

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.4
(236 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
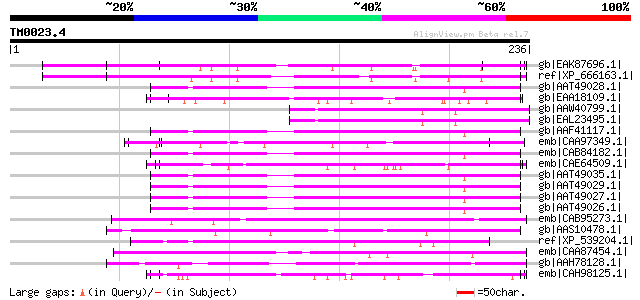
Score E
Sequences producing significant alignments: (bits) Value
gb|EAK87696.1| large low complexity coiled coil protien with lar... 65 1e-09
ref|XP_666163.1| myosin heavy chain [Cryptosporidium hominis] gi... 65 2e-09
gb|AAT49028.1| IgA1 protease [Neisseria meningitidis] gi|4903742... 58 2e-07
gb|EAA18109.1| erythrocyte binding protein [Plasmodium yoelii yo... 56 7e-07
gb|AAW40799.1| glycosyltransferase, putative [Cryptococcus neofo... 56 1e-06
gb|EAL23495.1| hypothetical protein CNBA1420 [Cryptococcus neofo... 56 1e-06
gb|AAF41117.1| IgA-specific serine endopeptidase [Neisseria meni... 55 2e-06
emb|CAA97349.1| SPAC17C9.03 [Schizosaccharomyces pombe] gi|19115... 55 2e-06
emb|CAB84182.1| IgA1 protease [Neisseria meningitidis Z2491] gi|... 55 2e-06
emb|CAE64509.1| Hypothetical protein CBG09244 [Caenorhabditis br... 55 2e-06
gb|AAT49035.1| IgA1 protease [Neisseria meningitidis] 55 2e-06
gb|AAT49029.1| IgA1 protease [Neisseria meningitidis] 55 2e-06
gb|AAT49027.1| IgA1 protease [Neisseria meningitidis] 55 2e-06
gb|AAT49026.1| IgA1 protease [Neisseria meningitidis] gi|4903743... 55 2e-06
emb|CAB95273.1| possible chromosome assembly protein [Leishmania... 55 2e-06
gb|AAS10478.1| merozoite surface protein 3b [Plasmodium vivax] 54 3e-06
ref|XP_539204.1| PREDICTED: similar to plectin 1 [Canis familiaris] 54 4e-06
emb|CAA87454.1| neurofilament protein M [Oryctolagus cuniculus] ... 54 4e-06
gb|AAH78128.1| LOC397995 protein [Xenopus laevis] 54 4e-06
emb|CAH98125.1| MAEBL, putative [Plasmodium berghei] 54 4e-06
>gb|EAK87696.1| large low complexity coiled coil protien with large repeat region
[Cryptosporidium parvum] gi|66356902|ref|XP_625629.1|
hypothetical protein cgd4_200 [Cryptosporidium parvum]
Length = 1833
Score = 65.5 bits (158), Expect = 1e-09
Identities = 56/197 (28%), Positives = 96/197 (48%), Gaps = 14/197 (7%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDA-S 103
E + K E A + E A+ K EK+E + KK E +AK++K +A +
Sbjct: 770 EEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEA 829
Query: 104 KGTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEI 163
K +A A A K E +A A+ + EEA+A + K +AE AK+ +
Sbjct: 830 KAKKEKEEAEAEAEAKAK-----KEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKE 884
Query: 164 -----AKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVK 218
AKK+ E+ A +K+ +EA+ AK K +E AEA ++++EEA + +K ++K
Sbjct: 885 EAEAKAKKEKEEAEAKAKKEKEEAE---AKAKKEKEEAEAKAKKEKEEADTKEKEKTEIK 941
Query: 219 VGGSGEVNQEEQAKNDQ 235
+ + EE +++ +
Sbjct: 942 ENSKIDSDSEENSESKE 958
Score = 59.3 bits (142), Expect = 9e-08
Identities = 49/160 (30%), Positives = 79/160 (48%), Gaps = 19/160 (11%)
Query: 69 AKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKG 128
AK ++ EK+E + KK E +AK++K +A L + A + K
Sbjct: 693 AKAKKEKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKAKKE------KE 746
Query: 129 VEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEI-----AKKQAEDTLAGVRKQLQEAD 183
E +A A+ + EEA+A + K +AE AK+ + AKK+ E+ A +K+ +EA+
Sbjct: 747 KEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAE 806
Query: 184 --------EVLAKDKAARESAEASDEEKEEEAKGEGVKKA 215
E AK K +E AEA ++++EEA+ E KA
Sbjct: 807 AKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAEAEAKA 846
Score = 58.2 bits (139), Expect = 2e-07
Identities = 59/211 (27%), Positives = 97/211 (45%), Gaps = 30/211 (14%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDA-S 103
E + K E A + E A+ K EK+E + KK E +AK++K +A +
Sbjct: 748 EEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEA 807
Query: 104 KGTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKA-AYSRLKSDAENAAKRAE 162
K +A A A K E +A A+ + EEA+A A ++ K + E A +A+
Sbjct: 808 KAKKEKEEAEAKAK---------KEKEEAEAKAKKEKEEAEAEAEAKAKKEKEEAEAKAK 858
Query: 163 I--------AKKQAEDTLAGVRKQLQEA--------DEVLAKDKAARESAEASDEEKEEE 206
AKK+ E+ A +K+ +EA +E AK K +E AEA ++++EE
Sbjct: 859 KEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEE 918
Query: 207 AKGEGVK---KADVKVGGSGEVNQEEQAKND 234
A+ + K +AD K E+ + + +D
Sbjct: 919 AEAKAKKEKEEADTKEKEKTEIKENSKIDSD 949
Score = 56.2 bits (134), Expect = 7e-07
Identities = 51/193 (26%), Positives = 87/193 (44%), Gaps = 12/193 (6%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASK 104
E K + E A + E A+ K EK+E + K E +AK++K
Sbjct: 645 EAAKVRAQKEKEEAEAKALKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKAKKEKEKEEA 704
Query: 105 GTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEI- 163
A + A + A + KA + + EAKA + K +AE AK+ +
Sbjct: 705 EAKAKKEKEEAEAKAKKEKEEAEA----KALKEKEEAEAKAKKEKEKEEAEAKAKKEKEE 760
Query: 164 ----AKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKV 219
AKK+ E+ A +K+ +EA+ AK K +E AEA ++++EEA+ + K+ +
Sbjct: 761 AEAKAKKEKEEAEAKAKKEKEEAE---AKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAE 817
Query: 220 GGSGEVNQEEQAK 232
+ + +E +AK
Sbjct: 818 AKAKKEKEEAEAK 830
Score = 48.1 bits (113), Expect = 2e-04
Identities = 59/230 (25%), Positives = 107/230 (45%), Gaps = 32/230 (13%)
Query: 16 SVSFNSVSRLKSSPTIFLFTALYSVFCAAEMVKNKGGVNPESMSAFIASQVETAKGHRKN 75
S S +S S + S P +A+ S E ++ P S+S +V++ G +
Sbjct: 552 SKSISSKSAISSLPKPKAKSAVKSAPATEENNGDQTISEPASIS-----EVKSKAGSKSM 606
Query: 76 EKKETKGVRK-KSPLV------------DEREAKRQKSDASKGTLAVNQALAAASCATGL 122
++K V K KS +V ++ E K + +A+K + A A
Sbjct: 607 PVSKSKSVIKAKSQVVAKSDMDSGQIVKEDNETKDNEDEAAKVRAQKEKEEAEAKA---- 662
Query: 123 LPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEA 182
LK E +A A+ + EEA+A + K +AE AK+ +K+ E+ A +K+ +EA
Sbjct: 663 ---LKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKAKK----EKEKEEAEAKAKKEKEEA 715
Query: 183 DEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ AK K +E AEA +++EEA+ + K+ + + + ++E+A+
Sbjct: 716 E---AKAKKEKEEAEAKALKEKEEAEAKAKKEKEKEEAEAKAKKEKEEAE 762
Score = 45.1 bits (105), Expect = 0.002
Identities = 51/214 (23%), Positives = 92/214 (42%), Gaps = 26/214 (12%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDAS- 103
E + K E A + E A+ K EK+E + KK E +AK++K +A
Sbjct: 873 EEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEADT 932
Query: 104 ----KGTLAVNQALAAAS---------CATGLLPHLKGVEIDKACAQLKLEEAKAAYSRL 150
K + N + + S +TGLL K+ + L + + ++
Sbjct: 933 KEKEKTEIKENSKIDSDSEENSESKEKKSTGLLAKTIAKAKAKSKSNSVLSKLPSKVNKE 992
Query: 151 KSDAENAAKRAEIAK---------KQAEDTLAGVRKQLQEADEVLAKDKAARESA--EAS 199
S AEN + EI K K+ ED++A ++ ++ L K K+ +E E
Sbjct: 993 TSKAENDLEN-EIEKDDESNKKETKKEEDSIAKLKAKVPVKPSPLLKSKSEKEKEKEEDK 1051
Query: 200 DEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKN 233
DEEK+E+ K + K + + G + N++++ ++
Sbjct: 1052 DEEKKEKDKEKKEKLKEKEEEGKEKSNEKDKGED 1085
Score = 41.6 bits (96), Expect = 0.019
Identities = 43/185 (23%), Positives = 78/185 (41%), Gaps = 25/185 (13%)
Query: 64 SQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLL 123
S+ E K + EKKE + KK L ++ E ++K + + + S +
Sbjct: 1148 SEKEKEKEDKDEEKKEKEDKEKKEKLKEKGEEGKEKEEGKEKEKEKEKDEKDKSKS---- 1203
Query: 124 PHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEAD 183
K + +K KL+E + + D+ + E KK+ ED +A ++ ++
Sbjct: 1204 ---KTKDFEKE----KLKETEKGEKEAEKDSSKKESKDE-EKKEKEDPIAKLKAKVPVKP 1255
Query: 184 EVLAKDKAARESA--EASDEEKEE-----------EAKGEGVKKADVKVGGSGEVNQEEQ 230
