

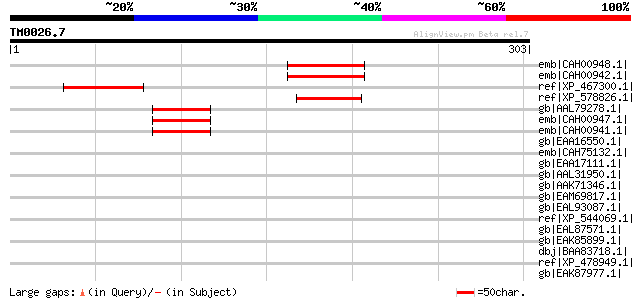
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.7
(303 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAH00948.1| unnamed protein product [Kluyveromyces lactis NR... 75 3e-12
emb|CAH00942.1| unnamed protein product [Kluyveromyces lactis NR... 75 3e-12
ref|XP_467300.1| hypothetical protein [Oryza sativa (japonica cu... 69 1e-10
ref|XP_578826.1| PREDICTED: similar to putative pheromone recept... 51 4e-05
gb|AAL79278.1| unknown [Saccharomyces cerevisiae] 50 8e-05
emb|CAH00947.1| unnamed protein product [Kluyveromyces lactis NR... 47 7e-04
emb|CAH00941.1| unnamed protein product [Kluyveromyces lactis NR... 47 7e-04
gb|EAA16550.1| hypothetical protein [Plasmodium yoelii yoelii] 44 0.006
emb|CAH75132.1| hypothetical protein PC100999.00.0 [Plasmodium c... 43 0.010
gb|EAA17111.1| hypothetical protein [Plasmodium yoelii yoelii] 43 0.010
gb|AAL31950.1| CDH1-D [Gallus gallus] 42 0.017
gb|AAK71346.1| ETS-related transcription factor ERF [Mus musculus] 39 0.19
gb|EAM69817.1| IMP dehydrogenase/GMP reductase:AsmA [Desulfuromo... 37 0.71
gb|EAL93087.1| APSES transcription factor (StuA), putative [Aspe... 37 0.71
ref|XP_544069.1| PREDICTED: similar to Zinc finger protein SLUG ... 37 0.71
gb|EAL87571.1| WD repeat protein [Aspergillus fumigatus Af293] 36 1.2
gb|EAK85899.1| hypothetical protein UM05039.1 [Ustilago maydis 5... 36 1.2
dbj|BAA83718.1| RNA binding protein [Homo sapiens] gi|19923466|r... 35 3.5
ref|XP_478949.1| Epstein-Barr virus EBNA-1-like protein [Oryza s... 35 3.5
gb|EAK87977.1| hypothetical protein cgd5_2570 [Cryptosporidium p... 35 3.5
>emb|CAH00948.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50307719|ref|XP_453852.1| unnamed protein product
[Kluyveromyces lactis]
Length = 114
Score = 74.7 bits (182), Expect = 3e-12
Identities = 34/45 (75%), Positives = 39/45 (86%)
Query: 163 FNRLRSRSLTELTTQIAPPNKNGHAPPHIESRKSSQSVNPYYVWT 207
F+RLRSRSL++L+ QI PP KNGHAPP +SRKSSQSVNPY VWT
Sbjct: 69 FSRLRSRSLSQLSRQITPPTKNGHAPPPTKSRKSSQSVNPYCVWT 113
>emb|CAH00942.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50312723|ref|XP_453846.1| unnamed protein product
[Kluyveromyces lactis]
Length = 119
Score = 74.7 bits (182), Expect = 3e-12
Identities = 34/45 (75%), Positives = 39/45 (86%)
Query: 163 FNRLRSRSLTELTTQIAPPNKNGHAPPHIESRKSSQSVNPYYVWT 207
F+RLRSRSL++L+ QI PP KNGHAPP +SRKSSQSVNPY VWT
Sbjct: 74 FSRLRSRSLSQLSRQITPPTKNGHAPPPTKSRKSSQSVNPYCVWT 118
>ref|XP_467300.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|41052959|dbj|BAD07869.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 109
Score = 69.3 bits (168), Expect = 1e-10
Identities = 33/47 (70%), Positives = 35/47 (74%)
Query: 32 NGALTLSGAPFQGTWARSATEDASLDYNSDALGDRFSWWALPGRRAL 78
+GALTL GAPFQGTWARS EDAS DYNSD RFS WA PG A+
