GVALACKFFGKDEICVSLYGDGAANQGQIFETYNMAALWKLPCIFICENNRYGMGTSVER 245
Query: 246 AAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEH 282
AA S YYKRGDY+PGL+VDGMDVL V++A KFA +H
Sbjct: 246 AAASTDYYKRGDYIPGLRVDGMDVLCVREATKFAADH 282
>gb|AAH80995.1| Pdha1-B-prov protein [Xenopus laevis]
Length = 400
Score = 284 bits (726), Expect = 2e-75
Identities = 148/268 (55%), Positives = 182/268 (67%), Gaps = 11/268 (4%)
Query: 23 KPLT-AAFSLRRSISSDSNPITIETSVPFTAHNCD-------PPSRSVETSTAELHSFFR 74
KP+T AA R + + N + F CD PP+++V T L ++R
Sbjct: 18 KPVTGAANEAVRVMMASRNYADFASEATFDIKKCDIHRLEEEPPTQAVLTREEGLQ-YYR 76
Query: 75 QMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLG 134
M T+RRME+ +D LYK K+IRGFCHLYDGQEA +G+E+GI D +ITAYR H
Sbjct: 77 TMQTIRRMELKSDQLYKQKIIRGFCHLYDGQEACCVGLESGINPTDHLITAYRAHGYTYT 136
Query: 135 RGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYS 194
RG ++ EI +EL GR GC+KGKGGSMH Y K NFYGG+GIVGAQVPLG GVA A K+
Sbjct: 137 RGVSVKEILAELTGRRGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGVALACKFF 194
Query: 195 KDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYK 254
+ +LYGDGAANQGQ+FE N+AALW LP I +CENN YGMGT+ RAA S YYK
Sbjct: 195 GKNEICVSLYGDGAANQGQIFETYNMAALWKLPCIFICENNRYGMGTSVERAAASTDYYK 254
Query: 255 RGDYVPGLKVDGMDVLAVKQACKFAKEH 282
RGDY+PGL+VDGMDVL V++A KFA +H
Sbjct: 255 RGDYIPGLRVDGMDVLCVREATKFAADH 282
>gb|EAA62343.1| hypothetical protein AN5162.2 [Aspergillus nidulans FGSC A4]
gi|67537984|ref|XP_662766.1| hypothetical protein
AN5162_2 [Aspergillus nidulans FGSC A4]
gi|49095676|ref|XP_409299.1| hypothetical protein
AN5162.2 [Aspergillus nidulans FGSC A4]
Length = 405
Score = 283 bits (723), Expect = 5e-75
Identities = 143/265 (53%), Positives = 187/265 (69%), Gaps = 5/265 (1%)
Query: 27 AAFSLRRSISSDSN-PITIETS-VPFTAHNCDPPSRSVETSTAELHSFFRQMATMRRMEI 84
AA S +I D N P T+ S F + DPP ++E + EL + M MRRME+
Sbjct: 31 AASSHAENIPEDENKPFTVRLSDESFETYEIDPPPYTLEVTKKELKQMYYDMVAMRRMEM 90
Query: 85 AADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFS 144
AAD LYK K IRGFCHL GQEAVA+G+E +T++D IITAYR H + RGGT+ I
Sbjct: 91 AADRLYKEKKIRGFCHLSTGQEAVAVGIEHALTREDKIITAYRCHGYAMMRGGTIRSIIG 150
Query: 145 ELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALY 204
EL+GR +G + GKGGSMH + NFYGG+GIVGAQVP+G G+AFAQ+Y+++++ + LY
Sbjct: 151 ELLGRREGIAYGKGGSMHMFAP--NFYGGNGIVGAQVPVGAGLAFAQQYNEEKSTSVVLY 208
Query: 205 GDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKV 264
GDGA+NQGQ+FEA N+A LW+LP + CENN YGMGT+ R++ YYKRG Y+PG+KV
Sbjct: 209 GDGASNQGQVFEAFNMAKLWNLPVLFGCENNKYGMGTSAARSSALTDYYKRGQYIPGIKV 268
Query: 265 DGMDVLAVKQACKFAKEHALK-NGP 288
+GMDVLA K A K+ K++A+ NGP
Sbjct: 269 NGMDVLATKAAVKYGKDYAISGNGP 293
>ref|NP_001017072.1| hypothetical protein LOC549826 [Xenopus tropicalis]
Length = 395
Score = 282 bits (722), Expect = 7e-75
Identities = 146/267 (54%), Positives = 181/267 (67%), Gaps = 14/267 (5%)
Query: 23 KPLTAAFSLRRSISSDSNPITIETSVPFTAHNCD-------PPSRSVETSTAELHSFFRQ 75
KP+TA R + + N + F CD PP+++V T L ++R