L K K+ +E E DEEK+E E + EG +K KV + +++E+
Sbjct: 1256 SPLLKSKSEKEKEKEEDKDEEKKEKEDKEKKEKLKEKEEEGKEKEKEKVKAQKKKDEKEE 1315
Query: 231 AKNDQ 235
D+
Sbjct: 1316 KDKDE 1320
Score = 38.5 bits (88), Expect = 0.16
Identities = 43/177 (24%), Positives = 80/177 (44%), Gaps = 32/177 (18%)
Query: 64 SQVETAKGHRKNEKKETKGVRKKSPLVD-EREAKRQKSDASKGTLAVNQALAAASCATGL 122
S+ E K K+E+K+ K KK L + E E K + ++ KG
Sbjct: 1041 SEKEKEKEEDKDEEKKEKDKEKKEKLKEKEEEGKEKSNEKDKGE---------------- 1084
Query: 123 LPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRA--EIAKKQAEDTLAGVRKQLQ 180
E DK+ +++K + + +L+ +N+ K+ + KK+ ED +A ++ ++
Sbjct: 1085 ----DKDEKDKSKSKIKDSQKE---EKLEETEKNSLKKESKDDEKKEKEDPIAKLKAKVP 1137
Query: 181 EADEVLAKDKAARESA--EASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQ 235
L K K+ +E + +E+KE+E K KK +K G E ++E+ K +
Sbjct: 1138 VKPSPLLKSKSEKEKEKEDKDEEKKEKEDK---EKKEKLKEKGE-EGKEKEEGKEKE 1190
Score = 34.7 bits (78), Expect = 2.3
Identities = 41/200 (20%), Positives = 81/200 (40%), Gaps = 24/200 (12%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASK 104
E +K KG E + E K + K +TK K+ E+ K + D+SK
Sbjct: 1171 EKLKEKGEEGKEKEEGK-EKEKEKEKDEKDKSKSKTKDFEKEKLKETEKGEKEAEKDSSK 1229
Query: 105 GTLAVNQ--------ALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAEN 156
+ A A P LK K+ + + EE K + K D E
Sbjct: 1230 KESKDEEKKEKEDPIAKLKAKVPVKPSPLLKS----KSEKEKEKEEDKDEEKKEKEDKE- 1284
Query: 157 AAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKK-- 214
K+ ++ +K+ E G K+ ++ KD+ + + +++KE+ + E ++K
Sbjct: 1285 --KKEKLKEKEEE----GKEKEKEKVKAQKKKDEKEEKDKDEKEDDKEKSKESENLEKKP 1338
Query: 215 --ADVKVGGSGEVNQEEQAK 232
++ + G++++++ K
Sbjct: 1339 TISEKVINPEGKIDEKDSEK 1358
>ref|XP_666163.1| myosin heavy chain [Cryptosporidium hominis]
gi|54657105|gb|EAL35936.1| myosin heavy chain
[Cryptosporidium hominis]
Length = 1604
Score = 65.1 bits (157), Expect = 2e-09
Identities = 53/188 (28%), Positives = 89/188 (47%), Gaps = 17/188 (9%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASK 104
E + K E A + E A+ K EK+E + KK E +AK++K +A
Sbjct: 667 EEAEAKANKEKEEAEAKALKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEA 726
Query: 105 GTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIA 164
L + A + E ++A A+ K E+ +A LK E AK A
Sbjct: 727 KALKEKEEAEAKAKK----------EKEEAEAKAKKEKEEAEAKALKEKEEAEAK----A 772
Query: 165 KKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGE 224
KK+ E+T A +K+ +EA+ AK K +E AEA ++++EEA+ + +K+ + + +
Sbjct: 773 KKEKEETEAKAKKEKEEAE---AKAKKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKAKK 829
Query: 225 VNQEEQAK 232
+E +AK
Sbjct: 830 EKEEAEAK 837
Score = 63.9 bits (154), Expect = 3e-09
Identities = 63/193 (32%), Positives = 90/193 (45%), Gaps = 23/193 (11%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDA-S 103
E + K E A + E A+ K EK+ET+ KK E +AK++K +A +
Sbjct: 744 EEAEAKAKKEKEEAEAKALKEKEEAEAKAKKEKEETEAKAKKEKEEAEAKAKKEKEEAEA 803
Query: 104 KGTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEI 163
K +A A A LK E +A A+ + EEA+A + K +AE AK+
Sbjct: 804 KAKKEKEEAEAKA---------LKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKE-- 852
Query: 164 AKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEA-SDEEKEEEAKGEGVK---KADVKV 219
K++AE K L+E +E AK K +E AEA + +EKEE E K K D K+
Sbjct: 853 -KEEAE------AKALKEKEEAEAKGKKEKEEAEAKAKKEKEETDAKEKEKTKIKEDSKI 905
Query: 220 GGSGEVNQEEQAK 232
E N E + K
Sbjct: 906 DSDSEENSESKEK 918
Score = 58.5 bits (140), Expect = 1e-07
Identities = 54/204 (26%), Positives = 91/204 (44%), Gaps = 21/204 (10%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASK 104
E + K E A + E A+ EK+E + KK E +AK++K +A
Sbjct: 700 EEAEAKAKKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKAKKEKEEAEAKAKKEKEEAEA 759
Query: 105 GTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEI- 163
L + A + K E +A A+ + EEA+A + K +AE AK+ +
Sbjct: 760 KALKEKEEAEAKA--------KKEKEETEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEE 811
Query: 164 ----AKKQAEDTLAGVRKQLQEAD--------EVLAKDKAARESAEASDEEKEEEAKGEG 211
A K+ E+ A +K+ +EA+ E AK K +E AEA +++EEA+ +G
Sbjct: 812 AEAKALKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKG 871
Query: 212 VKKADVKVGGSGEVNQEEQAKNDQ 235
K+ + + + +E AK +
Sbjct: 872 KKEKEEAEAKAKKEKEETDAKEKE 895
Score = 53.5 bits (127), Expect = 5e-06
Identities = 62/243 (25%), Positives = 111/243 (45%), Gaps = 38/243 (15%)
Query: 16 SVSFNSVSRLKSSPTIFLFTALYSVFCAAEMVKNKGGVNPESMSAFIASQVETAKGHRKN 75
S S +S S + S P +A+ S E ++ P S+S +V++ G +
Sbjct: 552 SKSISSKSAISSLPKSKAKSAVKSAPATEENNGDQTISEPASIS-----EVKSKAGSKSM 606
Query: 76 EKKETK-GVRKKSPLV------------DEREAKRQKSDASKGTLAVNQALAAASCATGL 122
++K G++ KS +V ++ E K + +A+K + A A
Sbjct: 607 PVSKSKSGIKAKSQVVAKSDMDSGKIVKEDNETKDNEDEAAKVRAQKEKEEAEAKA---- 662
Query: 123 LPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEI-----AKKQAEDTLAGVRK 177
LK E +A A + EEA+A + K +AE AK+ + AKK+ E+ A +K
Sbjct: 663 ---LKEKEEAEAKANKEKEEAEAKALKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKK 719
Query: 178 QLQEAD--------EVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEE 229
+ +EA+ E AK K +E AEA ++++EEA+ + +K+ + + + +E
Sbjct: 720 EKEEAEAKALKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKAKKEKEET 779
Query: 230 QAK 232
+AK
Sbjct: 780 EAK 782
Score = 44.7 bits (104), Expect = 0.002
Identities = 45/190 (23%), Positives = 80/190 (41%), Gaps = 30/190 (15%)
Query: 64 SQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLL 123
S+ E K K+EKKE + KK L ++ E ++K + + ++ S +
Sbjct: 1110 SEKEKEKEEDKDEKKEKEDKEKKEKLKEKEEEGKEKEEGKEKEKDKDKDEKDKSKS---- 1165
Query: 124 PHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEAD 183
K + +K KL+E + + D + E KK+ ED +A ++ ++
Sbjct: 1166 ---KTKDFEKE----KLKETEKGEKEAEKDTSKKESKDE-EKKEKEDPIAKLKAKVPVKP 1217
Query: 184 EVLAKDKAARESAEASD----------------EEKEEEA--KGEGVKKADVKVGGSGEV 225
L K K+ +E + D +EKEEE K EG +K KV +
Sbjct: 1218 SPLLKSKSEKEKEKEEDKDEKKEKEDKEKKEKLKEKEEEGKEKEEGKEKEKEKVKAQKKK 1277
Query: 226 NQEEQAKNDQ 235
+++E+ ND+
Sbjct: 1278 DEKEEKDNDE 1287
Score = 42.4 bits (98), Expect = 0.011
Identities = 39/193 (20%), Positives = 84/193 (43%), Gaps = 23/193 (11%)
Query: 47 VKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGT 106
+K K V P + + + + + + EKKE + KK L ++ E ++K + +
Sbjct: 984 LKAKVPVKPSPLLKSKSEKEKEKEEDKDEEKKEKEDKEKKEKLKEKEEEGKEKEEGKEKE 1043
Query: 107 LAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKK 166
++ E DK+ +++K + + + D+ + E KK
Sbjct: 1044 KEKDKD-----------------EKDKSKSKIKESQKEEKLEETEKDSSKKESKDE-EKK 1085
Query: 167 QAEDTLAGVRKQLQEADEVLAKDKAARESAEASD----EEKEEEAKGEGVKKADVKVGGS 222
+ ED +A ++ ++ L K K+ +E + D +EKE++ K E +K+ + + G
Sbjct: 1086 EKEDPIAKLKAKVPVKPSPLLKSKSEKEKEKEEDKDEKKEKEDKEKKEKLKEKE-EEGKE 1144
Query: 223 GEVNQEEQAKNDQ 235
E +E++ D+
Sbjct: 1145 KEEGKEKEKDKDK 1157
Score = 42.4 bits (98), Expect = 0.011
Identities = 46/182 (25%), Positives = 79/182 (43%), Gaps = 22/182 (12%)
Query: 67 ETAKGHRKNEKKETKGVRKKSPLV--------DEREAKRQKSDASKGTLAVNQALAAASC 118
E A+ K EK+ET K+ + D E K S G LA A A A
Sbjct: 876 EEAEAKAKKEKEETDAKEKEKTKIKEDSKIDSDSEENSESKEKKSTGLLAKTIAKAKAKS 935
Query: 119 ATGLLPHLKGVEIDKACAQLKLEEAKAAYS-RLKSDAENAAKRAEIAKKQAEDTLAGVRK 177
+ + + K +++ E +KA K + ++ + + EI K+ ED++A ++
Sbjct: 936 KSNSV-------LSKLPSKVNKETSKAENDLENKIEKDDESNKKEIKKE--EDSIAKLKA 986