Sbjct: 3 DGALTLPGAPFQGTWARSVAEDASPDYNSDGAAARFSSWASPGSLAV 49
>ref|XP_578826.1| PREDICTED: similar to putative pheromone receptor [Rattus
norvegicus]
Length = 374
Score = 51.2 bits (121), Expect = 4e-05
Identities = 25/38 (65%), Positives = 27/38 (70%)
Query: 168 SRSLTELTTQIAPPNKNGHAPPHIESRKSSQSVNPYYV 205
S SL+ELT IAPP KN H PP +SRKS QSVNP V
Sbjct: 4 SSSLSELTRHIAPPTKNSHEPPPTDSRKSYQSVNPVCV 41
>gb|AAL79278.1| unknown [Saccharomyces cerevisiae]
Length = 67
Score = 50.1 bits (118), Expect = 8e-05
Identities = 24/34 (70%), Positives = 27/34 (78%)
Query: 84 GATCVQRLNGSRDSAIHTKYRISLRSSSMRDDND 117
GA CVQR + SR+SAIH YRISLRSSSMR+ D
Sbjct: 8 GAMCVQRFDDSRNSAIHITYRISLRSSSMREPRD 41
>emb|CAH00947.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50307717|ref|XP_453851.1| unnamed protein product
[Kluyveromyces lactis]
Length = 54
Score = 47.0 bits (110), Expect = 7e-04
Identities = 23/34 (67%), Positives = 25/34 (72%)
Query: 84 GATCVQRLNGSRDSAIHTKYRISLRSSSMRDDND 117
GA CVQR + SR SAIH YR SLRSSSMR+ D
Sbjct: 8 GAMCVQRFDDSRKSAIHNTYRNSLRSSSMREPRD 41
>emb|CAH00941.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50307709|ref|XP_453845.1| unnamed protein product
[Kluyveromyces lactis]
Length = 103
Score = 47.0 bits (110), Expect = 7e-04
Identities = 23/34 (67%), Positives = 25/34 (72%)
Query: 84 GATCVQRLNGSRDSAIHTKYRISLRSSSMRDDND 117
GA CVQR + SR SAIH YR SLRSSSMR+ D
Sbjct: 57 GAMCVQRFDDSRKSAIHNTYRNSLRSSSMREPRD 90
>gb|EAA16550.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 99
Score = 43.9 bits (102), Expect = 0.006
Identities = 20/32 (62%), Positives = 25/32 (77%)
Query: 114 DDNDSSAGSPTETLLRLLLPLNDKVQWTSHNV 145
++ND SAGSPTETLLRLLLPL D++ S +
Sbjct: 38 NNNDPSAGSPTETLLRLLLPLKDRIYGPSKKI 69
>emb|CAH75132.1| hypothetical protein PC100999.00.0 [Plasmodium chabaudi]
Length = 43
Score = 43.1 bits (100), Expect = 0.010
Identities = 20/30 (66%), Positives = 23/30 (76%)
Query: 116 NDSSAGSPTETLLRLLLPLNDKVQWTSHNV 145
ND SAGSPTETLLRLLLPL D++ S +
Sbjct: 12 NDPSAGSPTETLLRLLLPLKDRIYGPSKKI 41
>gb|EAA17111.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 402
Score = 43.1 bits (100), Expect = 0.010
Identities = 20/30 (66%), Positives = 23/30 (76%)
Query: 116 NDSSAGSPTETLLRLLLPLNDKVQWTSHNV 145
ND SAGSPTETLLRLLLPL D++ S +
Sbjct: 27 NDPSAGSPTETLLRLLLPLKDRIYGPSKKI 56
>gb|AAL31950.1| CDH1-D [Gallus gallus]
Length = 451
Score = 42.4 bits (98), Expect = 0.017
Identities = 21/30 (70%), Positives = 25/30 (83%)
Query: 113 RDDNDSSAGSPTETLLRLLLPLNDKVQWTS 142
R ND SAGSPTETLLRLLLPL+ +V+ +S
Sbjct: 247 RPGNDPSAGSPTETLLRLLLPLDSQVRPSS 276
>gb|AAK71346.1| ETS-related transcription factor ERF [Mus musculus]
Length = 259
Score = 38.9 bits (89), Expect = 0.19
Identities = 18/22 (81%), Positives = 20/22 (90%)
Query: 116 NDSSAGSPTETLLRLLLPLNDK 137
ND SAGSPTETLLRLLLPL+ +
Sbjct: 214 NDPSAGSPTETLLRLLLPLDSQ 235