Sbjct: 18 KPVTAV----RVMVASRNYADFASEATFDIKKCDVHRLEEGPPTQAVLTREQGLQ-YYRT 72
Query: 76 MATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGR 135
M T+RRME+ +D LYK K+IRGFCHLYDGQEA +G+EA I D +ITAYR H R
Sbjct: 73 MQTIRRMELKSDQLYKQKIIRGFCHLYDGQEACCVGLEAAINPTDHLITAYRAHGYSYTR 132
Query: 136 GGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSK 195
G ++ EI +EL GR GC+KGKGGSMH Y K NFYGG+GIVGAQVPLG G+A A K+
Sbjct: 133 GVSVKEILAELTGRRGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGIALACKFFG 190
Query: 196 DEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKR 255
+ + ALYGDGAANQGQ+FE N+AALW LP I +CENN YGMGT+ RAA S YYKR
Sbjct: 191 KDEICVALYGDGAANQGQIFETYNMAALWKLPCIFICENNRYGMGTSVERAAASTDYYKR 250
Query: 256 GDYVPGLKVDGMDVLAVKQACKFAKEH 282
GDY+PGL+VDGMDVL V++A +FA +H
Sbjct: 251 GDYIPGLRVDGMDVLCVREATQFAADH 277
>gb|EAL20233.1| hypothetical protein CNBF0450 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57227932|gb|AAW44390.1| pyruvate
dehydrogenase e1 component alpha subunit, mitochondrial
precursor, putative [Cryptococcus neoformans var.
neoformans JEC21] gi|58269082|ref|XP_571697.1| pyruvate
dehydrogenase e1 component alpha subunit, mitochondrial
precursor, putative [Cryptococcus neoformans var.
neoformans JEC21]
Length = 413
Score = 282 bits (721), Expect = 9e-75
Identities = 139/249 (55%), Positives = 183/249 (72%), Gaps = 3/249 (1%)
Query: 36 SSDSNPITIETSVP-FTAHNCDPPSRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKL 94
+S S P ++ F ++ CD P + EL + +R M MRRME AAD+LYK KL
Sbjct: 47 ASGSEPFKVQLHADSFHSYRCDAPPPETTVTKDELINMYRTMVQMRRMEQAADALYKQKL 106
Query: 95 IRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCS 154
IRGFCHL GQEAV++GME IT +D +IT+YR H + RGGT+ + +ELMGR DG S
Sbjct: 107 IRGFCHLAIGQEAVSVGMETAITGQDRVITSYRCHTFAVLRGGTIKGVIAELMGRKDGMS 166
Query: 155 KGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQL 214
GKGGSMH + +F+GG+GIVGAQVP+G GVA AQKY+K++A TFALYGDGAANQGQ+
Sbjct: 167 FGKGGSMHIFTP--SFFGGNGIVGAQVPVGAGVALAQKYNKEKAATFALYGDGAANQGQV 224
Query: 215 FEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQ 274
FEA N+A LW+LP + VCENN YGMGT+ R++ + ++ RGD +PGL+V+GMD+LAV++
Sbjct: 225 FEAFNMAKLWNLPCVFVCENNKYGMGTSAERSSMNTQFFTRGDQIPGLQVNGMDILAVRE 284
Query: 275 ACKFAKEHA 283
A K+A+E A
Sbjct: 285 ATKWAREWA 293
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 481,223,799
Number of Sequences: 2540612
Number of extensions: 20394527
Number of successful extensions: 50137
Number of sequences better than 10.0: 799
Number of HSP's better than 10.0 without gapping: 635
Number of HSP's successfully gapped in prelim test: 164
Number of HSP's that attempted gapping in prelim test: 48678
Number of HSP's gapped (non-prelim): 944
length of query: 288
length of database: 863,360,394
effective HSP length: 127
effective length of query: 161
effective length of database: 540,702,670
effective search space: 87053129870
effective search space used: 87053129870
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0028b.3