Query: 178 QLQEADEVLAKDKAARESA--EASDEEKEEEAKGEGVKKADVK--VGGSGEVNQEEQAKN 233
++ L K K+ +E E DEEK+E+ E +K K G E +E++ +
Sbjct: 987 KVPVKPSPLLKSKSEKEKEKEEDKDEEKKEKEDKEKKEKLKEKEEEGKEKEEGKEKEKEK 1046
Query: 234 DQ 235
D+
Sbjct: 1047 DK 1048
Score = 34.3 bits (77), Expect = 3.0
Identities = 40/176 (22%), Positives = 76/176 (42%), Gaps = 16/176 (9%)
Query: 67 ETAKGHRKNEKKETKGVRKKSPLVDERE------AKRQKSDASKGTLAVNQALAAASCAT 120
E K K+EK ++K K+S ++ E +K++ D K A A
Sbjct: 1041 EKEKEKDKDEKDKSKSKIKESQKEEKLEETEKDSSKKESKDEEKKEKEDPIAKLKAKVPV 1100
Query: 121 GLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQ 180
P LK K+ + + EE K K + E+ K+ ++ +K+ E K+ +
Sbjct: 1101 KPSPLLKS----KSEKEKEKEEDKDE----KKEKEDKEKKEKLKEKEEEGKEKEEGKEKE 1152
Query: 181 EADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQV 236
+ + KDK+ ++ + E+ +E KGE K+A+ ++E++ K D +
Sbjct: 1153 KDKDKDEKDKSKSKTKDFEKEKLKETEKGE--KEAEKDTSKKESKDEEKKEKEDPI 1206
>gb|AAT49028.1| IgA1 protease [Neisseria meningitidis] gi|49037422|gb|AAT49018.1|
IgA1 protease [Neisseria meningitidis]
Length = 396
Score = 58.2 bits (139), Expect = 2e-07
Identities = 47/171 (27%), Positives = 83/171 (48%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + HRK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 209 EAQALAAHRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKAAAEAQALAA------- 259
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 260 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 314
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 315 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 365
Score = 37.7 bits (86), Expect = 0.27
Identities = 37/148 (25%), Positives = 68/148 (45%), Gaps = 11/148 (7%)
Query: 70 KGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGV 129
+GHR ++ + + ++ L A+RQ+ + L Q A A L H K
Sbjct: 166 RGHRSVQQNNVEIAQAQAEL-----ARRQQEERKAAELLAKQRAEAEREAQALAAHRKAE 220
Query: 130 --EIDKACAQLK-LEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVL 186
E + A+L +EA+ + L ++ + AA+ +A +Q + LA +++ ++A E+
Sbjct: 221 AEEAKRQAAELAHRQEAERKAAELSANQKAAAEAQALAARQ-QKALARQQEEARKAAELA 279
Query: 187 AKDKAARESAEASDEEKEEEAKGEGVKK 214
K KA E A E ++ A E K+
Sbjct: 280 VKQKAETERKTA--ELAKQRAAAEAAKR 305
Score = 33.1 bits (74), Expect = 6.6
Identities = 30/88 (34%), Positives = 47/88 (53%), Gaps = 11/88 (12%)
Query: 137 QLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDT-----LAGVRKQLQEAD--EVLAKD 189
+ K ++AKA + + AE K AE+AK++AE LA +K QE E+ +
Sbjct: 11 EAKRQQAKAEQVK-RQQAEAERKSAELAKQKAEAEREARELATRQKAEQERSSAELARRH 69
Query: 190 KAARESAEASDEEK---EEEAKGEGVKK 214
K RE+AE S ++K E EA+ V++
Sbjct: 70 KKEREAAELSAKQKAEAEREAQALAVRR 97
>gb|EAA18109.1| erythrocyte binding protein [Plasmodium yoelii yoelii]
Length = 1701
Score = 56.2 bits (134), Expect = 7e-07
Identities = 54/177 (30%), Positives = 84/177 (46%), Gaps = 17/177 (9%)
Query: 65 QVETAKGHRKNEKK-----ETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCA 119
++E AK + KK + + +KK+ + E +++K++A+K L + AA A
Sbjct: 1201 KIEAAKKAEEERKKAEAVKKAEEAKKKAEAAKKAEERKKKAEAAKKALERKKKAEAAKKA 1260
Query: 120 TGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQL 179
L K E K + K EA K AE AAK+AE KK+AE +K
Sbjct: 1261 ---LERKKKAEAAKKAEEKKKAEAAKKAEEEKKKAE-AAKKAEEEKKKAE----AAKKAE 1312
Query: 180 QEADEVLAKDKAARES--AEASDEEKEEEAKGEGVKKA--DVKVGGSGEVNQEEQAK 232
+E + A KA E AEA+ + +EE+ K E KKA + K + + +EE+ K
Sbjct: 1313 EEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEERKK 1369
Score = 49.7 bits (117), Expect = 7e-05
Identities = 50/172 (29%), Positives = 74/172 (42%), Gaps = 17/172 (9%)
Query: 73 RKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEID 132
R E K+ + RKK + E +R+K++A K +A A A K E
Sbjct: 1188 RIEEAKKAEEERKKIEAAKKAEEERKKAEAVK---KAEEAKKKAEAAKKAEERKKKAEAA 1244
Query: 133 KACAQLK--LEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDK 190
K + K E AK A R K AAK+AE KK A K+ EA + ++K
Sbjct: 1245 KKALERKKKAEAAKKALERKKK--AEAAKKAEEKKKAEAAKKAEEEKKKAEAAKKAEEEK 1302
Query: 191 AARESAEASDEEK----------EEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
E+A+ ++EEK EE+ K E KKA+ + + + E+ K
Sbjct: 1303 KKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEEK 1354
Score = 48.9 bits (115), Expect = 1e-04
Identities = 53/180 (29%), Positives = 85/180 (46%), Gaps = 17/180 (9%)
Query: 63 ASQVETAKGHRKNEKKETKGVRKKSPLVDEREAK--RQKSDASKGTLAVNQALAAASCAT 120
A V+ A+ RK + E K ++ + E++A+ R++ +A K + + AA A
Sbjct: 1124 AEAVKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKIIEAAKKAE 1183
Query: 121 GLLPHLKGVEIDKACAQLKLEEA--KAAYSRLKSDA----ENAAKRAEIAKKQAEDTLAG 174
++ E KA + K EA KA R K++A E A K+AE AKK E
Sbjct: 1184 EERKRIE--EAKKAEEERKKIEAAKKAEEERKKAEAVKKAEEAKKKAEAAKKAEER---- 1237
Query: 175 VRKQLQEADEVLAKDKAARESAEASDEEKEEEA--KGEGVKKADVKVGGSGEVNQEEQAK 232
+K+ + A + L + K A + +A + +K+ EA K E KKA+ E + E AK
Sbjct: 1238 -KKKAEAAKKALERKKKAEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKAEAAK 1296
Score = 47.0 bits (110), Expect = 4e-04
Identities = 47/177 (26%), Positives = 79/177 (44%), Gaps = 15/177 (8%)
Query: 63 ASQVETAKGHRKNEKKETKGV--RKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCAT 120
A + A+ RK + E K RKK+ + E +R++ +A K + + A A
Sbjct: 1370 AEAAKKAEEERKRIEAEKKAEEERKKAEAAKKAEEERKRIEAEKKAEEERKRIEAEKKAE 1429
Query: 121 GLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDT--LAGVRKQ 178
K +E +K K EE + +K AE KR E KK E+ + V+K
Sbjct: 1430 ---EERKRIEAEK-----KAEEERKRIEAVKK-AEEERKRIEAEKKAEEERKRIEAVKKA 1480
Query: 179 LQEADEVLAKDKAARESA--EASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKN 233
+E + A+ KA E EA+ + +EE K E VKK + + + +++ + + N
Sbjct: 1481 EEERKRIEAEKKAEEERKIIEAAKKAEEERIKAEAVKKEEEVIKKNSNLSETKISNN 1537
Score = 44.7 bits (104), Expect = 0.002
Identities = 47/163 (28%), Positives = 75/163 (45%), Gaps = 23/163 (14%)
Query: 65 QVETAKGHRKNEK----KETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCAT 120
+ E AK + +K K+ + +KK+ + E +++K++A+K + AA A
Sbjct: 1266 KAEAAKKAEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAE 1325
Query: 121 GLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAEN-----AAKRAEIAKKQAEDTLAGV 175
E KA A K EE K K E AAK+AE +K+AE
Sbjct: 1326 E--------EKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEERKKAE----AA 1373
Query: 176 RKQLQEADEVLAKDKA--ARESAEASDEEKEEEAKGEGVKKAD 216
+K +E + A+ KA R+ AEA+ + +EE + E KKA+
Sbjct: 1374 KKAEEERKRIEAEKKAEEERKKAEAAKKAEEERKRIEAEKKAE 1416
Score = 44.3 bits (103), Expect = 0.003
Identities = 43/149 (28%), Positives = 66/149 (43%), Gaps = 15/149 (10%)
Query: 73 RKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEID 132
+K E + +KK+ + E +++K++A+K + AA A E
Sbjct: 1265 KKAEAAKKAEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEE--------EKK 1316
Query: 133 KACAQLKLEEAKAAYSRLKSDAEN-----AAKRAEIAKKQAEDTLAGVRKQLQEADEVLA 187
KA A K EE K K E AAK+AE KK+AE A ++ ++ E
Sbjct: 1317 KAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAE--AAKKAEEERKKAEAAK 1374
Query: 188 KDKAARESAEASDEEKEEEAKGEGVKKAD 216
K + R+ EA + +EE K E KKA+
Sbjct: 1375 KAEEERKRIEAEKKAEEERKKAEAAKKAE 1403
Score = 42.7 bits (99), Expect = 0.008
Identities = 47/162 (29%), Positives = 75/162 (46%), Gaps = 19/162 (11%)
Query: 56 ESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAA 115
E A A + E K + KK + +KK+ + E +++K++A+K + A
Sbjct: 1275 EKKKAEAAKKAEEEKKKAEAAKKAEEE-KKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEA 1333