>gb|EAM69817.1| IMP dehydrogenase/GMP reductase:AsmA [Desulfuromonas acetoxidans
DSM 684]
Length = 798
Score = 37.0 bits (84), Expect = 0.71
Identities = 35/120 (29%), Positives = 56/120 (46%), Gaps = 10/120 (8%)
Query: 159 FTGPFNRLRSRSLTELTTQIAPPNKNGHAPPHIESRKSSQSVNPYYVWTCCSSFINPRIS 218
F+G +RL+S+ +LT +IA AP +E SV Y V S R+
Sbjct: 675 FSGSRSRLKSQGTVQLTKEIASTLDVYLAPALVEKLNPGDSVRKYMV----SQDGWGRV- 729
Query: 219 PLTM--KYECPQLSLSIITPMPKAKTIGKERIASKRDEPTGEHQRQTDRPNPRSNYELFN 276
PLT+ +Y+ PQLS ++ KA + ++ K + TG+ +T P + +L N
Sbjct: 730 PLTISGRYDRPQLSYNMTVVGQKASEVVTRKLQEKLQKHTGD---ETAEPLVKPAADLLN 786
>gb|EAL93087.1| APSES transcription factor (StuA), putative [Aspergillus fumigatus
Af293]
Length = 635
Score = 37.0 bits (84), Expect = 0.71
Identities = 36/138 (26%), Positives = 48/138 (34%), Gaps = 17/138 (12%)
Query: 140 WTSHNVASSEPPTLPQSEHFTGPFNRLRSRSLTELTTQIAPPNKNGHAPPHIESRKSSQS 199
W + + S P T P S T + RS T TT PP N +S+ +
Sbjct: 328 WNNQGMNSGVPNTQPLSIDTT--LSNTRSMPTTPATT---PPGNNLQGMQSYQSQSGYDT 382
Query: 200 VNPYYVWTCCSSFINPRISPLTMKYECPQLSLSII-TPMPKAKTIGKERIASKRDEPTGE 258
PYY +P + + PQ SLS PMP I E P G+
Sbjct: 383 SKPYY-----------STAPPSHPHYAPQYSLSQYGQPMPPHSYIKNEMAPPAGRAPGGQ 431
Query: 259 HQRQTDRPNPRSNYELFN 276
+ +T P Y N
Sbjct: 432 SETETSDVKPADRYSQSN 449
>ref|XP_544069.1| PREDICTED: similar to Zinc finger protein SLUG (Neural crest
transcription factor Slug) (Snail homolog 2) [Canis
familiaris]
Length = 268
Score = 37.0 bits (84), Expect = 0.71
Identities = 37/138 (26%), Positives = 59/138 (41%), Gaps = 7/138 (5%)
Query: 153 LPQSEHFTGPFNRLRSRSLTELTTQ---IAPPNKNGHAPPHIESRK--SSQSVNPYYVWT 207
+P+S FN + + EL T I+P + P I + SS + +P VWT
Sbjct: 1 MPRSFLVKKHFNASKKPNYGELDTHTVIISPYLYESYPMPVIPQPEILSSGAYSPITVWT 60
Query: 208 CCSSFINPRISPLTMKYECPQLSLSIITPMPKAKTIGKERIASKRDEPTGEHQRQTDRPN 267
+ F +P + L+ P SL ++P P + T K+ S+ E + Q+ +
Sbjct: 61 TAAPFHSPLPNGLSSLSGYPS-SLGRVSPPPPSDTSSKDHSGSESPISDEEERLQSKLSD 119
Query: 268 PRS-NYELFNCNNLNIRY 284
P + E F CN N Y
Sbjct: 120 PHAIEAEKFQCNLCNKTY 137
>gb|EAL87571.1| WD repeat protein [Aspergillus fumigatus Af293]
Length = 721
Score = 36.2 bits (82), Expect = 1.2
Identities = 24/78 (30%), Positives = 36/78 (45%), Gaps = 2/78 (2%)
Query: 110 SSMRDDNDSSAGSPTETLLRLLLPLNDKVQWTSHNVASSEPPTLPQSEHFTGPFNRLRS- 168
SS DD+DSS+ S E L+R P + K+ +S+ + PP + S T R RS
Sbjct: 598 SSSDDDDDSSSASEAERLVRSRSPHSGKISTSSYKRTNRPPPQMRVSSSTTSHSTRDRSF 657
Query: 169 -RSLTELTTQIAPPNKNG 185
+ T+ P + G
Sbjct: 658 ASRAQNMRTKTKPARRGG 675
>gb|EAK85899.1| hypothetical protein UM05039.1 [Ustilago maydis 521]
gi|49077634|ref|XP_402654.1| hypothetical protein
UM05039.1 [Ustilago maydis 521]
Length = 1530
Score = 36.2 bits (82), Expect = 1.2
Identities = 31/126 (24%), Positives = 51/126 (39%), Gaps = 10/126 (7%)
Query: 7 DARSTQVSKHAETARAAIHNHGSGHNGALTLSGAPFQ---GTWARSATEDASLDYNSDAL 63