Query: 116 ASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGV 175
A A E KA A K EE K K AE K+AE AKK E+
Sbjct: 1334 AKKAEE--------EKKKAEAAKKAEEEKKKAEAAKK-AEEERKKAEAAKKAEEE----- 1379
Query: 176 RKQLQEADEVLAKDKAARESAEASDEEK---EEEAKGEGVKK 214
RK++ EA++ +++ E+A+ ++EE+ E E K E +K
Sbjct: 1380 RKRI-EAEKKAEEERKKAEAAKKAEEERKRIEAEKKAEEERK 1420
Score = 41.2 bits (95), Expect = 0.024
Identities = 43/149 (28%), Positives = 67/149 (44%), Gaps = 15/149 (10%)
Query: 73 RKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEID 132
+K E + RKK ++ +++K++A+K + AA A E
Sbjct: 1252 KKAEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKAEAAKKAEE--------EKK 1303
Query: 133 KACAQLKLEEAKAAYSRLKSDAEN-----AAKRAEIAKKQAEDTLAGVRKQLQEADEVLA 187
KA A K EE K K E AAK+AE KK+AE A ++ ++ E
Sbjct: 1304 KAEAAKKAEEEKKKAEAAKKAEEEKKKAEAAKKAEEEKKKAE--AAKKAEEEKKKAEAAK 1361
Query: 188 KDKAARESAEASDEEKEEEAKGEGVKKAD 216
K + R+ AEA+ + +EE + E KKA+
Sbjct: 1362 KAEEERKKAEAAKKAEEERKRIEAEKKAE 1390
Score = 40.8 bits (94), Expect = 0.032
Identities = 51/188 (27%), Positives = 85/188 (45%), Gaps = 18/188 (9%)
Query: 56 ESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAA 115
E A A + E K + KK + RKK+ + E +R++ +A K + A
Sbjct: 1340 EKKKAEAAKKAEEEKKKAEAAKKAEEE-RKKAEAAKKAEEERKRIEAEKKAEEERKKAEA 1398
Query: 116 ASCATGLLPHLKGVEIDKACAQLKLE---EAKAAYSRLKSDAENAAKRAEIAKKQAEDTL 172
A A K +E +K + + E KA R + +AE K+AE +K+ E
Sbjct: 1399 AKKAE---EERKRIEAEKKAEEERKRIEAEKKAEEERKRIEAE---KKAEEERKRIE--- 1449
Query: 173 AGVRKQLQEADEVLAKDKA--ARESAEASDEEKEEEAKGEGVKKADV--KVGGSGEVNQE 228
V+K +E + A+ KA R+ EA + +EE + E KKA+ K+ + + +E
Sbjct: 1450 -AVKKAEEERKRIEAEKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKIIEAAKKAEE 1508
Query: 229 EQAKNDQV 236
E+ K + V
Sbjct: 1509 ERIKAEAV 1516
Score = 40.0 bits (92), Expect = 0.054
Identities = 47/171 (27%), Positives = 77/171 (44%), Gaps = 26/171 (15%)
Query: 56 ESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAA 115
E A A + E K + KK + +KK+ + E +++K++A+K + A
Sbjct: 1301 EKKKAEAAKKAEEEKKKAEAAKKAEEE-KKKAEAAKKAEEEKKKAEAAKKAEEEKKKAEA 1359
Query: 116 ASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGV 175
A A E KA A K EE + K AE K+AE AKK E+
Sbjct: 1360 AKKAEE--------ERKKAEAAKKAEEERKRIEAEKK-AEEERKKAEAAKKAEEE----- 1405
Query: 176 RKQLQEADEVLAKDKAARESAEASDEEK----------EEEAKGEGVKKAD 216
RK++ EA++ +++ E+ + ++EE+ EE + E VKKA+
Sbjct: 1406 RKRI-EAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKRIEAVKKAE 1455
Score = 38.9 bits (89), Expect = 0.12
Identities = 46/176 (26%), Positives = 78/176 (44%), Gaps = 23/176 (13%)
Query: 56 ESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAA 115
E A A + E K + KK + +KK+ + E +++K++A+K + A
Sbjct: 1314 EKKKAEAAKKAEEEKKKAEAAKKAEEE-KKKAEAAKKAEEEKKKAEAAKKAEEERKKAEA 1372
Query: 116 ASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAEN----AAKRAEIAKK--QAE 169
A A K +E +K EE K A + K++ E A K+AE +K +AE
Sbjct: 1373 AKKAE---EERKRIEAEKKAE----EERKKAEAAKKAEEERKRIEAEKKAEEERKRIEAE 1425
Query: 170 DTLAGVRKQLQ---------EADEVLAKDKAARESAEASDEEKEEEAKGEGVKKAD 216
RK+++ + E + K + R+ EA + +EE + E VKKA+
Sbjct: 1426 KKAEEERKRIEAEKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKRIEAVKKAE 1481
Score = 36.6 bits (83), Expect = 0.60
Identities = 42/176 (23%), Positives = 70/176 (38%), Gaps = 33/176 (18%)
Query: 67 ETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHL 126
E+ +E K + +RK+ E R+ +A K A
Sbjct: 1073 ESDSTRNTDESKMDEVIRKREEAAKNAEIIRKFEEAQKAAWAK----------------- 1115
Query: 127 KGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVL 186
K E K +K E + + AE KR E KK E+ RK++ EA++
Sbjct: 1116 KAEEERKKAEAVKKAEEERKRIEAEKKAEEERKRIEAEKKAEEE-----RKRI-EAEKKA 1169
Query: 187 AKDKAARESAEASDEEK----------EEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
+++ E+A+ ++EE+ EE K E KKA+ + + V + E+AK
Sbjct: 1170 EEERKIIEAAKKAEEERKRIEEAKKAEEERKKIEAAKKAEEERKKAEAVKKAEEAK 1225
>gb|AAW40799.1| glycosyltransferase, putative [Cryptococcus neoformans var.
neoformans JEC21] gi|58258411|ref|XP_566618.1|
glycosyltransferase, putative [Cryptococcus neoformans
var. neoformans JEC21]
Length = 640
Score = 55.8 bits (133), Expect = 1e-06
Identities = 38/116 (32%), Positives = 68/116 (57%), Gaps = 8/116 (6%)
Query: 128 GVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVL- 186
GV+ + +LK +EA+ A + K +AE A ++A+ A++QA +T K Q+A+E
Sbjct: 524 GVQEFERLKKLK-KEAEEAEKKAKEEAEEAERKAKEAEQQANETSEQADKSAQDAEEKAK 582
Query: 187 -AKDKAARESAEASDE-----EKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQV 236
A ++AAR++ EASD+ E+ E+AK K+D + GG+GE + + +++
Sbjct: 583 EAAEEAARKAQEASDKVEALGEEVEKAKEVAEGKSDGQDGGNGEFTESREETEEEM 638
>gb|EAL23495.1| hypothetical protein CNBA1420 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 640
Score = 55.8 bits (133), Expect = 1e-06
Identities = 38/116 (32%), Positives = 68/116 (57%), Gaps = 8/116 (6%)
Query: 128 GVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVL- 186
GV+ + +LK +EA+ A + K +AE A ++A+ A++QA +T K Q+A+E
Sbjct: 524 GVQEFERLKKLK-KEAEEAEKKAKEEAEEAERKAKEAEQQANETSEQADKSAQDAEEKAK 582
Query: 187 -AKDKAARESAEASDE-----EKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQV 236
A ++AAR++ EASD+ E+ E+AK K+D + GG+GE + + +++
Sbjct: 583 EAAEEAARKAQEASDKVEALGEEVEKAKEVAEGKSDGQDGGNGEFTESREETEEEM 638
>gb|AAF41117.1| IgA-specific serine endopeptidase [Neisseria meningitidis MC58]
gi|11353752|pir||C81169 IgA-specific metalloendopeptidase
(EC 3.4.24.13) NMB0700 [imported] - Neisseria
meningitidis (strain MC58 serogroup B)
gi|15676598|ref|NP_273742.1| IgA-specific serine
endopeptidase [Neisseria meningitidis MC58]
Length = 1815
Score = 55.1 bits (131), Expect = 2e-06
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + RK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 1214 EAQALAARRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKAAAEAQALAA------- 1264
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 1265 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 1319
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 1320 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 1370
Score = 35.4 bits (80), Expect = 1.3
Identities = 52/193 (26%), Positives = 80/193 (40%), Gaps = 31/193 (16%)
Query: 51 GGVNPESMSAFIASQVET--------AKGH------RKNEKKETKGVRKKSPLVDEREAK 96
G N E ++ F AS V+ A H R K E R +P + R +
Sbjct: 912 GEPNQEGLNLFDASSVQDRSRLSVSLANNHVDLGALRYTIKTENGITRLYNPYAENR--R 969
Query: 97 RQKSDASKGTLAVNQALAAASCATGLLPHLKGVEIDKACAQL-KLEEAKAAYSRL----K 151
R K S T +QA A + + + + Q + EEAK ++ +
Sbjct: 970 RVKPAPSPATNTASQAQKATQTDGAQIAKPQNIVVAPPSPQANQAEEAKRQQAKAEQVKR 1029
Query: 152 SDAENAAKRAEIAKKQAEDT-----LAGVRKQLQEAD--EVLAKDKAARESAEASDEEK- 203
AE K AE+AK++AE LA +K QE E+ + + RE+AE S ++K
Sbjct: 1030 QQAEAERKSAELAKQKAEAEREARELATRQKAEQERSSAELARRHEKEREAAELSAKQKV 1089
Query: 204 --EEEAKGEGVKK 214
E EA+ V++
Sbjct: 1090 EAEREAQALAVRR 1102
>emb|CAA97349.1| SPAC17C9.03 [Schizosaccharomyces pombe]
gi|19115514|ref|NP_594602.1| hypothetical protein
SPAC17C9.03 [Schizosaccharomyces pombe 972h-]
gi|1723562|sp|Q10475|IF4G_SCHPO Eukaryotic translation
initiation factor 4 gamma (eIF-4-gamma) (eIF-4G)
Length = 1403
Score = 55.1 bits (131), Expect = 2e-06
Identities = 52/187 (27%), Positives = 89/187 (46%), Gaps = 11/187 (5%)
Query: 53 VNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQA 112
VNP + + + Q + + + P + E EA ++K DA K LA+ Q
Sbjct: 480 VNPVTHTEVVVPQKNASSPNPSETNSRAETPTAAPPQISEEEASQRK-DAIK--LAIQQR 536
Query: 113 LA--AASCATGLLPHLKGVEIDKACAQLKLEEAKA-AYSRLKSDAENAAKRA--EIAKKQ 167
+ A + A +E ++ + E+AK A + K +AE AKR E AK++
Sbjct: 537 IQEKAEAEAKRKAEEKARLEAEENAKREAEEQAKREAEEKAKREAEEKAKREAEEKAKRE 596
Query: 168 AEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQ 227
AE+ +++ +E + A++KA RE+ E + E EE+AK E +KA + +
Sbjct: 597 AEEN---AKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREA 653
Query: 228 EEQAKND 234
EE+AK +
Sbjct: 654 EEKAKRE 660
Score = 53.1 bits (126), Expect = 6e-06
Identities = 54/189 (28%), Positives = 86/189 (44%), Gaps = 19/189 (10%)
Query: 55 PESMSAFIASQ----VETAKGHRKNEKKETKGVRK---KSPLVDEREAKRQKSDASKGTL 107
P +S ASQ ++ A R EK E + RK K+ L E AKR+ + +K
Sbjct: 514 PPQISEEEASQRKDAIKLAIQQRIQEKAEAEAKRKAEEKARLEAEENAKREAEEQAKREA 573
Query: 108 AVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRA--EIAK 165
A A K +KA + + + A + K +AE AKR E AK
Sbjct: 574 EEKAKREAEEKA-------KREAEEKAKREAEENAKREAEEKAKREAEEKAKREAEEKAK 626
Query: 166 KQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEV 225
++AE+ +++ +E + A++KA RE+ E + E EE AK E +KA + + +
Sbjct: 627 REAEEK---AKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEENAKR 683
Query: 226 NQEEQAKND 234
EE+ K +
Sbjct: 684 EAEEKVKRE 692
Score = 48.1 bits (113), Expect = 2e-04
Identities = 44/169 (26%), Positives = 76/169 (44%), Gaps = 6/169 (3%)
Query: 69 AKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKG 128
A+ RK E+K + + E +AKR+ + +K A A
Sbjct: 543 AEAKRKAEEKARLEAEENAKREAEEQAKREAEEKAKREAEEKAKREAEEKAKREAEENAK 602
Query: 129 VEIDKACAQLKLEEAKA-AYSRLKSDAENAAKRA--EIAKKQAEDTLAGVRKQLQEADEV 185
E ++ + E+AK A + K +AE AKR E AK++AE+ +++ +E +
Sbjct: 603 REAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEKAKREAEEK---AKREAEEKAKR 659
Query: 186 LAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKND 234
A++ A RE+ E + E EE AK E +K + + + EE+ K +
Sbjct: 660 EAEENAKREAEEKAKREAEENAKREAEEKVKRETEENAKRKAEEEGKRE 708
Score = 47.0 bits (110), Expect = 4e-04
Identities = 40/151 (26%), Positives = 70/151 (45%), Gaps = 12/151 (7%)
Query: 70 KGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGV 129
K R+ E+K + +K+ E AKR+ + +K A A K
Sbjct: 576 KAKREAEEKAKREAEEKAKREAEENAKREAEEKAKREAEEKAKREAEEKA-------KRE 628
Query: 130 EIDKACAQLKLEEAKAAYSRLKSDAENAAKRA--EIAKKQAEDTLAGVRKQLQEADEVLA 187
+KA + + + + A + K +AE AKR E AK++AE+ +++ +E + A
Sbjct: 629 AEEKAKREAEEKAKREAEEKAKREAEEKAKREAEENAKREAEEK---AKREAEENAKREA 685
Query: 188 KDKAARESAEASDEEKEEEAKGEGVKKADVK 218
++K RE+ E + + EEE K E K ++K
Sbjct: 686 EEKVKRETEENAKRKAEEEGKREADKNPEIK 716
>emb|CAB84182.1| IgA1 protease [Neisseria meningitidis Z2491]
gi|15793871|ref|NP_283693.1| IgA1 protease [Neisseria
meningitidis Z2491] gi|11353751|pir||A81937 IgA-specific
metalloendopeptidase (EC 3.4.24.13) NMA0905 [imported] -
Neisseria meningitidis (strain Z2491 serogroup A and
others)
Length = 1773
Score = 55.1 bits (131), Expect = 2e-06
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + RK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 1210 EAQALAARRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKAAAEAQALAA------- 1260
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 1261 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 1315
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 1316 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 1366
>emb|CAE64509.1| Hypothetical protein CBG09244 [Caenorhabditis briggsae]
Length = 3376
Score = 55.1 bits (131), Expect = 2e-06
Identities = 48/174 (27%), Positives = 83/174 (47%), Gaps = 13/174 (7%)
Query: 67 ETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHL 126
+T KG + KKE + KK + +AK++ D +K + +A A T
Sbjct: 863 KTKKGSDEKAKKEAEAKTKKEA---DDKAKKEADDKTKKE-SDEKAKKEAEAKTKKEADD 918
Query: 127 KGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLA--------GVRKQ 178
K +A + + + K A ++ K +A++ AK+ +AKK+A+D K
Sbjct: 919 KARNEAEAKKEAEDKAEKGADAKTKKEADDKAKKEAVAKKEADDKTKKEAEAEKEAEAKT 978
Query: 179 LQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
+EAD+ + KD A+ EA DE+ ++EA+ + K+AD K E +E + K
Sbjct: 979 KKEADDKVKKDLEAKTKKEA-DEKAKKEAEAKIKKEADDKARNEAEAKKEAEDK 1031
Score = 47.0 bits (110), Expect = 4e-04
Identities = 46/173 (26%), Positives = 81/173 (46%), Gaps = 4/173 (2%)
Query: 63 ASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGL 122
A + + K +K+ RK++ E EAK++ D +K AV + +A
Sbjct: 1164 ADEKAKKEAEAKTKKEADDKARKEAEAKKEAEAKKEADDKAKKE-AVAKKVADDKTKKEA 1222
Query: 123 LPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAE-DTLAGVR-KQLQ 180
+ K A+ K ++ A ++ ++D + K E A K+AE T V K +
Sbjct: 1223 DDKARNEAEAKKEAEDKAKKEDDAKTKKEADDKAKKKADEKANKEAEAKTKKDVEAKTKK 1282
Query: 181 EADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKN 233
EAD+ KD A+ EA DE+ ++EA+ + K+A+ K + ++ +AKN
Sbjct: 1283 EADDKAKKDLEAKTKKEA-DEKAKKEAEAKIKKEAEAKKEADDKAKKDSEAKN 1334
Score = 46.6 bits (109), Expect = 6e-04
Identities = 46/171 (26%), Positives = 80/171 (45%), Gaps = 8/171 (4%)
Query: 69 AKGHRKNEKKETKGVRKKSPLVDEREAKRQ---KSDASKGTLAVNQALAAASCATGLLPH 125
A+ ++ E K KG K+ + +AK++ K +A T +A A T
Sbjct: 924 AEAKKEAEDKAEKGADAKTKKEADDKAKKEAVAKKEADDKTKKEAEAEKEAEAKTKKEAD 983
Query: 126 LKGVEIDKACAQLKLEEA--KAAYSRLKSDAENAAKRAEIAKKQAEDTLA--GVRKQLQE 181
K + +A + + +E K A +++K +A++ A+ AKK+AED K +E
Sbjct: 984 DKVKKDLEAKTKKEADEKAKKEAEAKIKKEADDKARNEAEAKKEAEDKAKKEADAKTKKE 1043
Query: 182 ADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
AD+ KD A EA DE+ ++EA+ + K+AD K E ++ +
Sbjct: 1044 ADDKAKKDLEAMTKKEA-DEKAKKEAEAKTKKEADEKAKKEAEAKTKKDVE 1093
Score = 46.6 bits (109), Expect = 6e-04
Identities = 51/190 (26%), Positives = 86/190 (44%), Gaps = 24/190 (12%)
Query: 63 ASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGL 122
A + AK ++ + K K + K+ E +AK++ +K A A A T
Sbjct: 754 AEKEAEAKTKKEADDKAKKDLEAKTKKEAEEKAKKEAEVKTKKE-ADETAKKEADAKT-- 810
Query: 123 LPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAE--------NAAKRAEIAKKQAED-TLA 173
K DKA L+ + K A + K +AE + AK+ AKK+A+D T
Sbjct: 811 ----KKEADDKAKKNLEAKTKKEAEEKAKKEAEVKTRKEADDKAKKEAEAKKEADDKTKK 866
Query: 174 G----VRKQLQEADEVLAKDKAARESAEA----SDEEKEEEAKGEGVKKADVKVGGSGEV 225
G +K+ + + A DKA +E+ + SDE+ ++EA+ + K+AD K E
Sbjct: 867 GSDEKAKKEAEAKTKKEADDKAKKEADDKTKKESDEKAKKEAEAKTKKEADDKARNEAEA 926
Query: 226 NQEEQAKNDQ 235
+E + K ++
Sbjct: 927 KKEAEDKAEK 936
Score = 44.3 bits (103), Expect = 0.003
Identities = 43/154 (27%), Positives = 67/154 (42%), Gaps = 23/154 (14%)
Query: 77 KKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEIDKACA 136
KKE KK DE+ K ++ K A + A DKA
Sbjct: 1249 KKEADDKAKKK--ADEKANKEAEAKTKKDVEAKTKKEAD----------------DKAKK 1290
Query: 137 QLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESA 196
L+ + K A + K +AE K+ AKK+A+D K+ EA D+ AR+ A
Sbjct: 1291 DLEAKTKKEADEKAKKEAEAKIKKEAEAKKEADDK----AKKDSEAKNKKEADEKARKEA 1346
Query: 197 EASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQ 230
E + +E E++AK E K + ++G S E +++
Sbjct: 1347 E-TKKEAEDKAKKEAETKKEAELGKSDETKPKKR 1379
Score = 44.3 bits (103), Expect = 0.003
Identities = 52/174 (29%), Positives = 78/174 (43%), Gaps = 23/174 (13%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSP---LVDEREAKRQKSDASKGTLAVNQALAAASCATG 121