DA + + T+ A I + H + F+ +W R A L+ S ++
Sbjct: 74 DAANVKQKLSIRTSTAQIRSSEPSHEAIFERASFGFRHGGNSWLREKPSKARLNAPSSSV 133
Query: 64 GDRFSWWALPGRRALNLMASGATCVQRLNGSRDSAIHTKYRISLRSSSMRDDNDSSAGSP 123
G W +RA+ + A+ + G +S HT Y +L SS +R +DS G P
Sbjct: 134 GPLGQW-----KRAILIFRENASL--SIFGEANSLAHTVYCDALSSSDVRSVDDSLFGRP 186
Query: 124 TETLLR 129
+R
Sbjct: 187 HVLFIR 192
>dbj|BAA83718.1| RNA binding protein [Homo sapiens] gi|19923466|ref|NP_057417.2|
splicing coactivator subunit SRm300 [Homo sapiens]
Length = 2752
Score = 34.7 bits (78), Expect = 3.5
Identities = 27/95 (28%), Positives = 37/95 (38%), Gaps = 11/95 (11%)
Query: 189 PHIESRKSSQSVNPYYVWTCCSSFINPRISPLTMKYECPQLSLSIITPMPKAKTIGKERI 248
P ++SR S+ CS PR+ T + P S S P PK K I R
Sbjct: 879 PELKSRTPSRH--------SCSGSSPPRVKSSTPPRQSPSRSSS---PQPKVKAIISPRQ 927
Query: 249 ASKRDEPTGEHQRQTDRPNPRSNYELFNCNNLNIR 283
S + R T R PR + + C+N+ R
Sbjct: 928 RSHSGSSSPSPSRVTSRTTPRRSRSVSPCSNVESR 962
>ref|XP_478949.1| Epstein-Barr virus EBNA-1-like protein [Oryza sativa (japonica
cultivar-group)] gi|34394490|dbj|BAC83754.1|
Epstein-Barr virus EBNA-1-like protein [Oryza sativa
(japonica cultivar-group)] gi|34393443|dbj|BAC82982.1|
Epstein-Barr virus EBNA-1-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 482
Score = 34.7 bits (78), Expect = 3.5
Identities = 28/78 (35%), Positives = 35/78 (43%), Gaps = 9/78 (11%)
Query: 2 GSPQADARSTQVSKHAETARA--AIHNHGSGHNGALTLSGAPFQGTWARSATEDASLDYN 59
GSP+A T++S T R+ A H HG G + LT G GT AR T+ LD
Sbjct: 266 GSPRAVPGGTRLSVTRFTVRSEHAAHAHGRGRDAVLTARG----GTRARGGTDRGRLD-- 319
Query: 60 SDALGDRFSWWALPGRRA 77
L + W L G A
Sbjct: 320 -PVLAELAPTWRLRGCHA 336
>gb|EAK87977.1| hypothetical protein cgd5_2570 [Cryptosporidium parvum]
gi|66358032|ref|XP_626194.1| hypothetical protein
cgd5_2570 [Cryptosporidium parvum]
Length = 368
Score = 34.7 bits (78), Expect = 3.5
Identities = 22/89 (24%), Positives = 37/89 (40%), Gaps = 1/89 (1%)
Query: 138 VQWTSHNVASSEPPTLPQSEHFTGPFNRLRSRSLTELTTQIAPPNKNGHAPPHIESRKSS 197
+ + ++ P++ ++ N L S +L E + N H+PP I S K S
Sbjct: 87 LSYNGSSINEQNSPSVTSNKRLASLKNELYSENLIERNVTASNNNAANHSPPSISSVKYS 146
Query: 198 QSVNPYYVWTCCSSFINPRISP-LTMKYE 225
Y SS ++P +SP K+E
Sbjct: 147 SLKGHYNNIDSSSSPVSPEVSPNFVFKFE 175
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.128 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 552,054,019
Number of Sequences: 2540612
Number of extensions: 22831401
Number of successful extensions: 60593
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 60551
Number of HSP's gapped (non-prelim): 62
length of query: 303
length of database: 863,360,394
effective HSP length: 127
effective length of query: 176
effective length of database: 540,702,670
effective search space: 95163669920
effective search space used: 95163669920
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0026.7