+ +T K + KKE + KK +E EAK++ D +K A T
Sbjct: 991 EAKTKKEADEKAKKEAEAKIKKEADDKARNEAEAKKEAEDKAK---------KEADAKT- 1040
Query: 122 LLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRA--EIAKKQAE-DTLAGVR-K 177
K DKA L+ K A + K +AE K+ E AKK+AE T V K
Sbjct: 1041 -----KKEADDKAKKDLEAMTKKEADEKAKKEAEAKTKKEADEKAKKEAEAKTKKDVEAK 1095
Query: 178 QLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQA 231
+EAD+ KD A+ EA DE+ ++EA+ + K+AD K + +++A
Sbjct: 1096 TKKEADDKAKKDLEAKTKKEA-DEKAKKEAEAKIKKEADDKTKKEADAKTKKEA 1148
Score = 40.4 bits (93), Expect = 0.041
Identities = 32/107 (29%), Positives = 57/107 (52%), Gaps = 11/107 (10%)
Query: 136 AQLKLEEAKAAYSRLKSDAENAAKRAEIA--KKQAEDTL-----AGVRKQL---QEADEV 185
++ +++ K ++ K +A+N AK+ A KK+A+D A ++K+ +EADE
Sbjct: 678 SETEVKAKKDLEAKTKKEADNKAKKEAEAKTKKEADDKAKKKADAEIKKEAVTKKEADEK 737
Query: 186 LAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
KD A+ E DE E+EA+ + K+AD K E +++A+
Sbjct: 738 AKKDLEAKTKKEV-DENAEKEAEAKTKKEADDKAKKDLEAKTKKEAE 783
Score = 38.5 bits (88), Expect = 0.16
Identities = 42/176 (23%), Positives = 73/176 (40%), Gaps = 14/176 (7%)
Query: 70 KGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGV 129
K ++ E K K K+ + + K++ D +K L A A
Sbjct: 1119 KAKKEAEAKIKKEADDKTKKEADAKTKKEADDKAKKDLEAKTKKEADEKAKKEAEAKTKK 1178
Query: 130 EIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKK--------QAEDTLAGVRKQLQE 181
E D A+ + E K A + K +A++ AK+ +AKK +A+D + +E
Sbjct: 1179 EADDK-ARKEAEAKKEAEA--KKEADDKAKKEAVAKKVADDKTKKEADDKARNEAEAKKE 1235
Query: 182 ADEVLAKDKAARESAEASDEEK---EEEAKGEGVKKADVKVGGSGEVNQEEQAKND 234
A++ K+ A+ EA D+ K +E+A E K V + +++AK D
Sbjct: 1236 AEDKAKKEDDAKTKKEADDKAKKKADEKANKEAEAKTKKDVEAKTKKEADDKAKKD 1291
Score = 36.6 bits (83), Expect = 0.60
Identities = 42/151 (27%), Positives = 67/151 (43%), Gaps = 20/151 (13%)
Query: 88 PLVDEREAKRQKSDASKGTL-------AVNQALAAASCATGLLPHLKGVEIDKACAQLKL 140
PL D++E K + +K L A N+A A T K DKA +
Sbjct: 669 PLKDQKEVKSETEVKAKKDLEAKTKKEADNKAKKEAEAKT------KKEADDKAKKKADA 722
Query: 141 EEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASD 200
E K A ++ ++D E A K E K+ D A K+ + + A DKA ++ +
Sbjct: 723 EIKKEAVTKKEAD-EKAKKDLEAKTKKEVDENA--EKEAEAKTKKEADDKAKKDLEAKTK 779
Query: 201 EEKEEEAKGEGVKKADVKVGGSGEVNQEEQA 231
+E EE+AK K+A+VK + +++A
Sbjct: 780 KEAEEKAK----KEAEVKTKKEADETAKKEA 806
Score = 33.5 bits (75), Expect = 5.1
Identities = 30/110 (27%), Positives = 52/110 (47%), Gaps = 15/110 (13%)
Query: 127 KGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQA-----EDTLAGVRKQLQE 181
K +D + + E+ K + + L S+A K ++ KK+ E+TL V+++L++
Sbjct: 2756 KSEAVDDSKTTVSTEKPKDS-ADLASEAVEEKKTKKVTKKKVPENKGEETLQEVKEKLKK 2814
Query: 182 ADEV-LAKDKAARESAEASD--------EEKEEEAKGEGVKKADVKVGGS 222
V +D++ R S + SD E+K E + KK D K G S
Sbjct: 2815 GKAVEKVQDESRRGSLQTSDAESVTTASEKKAESEAEKSAKKTDEKTGKS 2864
>gb|AAT49035.1| IgA1 protease [Neisseria meningitidis]
Length = 396
Score = 55.1 bits (131), Expect = 2e-06
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + RK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 209 EAQALAARRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKATAEAQALAA------- 259
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 260 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 314
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 315 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 365
>gb|AAT49029.1| IgA1 protease [Neisseria meningitidis]
Length = 396
Score = 55.1 bits (131), Expect = 2e-06
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + RK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 209 EAQALAARRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKAAAEAQALAA------- 259
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 260 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 314
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 315 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 365
>gb|AAT49027.1| IgA1 protease [Neisseria meningitidis]
Length = 396
Score = 55.1 bits (131), Expect = 2e-06
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + RK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 209 EAQALAARRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKAAAEAQALAA------- 259
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 260 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 314
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 315 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 365
>gb|AAT49026.1| IgA1 protease [Neisseria meningitidis] gi|49037434|gb|AAT49024.1|
IgA1 protease [Neisseria meningitidis]
gi|49037432|gb|AAT49023.1| IgA1 protease [Neisseria
meningitidis]
Length = 396
Score = 55.1 bits (131), Expect = 2e-06
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 17/171 (9%)
Query: 65 QVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLP 124
+ + RK E +E K R+ + L +EA+R+ ++ S A +A A A+
Sbjct: 209 EAQALAARRKAEAEEAK--RQAAELAHRQEAERKAAELSANQKAAAEAQALAA------- 259
Query: 125 HLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADE 184
KA A+ + E KAA +K AE K AE+AK++A A +++ ++ E
Sbjct: 260 -----RQQKALARQQEEARKAAELAVKQKAETERKTAELAKQRAAAEAAKRQQEARQTAE 314
Query: 185 VLAKDKAARESAEASDEEKEE---EAKGEGVKKADVKVGGSGEVNQEEQAK 232
+ + +A R++AE S ++K E EA +KA+ + +Q ++ K
Sbjct: 315 LARRQEAERQAAELSAKQKAETDREAAESAKRKAEEEEHRQAAQSQPQRRK 365
>emb|CAB95273.1| possible chromosome assembly protein [Leishmania major]
Length = 1355
Score = 54.7 bits (130), Expect = 2e-06
Identities = 49/198 (24%), Positives = 87/198 (43%), Gaps = 13/198 (6%)
Query: 47 VKNKGGVNPESMSAFIASQVETAKGH--------RKNEKKETKGVRKKSPLVD-EREAKR 97
V GG ++++A + +Q E A R+ +K RK + +VD E++ K+
Sbjct: 760 VDEAGGARFKAVAASLTAQQERADAEEASLRACRRQAQKHRATQERKAADIVDYEQQLKK 819
Query: 98 QKSDASKGTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENA 157
+ +A A Q GL+ L G E A AQ E+A+AA A NA
Sbjct: 820 LEEEAQDN--AEEQVRTCVKVLEGLVRELNGAEAQLAKAQTSAEDARAAVPVANGAALNA 877
Query: 158 AKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADV 217
K+ + ++ + A + LQE + K + E + E +E G+G A+
Sbjct: 878 QKKLDDEQRYRQTEAAKIADALQELSKFQQKIEGCEEKIRENVELYGKETLGQG--DAED 935
Query: 218 KVGGSGEVNQEEQAKNDQ 235
+ + EV++E+ ++D+
Sbjct: 936 EEDQAAEVDEEDGDEDDE 953
>gb|AAS10478.1| merozoite surface protein 3b [Plasmodium vivax]
Length = 810
Score = 54.3 bits (129), Expect = 3e-06
Identities = 58/200 (29%), Positives = 90/200 (45%), Gaps = 19/200 (9%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDE---REAKRQKSD 101
E +K G S S + V TA + +KE + +K+ E +E + D
Sbjct: 193 ETIKETG----TSESEIVTKAVATATAEEEKTQKEAQTASEKADKAVEETQKEVDKGIED 248
Query: 102 ASKGTLAVNQALAAAS-CATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKR 160
SK T + L + A+ K + + A AQ+ E AKA + K +AENA
Sbjct: 249 ESKETSDLEDILKSVKELASSAEDASKNAKKEMAKAQIAAEVAKA--EKAKIEAENAKLI 306
Query: 161 AEIAKKQAEDTLAGVRKQLQEADEVLAK--------DKAARESAEASDEEKEEEAKGEGV 212
A+ A K AED +A K Q A V AK +AA E+A A +E + E+K + V
Sbjct: 307 ADTASKAAED-IAKSSKAAQIAKNVSAKAEEKSKVATEAADEAANALNEAENPESKIDDV 365
Query: 213 KKADVKVGGSGEVNQEEQAK 232
+K + + E ++E++K
Sbjct: 366 RKKATEAVNAAEEAKKEKSK 385
Score = 40.4 bits (93), Expect = 0.041
Identities = 53/184 (28%), Positives = 86/184 (45%), Gaps = 28/184 (15%)
Query: 57 SMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSD--ASKGTLAVNQALA 114
S +A IA V +AK K+ K T+ + + ++E E K D K T AVN A
Sbjct: 321 SKAAQIAKNV-SAKAEEKS-KVATEAADEAANALNEAENPESKIDDVRKKATEAVNAAEE 378
Query: 115 AASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAG 174
A +K+ A++ +E AKA +A+ A +A++A KQ D +
Sbjct: 379 AKK--------------EKSKAEIAVEVAKA------EEAKKEAGKAKVAAKQVADK-SK 417
Query: 175 VRKQLQEADEVLAK-DKAARESAEA-SDEEKEEEAKGE-GVKKADVKVGGSGEVNQEEQA 231
+ K +Q AD+ K D+A++ + EA SD E E+ GE K ++K +N +A
Sbjct: 418 LEKAIQAADKASKKTDEASKLAEEALSDLESLEKETGEIKTKVNEIKEKVQNAINAALEA 477
Query: 232 KNDQ 235
++
Sbjct: 478 HKEK 481
Score = 35.8 bits (81), Expect = 1.0
Identities = 51/179 (28%), Positives = 74/179 (40%), Gaps = 33/179 (18%)
Query: 84 RKKSPLVDEREAKRQKSDASKGTLAVNQA----LAAASCATGLLPHLKGV----EIDK-- 133
+ P V +E + ++S SK A +A AAAS L L+ V E+DK
Sbjct: 41 KSAEPKVHAQEEETKESLKSKAQNAKEEAEKAAKAAASAKDRTLVALEKVDVPTELDKVK 100
Query: 134 -----ACAQLKLEEA---KAAYS-----------RLKSDAENAAKRAEIAKK---QAEDT 171
A Q K +E KAA S LKS+ ENA K A+ AK+ +AE
Sbjct: 101 EFAVSAATQAKNQETIATKAAESAEAIGGDGDLGNLKSEVENAEKAAKKAKQLQIKAEIA 160
Query: 172 LAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQ 230
V Q + + A+ A + EA+ KEE K G ++++ EE+
Sbjct: 161 EQAVMAQAAKTEAKKAQKDAEKAKTEAT-TAKEETIKETGTSESEIVTKAVATATAEEE 218
Score = 35.4 bits (80), Expect = 1.3
Identities = 26/94 (27%), Positives = 43/94 (45%), Gaps = 4/94 (4%)
Query: 141 EEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASD 200
EE K + LKS A+NA + AE A K A L++ D DK + A+
Sbjct: 52 EETKES---LKSKAQNAKEEAEKAAKAAASAKDRTLVALEKVDVPTELDKVKEFAVSAAT 108
Query: 201 EEKEEEAKGEGVKKADVKVGGSGEV-NQEEQAKN 233
+ K +E ++ +GG G++ N + + +N
Sbjct: 109 QAKNQETIATKAAESAEAIGGDGDLGNLKSEVEN 142
Score = 34.3 bits (77), Expect = 3.0
Identities = 56/213 (26%), Positives = 96/213 (44%), Gaps = 49/213 (23%)
Query: 43 AAEMVKNKGGVNPESMSAFIASQ--------VETAKGHRKNEKKETKGVRKKSPLVDERE 94
AA+ V +K + +A AS+ E A ++ +KET ++ K V+E +
Sbjct: 408 AAKQVADKSKLEKAIQAADKASKKTDEASKLAEEALSDLESLEKETGEIKTK---VNEIK 464
Query: 95 AKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDA 154
K Q A+N AL A +K A++ +E AKA K +A
Sbjct: 465 EKVQN--------AINAALEAHK--------------EKTIAEITVEVAKA--EEAKKEA 500
Query: 155 ENAAKRAEIAKKQAE------DTLAGVRKQLQEADEVL-----AKDKAARESA-EASDEE 202
+NA AE AK+ AE + + +++++A E A +AA E+A + S EE
Sbjct: 501 DNAKVAAEKAKETAEKIAKTSKSTEKITEEVRKATEFAKTAGDATTQAATEAAGDVSSEE 560
Query: 203 KEEEAKGEGVK-KADVKVGGSGE-VNQEEQAKN 233
++++ E +K KA+ S E + + +A+N
Sbjct: 561 QKQKVLLESIKQKAESAFQASQEAIKAKTEAEN 593
>ref|XP_539204.1| PREDICTED: similar to plectin 1 [Canis familiaris]
Length = 5085
Score = 53.9 bits (128), Expect = 4e-06
Identities = 46/169 (27%), Positives = 90/169 (53%), Gaps = 9/169 (5%)
Query: 56 ESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAA 115
E + +A++ E A+ RK+ +E + R K+ + + R + + S L + Q A
Sbjct: 2559 ERVQKSLAAEEEAAR-QRKSALEEVE--RLKAKVEEARRLRERAEQESARQLQLAQEAAQ 2615
Query: 116 ASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAE---NAAKRAEIAKKQAEDTL 172
H V+ + Q L++ ++ RL+++AE AA+ AE A+++AE
Sbjct: 2616 KRLQAEEKAHAFAVQQKEQELQQTLQQEQSMLERLRAEAEAARRAAEEAEEARERAEREA 2675
Query: 173 AGVRKQLQEADEV--LAKDKA-ARESAEASDEEKEEEAKGEGVKKADVK 218
A R+Q++EA+ + LA+++A A+ A+A+ E+ +EA+ E V++A +
Sbjct: 2676 AQSRRQVEEAERLKQLAEEQAQAQAQAQAAAEKLRKEAEQEAVRRAQAE 2724
Score = 45.1 bits (105), Expect = 0.002
Identities = 47/181 (25%), Positives = 82/181 (44%), Gaps = 23/181 (12%)
Query: 77 KKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEID---- 132
K E + R+K + E R +++ ++ L +A A L + E++
Sbjct: 2093 KAEAEAAREKQRALQALEDVRLQAEEAERRLRQAEADRARQVQVALETAQRSAEVELQSK 2152
Query: 133 -----KACAQLK--LEEAKAAYSRLKSDAENAAKR---AEIAKKQAEDTLAGVRKQLQE- 181
+ AQL+ L+E A ++L+ +AE A++ AE A+++AE L R + E
Sbjct: 2153 RASFAEKTAQLERTLQEEHVAVAQLREEAERRAQQQAEAERAREEAEQELERWRLKANEA 2212
Query: 182 ------ADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQ 235
A+EV + A+ AE EE E EA+ G KA+ + E+ ++E K Q
Sbjct: 2213 LRLRLQAEEVAQQKSLAQAEAEKQKEEAEREARRRG--KAEEQAVRQRELAEQELEKQRQ 2270
Query: 236 V 236
+
Sbjct: 2271 L 2271
Score = 38.9 bits (89), Expect = 0.12
Identities = 38/150 (25%), Positives = 68/150 (45%), Gaps = 12/150 (8%)
Query: 77 KKETKGVRKKSPLVDEREAKRQKSDAS----KGTLAVNQALAAASCATGLLPHLKGVEID 132
+ ET+ ++ L++E A+ Q+ A+ + L A A L + E
Sbjct: 2290 RAETEQGEQQRQLLEEELARLQREAAAATHKRQELEAELAKVRAEMEVLLASKARAEEES 2349
Query: 133 KACAQLKLEEAKAAYSRLKSDAENAAKR---AEIAKKQAEDTLAGVRKQLQEADEVLAK- 188
++ ++ + +A SR + AE AA+ AE K+Q + +Q EA+ VLA+
Sbjct: 2350 RSTSEKSKQRLEAEASRFRELAEEAARLRALAEETKRQRQLAEEDAARQRAEAERVLAEK 2409
Query: 189 ----DKAARESAEASDEEKEEEAKGEGVKK 214
+A R EA KE+EA+ E +++
Sbjct: 2410 LAAISEATRLKTEAEIALKEKEAENERLRR 2439
Score = 37.4 bits (85), Expect = 0.35
Identities = 44/176 (25%), Positives = 77/176 (43%), Gaps = 25/176 (14%)
Query: 73 RKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKGVEID 132
R E+ E + R++ +E + +R+ + + +LA + A + L+ VE
Sbjct: 2531 RSKEQAELEATRQRQLAAEEEQRRREAEERVQKSLAAEEEAARQRKSA-----LEEVERL 2585
Query: 133 KACAQLKLEEAKAAYSRLKSDA--------ENAAKRAEIAKK-------QAEDTLAGVRK 177
KA K+EEA+ R + ++ E A KR + +K Q E L +
Sbjct: 2586 KA----KVEEARRLRERAEQESARQLQLAQEAAQKRLQAEEKAHAFAVQQKEQELQQTLQ 2641
Query: 178 QLQEADEVL-AKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
Q Q E L A+ +AAR +AE ++E +E + + V+ + EEQA+
Sbjct: 2642 QEQSMLERLRAEAEAARRAAEEAEEARERAEREAAQSRRQVEEAERLKQLAEEQAQ 2697
>emb|CAA87454.1| neurofilament protein M [Oryctolagus cuniculus]
gi|1083164|pir||S55395 neurofilament protein M - rabbit
(fragment) gi|1709261|sp|P54938|NFM_RABIT Neurofilament
triplet M protein (160 kDa neurofilament protein)
(Neurofilament medium polypeptide) (NF-M)
Length = 644
Score = 53.9 bits (128), Expect = 4e-06
Identities = 48/209 (22%), Positives = 88/209 (41%), Gaps = 29/209 (13%)
Query: 48 KNKGGVNPESMSAFIASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTL 107
KN+G A + AK +K E+K + K+ P+ + + K +K+ +
Sbjct: 347 KNEGEQEEGETEAEGEVEEAEAKEEKKTEEKSEEVAAKEEPVTEAKVGKPEKAKSPVPKS 406
Query: 108 AVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAE----I 163
V + A G KG + ++ + K+EE K ++ E K+ E +
Sbjct: 407 PVEEVKPKAEATAG-----KGEQKEE---EEKVEEEKKKAAKESPKEEKVEKKEEKPKDV 458
Query: 164 AKKQAED----------------TLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEA 207
KK+AE T G+ K+ +E ++ L ++K ++ E E+E
Sbjct: 459 PKKKAESPVKEEAAEEAATITKPTKVGLEKETKEGEKPLQQEKEKEKAGEEGGSEEEGSD 518
Query: 208 KG-EGVKKADVKVGGSGEVNQEEQAKNDQ 235
+G + KK D+ V G GE +EE+ + +
Sbjct: 519 QGSKRAKKEDIAVNGEGEGKEEEEPETKE 547
Score = 38.1 bits (87), Expect = 0.21
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 5/76 (6%)
Query: 160 RAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKV 219
+ E K + ED L + ++L V K++ E AE +EE+E E + KK+ VK
Sbjct: 247 KVEDEKSEMEDALTAIAEEL----AVSVKEEEKEEEAEGKEEEQEAEEEVAAAKKSPVK- 301
Query: 220 GGSGEVNQEEQAKNDQ 235
+ E+ +EE K ++
Sbjct: 302 ATTPEIKEEEGEKEEE 317
Score = 37.0 bits (84), Expect = 0.46
Identities = 27/94 (28%), Positives = 45/94 (47%), Gaps = 8/94 (8%)
Query: 141 EEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASD 200
EE + + + E K E ++AE+ +A +K +A K++ + E +
Sbjct: 264 EELAVSVKEEEKEEEAEGKEEE---QEAEEEVAAAKKSPVKATTPEIKEEEGEKEEEGQE 320
Query: 201 EEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKND 234
EE+EEE EGVK + GGS +E +KN+
Sbjct: 321 EEEEEE--DEGVKSDQAEEGGS---EKEGSSKNE 349
Score = 35.8 bits (81), Expect = 1.0
Identities = 40/167 (23%), Positives = 71/167 (41%), Gaps = 16/167 (9%)
Query: 64 SQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQAL------AAAS 117
++ KG +K E+++ + +KK+ +E K +K + + +A AA
Sbjct: 415 AEATAGKGEQKEEEEKVEEEKKKAAKESPKEEKVEKKEEKPKDVPKKKAESPVKEEAAEE 474
Query: 118 CATGLLPHLKGVEIDKACAQLKLEEAKA---AYSRLKSDAENAAKRAEIAKKQAEDTLAG 174
AT P G+E + + L++ K A S+ E + + ++ AKK ED
Sbjct: 475 AATITKPTKVGLEKETKEGEKPLQQEKEKEKAGEEGGSEEEGSDQGSKRAKK--EDIAVN 532
Query: 175 VRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGG 221
+ +E +E K+K + +EEK G + AD K GG
Sbjct: 533 GEGEGKEEEEPETKEKGS-----GREEEKGVVTNGLDLSPADEKKGG 574
Score = 35.8 bits (81), Expect = 1.0
Identities = 46/187 (24%), Positives = 75/187 (39%), Gaps = 22/187 (11%)
Query: 67 ETAKG-HRKNEKKETKGVRKKSPLV------------DEREAKRQKSDASKGTLAVNQAL 113
E A+G + E +E KKSP+ E E + ++ + + +QA
Sbjct: 277 EEAEGKEEEQEAEEEVAAAKKSPVKATTPEIKEEEGEKEEEGQEEEEEEEDEGVKSDQAE 336
Query: 114 AAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLA 173
S G + E + A+ ++EEA+A K + + K E+A K+ T A
Sbjct: 337 EGGSEKEGSSKNEGEQEEGETEAEGEVEEAEA-----KEEKKTEEKSEEVAAKEEPVTEA 391
Query: 174 GVRKQLQEADEV----LAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEE 229
V K + V + + K E+ E+KEEE K E KK K E +++
Sbjct: 392 KVGKPEKAKSPVPKSPVEEVKPKAEATAGKGEQKEEEEKVEEEKKKAAKESPKEEKVEKK 451
Query: 230 QAKNDQV 236
+ K V
Sbjct: 452 EEKPKDV 458
>gb|AAH78128.1| LOC397995 protein [Xenopus laevis]
Length = 913
Score = 53.9 bits (128), Expect = 4e-06
Identities = 47/194 (24%), Positives = 83/194 (42%), Gaps = 26/194 (13%)
Query: 45 EMVKNKGGVNPESMSAFIASQVETAKGHRKN---EKKETKGVRKKSPLVDEREAKRQKSD 101
E K +GG E +A+ E +G + EK E KG +E+E + + +
Sbjct: 598 EAEKEEGGEEEEKEE--VAADEEGGEGEEEGGEGEKDEDKGE-------EEKEGEAEAEE 648
Query: 102 ASKGTLAVNQALAAASCATGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRA 161
A G + A +++ + K+ KA +++ E + A
Sbjct: 649 AEGGEEETTEEAQAEEV------------VEETMTETKIVREKAEVQETEAEEEET-QDA 695
Query: 162 EIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGG 221
E K + ED G K+ +E +E + + A E E +EEK EE KGE + + + K G
Sbjct: 696 EEQKDEGEDEETGEGKETEEKEEGGEEKEDADEGTEQQEEEKAEEEKGE-IDEGETKEGA 754
Query: 222 SGEVNQEEQAKNDQ 235
+ +++QAK+ +
Sbjct: 755 EEDAGEDKQAKSPE 768
Score = 36.6 bits (83), Expect = 0.60
Identities = 25/96 (26%), Positives = 44/96 (45%), Gaps = 3/96 (3%)
Query: 141 EEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKA-ARESAEAS 199
EE + + + E + + + G + +E +E +++A A E E
Sbjct: 474 EEEEQGEETQEGEEEAEEEIVAAVESSVQAAAPGEEAEGEEEEEEKGEEEAEAEEEGEKE 533
Query: 200 DEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQ 235
+EEKEEE + EG K + + G GE +EE K ++
Sbjct: 534 EEEKEEEGEDEGEKVDEEEEG--GEQGEEEDVKAEE 567
Score = 33.5 bits (75), Expect = 5.1
Identities = 30/106 (28%), Positives = 49/106 (45%), Gaps = 13/106 (12%)
Query: 140 LEEAKAAYSRLKSDAENAAKRAEIA---------KKQAEDTLAGVRKQLQE-ADEVLAKD 189
+EE K R +A AA E A ++Q E+T G + +E V +
Sbjct: 442 IEETKVEDDRSDMEAALAAMADEFALGFEEGGEEEEQGEETQEGEEEAEEEIVAAVESSV 501
Query: 190 KAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQ 235
+AA EA EE+EEE +G ++A+ + G E ++E+ D+
Sbjct: 502 QAAAPGEEAEGEEEEEE---KGEEEAEAEEEGEKEEEEKEEEGEDE 544
>emb|CAH98125.1| MAEBL, putative [Plasmodium berghei]
Length = 1712
Score = 53.9 bits (128), Expect = 4e-06
Identities = 49/177 (27%), Positives = 82/177 (45%), Gaps = 12/177 (6%)
Query: 65 QVETAKGHRKNEKKE-----TKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCA 119
++E AK + KK+ + RKK + E KR+K +A+K V + A
Sbjct: 1227 RIEEAKKVEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAK---KVEEERKRIEEA 1283
Query: 120 TGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQL 179
+ K E K + + ++ A + K + AAK+AE +K+AE A + +
Sbjct: 1284 KKVEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKAE---AAKKAER 1340
Query: 180 QEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQ-EEQAKNDQ 235
+E DE K + R+ EA+ + +EE K E KKA+ + E + EE+ K D+
Sbjct: 1341 KEKDEAAKKAEEKRKKDEAAKKVEEERKKAEEAKKAEEERKRIEEAKKVEEKRKKDE 1397
Score = 53.5 bits (127), Expect = 5e-06
Identities = 53/172 (30%), Positives = 81/172 (46%), Gaps = 11/172 (6%)
Query: 63 ASQVETAKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGL 122
A +VE + R E K+ + RKK + E KR+K +A+K + AA A
Sbjct: 1270 AKKVEEER-KRIEEAKKVEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAKKAEE- 1327
Query: 123 LPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGV--RKQLQ 180
K KA + K E AK A + K D AAK+ E +K+AE+ RK+++
Sbjct: 1328 -KRKKAEAAKKAERKEKDEAAKKAEEKRKKD--EAAKKVEEERKKAEEAKKAEEERKRIE 1384
Query: 181 EADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
EA +V K R+ EA+ + +EE K E KKA+ + E + E+ +
Sbjct: 1385 EAKKVEEK----RKKDEAAKKAEEERKKAEAAKKAEEERKRIEEAKKAEEER 1432
Score = 52.0 bits (123), Expect = 1e-05
Identities = 52/179 (29%), Positives = 79/179 (44%), Gaps = 14/179 (7%)
Query: 69 AKGHRKNEK--KETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHL 126
A+ RK ++ K+ + RKK + E KR+K +A+K + + A
Sbjct: 1182 AEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAKKAEEERKRIEEAKKVEE--KRK 1239
Query: 127 KGVEIDKACAQLKLEEA--KAAYSRLKSDA----ENAAKRAEIAKKQAE----DTLAGVR 176
K KA + K +EA KA R K +A E KR E AKK E D A
Sbjct: 1240 KDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAKKVEEERKRIEEAKKVEEKRKKDEAAKKA 1299
Query: 177 KQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAKNDQ 235
++ ++ DE K + R+ EA+ + +E+ K E KKA+ K EE+ K D+
Sbjct: 1300 EEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKAEAAKKAERKEKDEAAKKAEEKRKKDE 1358
Score = 47.4 bits (111), Expect = 3e-04
Identities = 52/182 (28%), Positives = 82/182 (44%), Gaps = 22/182 (12%)
Query: 63 ASQVETAKGHRKN--EKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCAT 120
A + + A+ RK E K+ + RKK + E +R+K++A+K + + A A
Sbjct: 1370 AEEAKKAEEERKRIEEAKKVEEKRKKDEAAKKAEEERKKAEAAKKAEEERKRIEEAKKAE 1429
Query: 121 GLLPHLKGVEIDKACAQ--LKLEEAKAAYSRLK-----SDAENAAKRAEIAKKQAEDTLA 173
K +E K + ++EEAK K AE KR E AKK E+
Sbjct: 1430 ---EERKRIEAAKKVEEERKRIEEAKKVEEERKRIEEAKKAEEERKRIEAAKKVEEE--- 1483
Query: 174 GVRKQLQEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQ-EEQAK 232
RK+++EA + + K E+ +A +E K EA KKA+ + E + EE+ K
Sbjct: 1484 --RKRIEEAKKSEEERKRIEEAKKAEEERKRIEA----AKKAEEERKRIEEAKKAEEEIK 1537
Query: 233 ND 234
D
Sbjct: 1538 KD 1539
Score = 45.4 bits (106), Expect = 0.001
Identities = 48/177 (27%), Positives = 80/177 (45%), Gaps = 21/177 (11%)
Query: 62 IASQVETAK--GHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCA 119
IA ++ K G E K+ + RK+ + E +R+K++A+K + + A
Sbjct: 1110 IADMIKKRKEAGENAEEVKKAEEERKRIEAAKKVEEERKKAEAAKKAEEERKRIEEA--- 1166
Query: 120 TGLLPHLKGVEIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGVRKQL 179
K VE + K E AK A + K D AAK+AE +K+ +D A ++
Sbjct: 1167 -------KKVEEKRK----KDEAAKKAEEKRKKD--EAAKKAE--EKRKKDEAAKKAEEK 1211
Query: 180 QEADEVLAKDKAARESAEASDEEKEEEAKGEGVKKADVK-VGGSGEVNQEEQAKNDQ 235
++ DE K + R+ E + + +E+ K E KKA+ K EE+ K D+
Sbjct: 1212 RKKDEAAKKAEEERKRIEEAKKVEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDE 1268
Score = 42.0 bits (97), Expect = 0.014
Identities = 43/152 (28%), Positives = 66/152 (43%), Gaps = 15/152 (9%)
Query: 65 QVETAKGHRKNEK----KETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCAT 120
+ E AK + EK K+ + RKK + E +R+K++ +K + + A
Sbjct: 1331 KAEAAKKAERKEKDEAAKKAEEKRKKDEAAKKVEEERKKAEEAKKAEEERKRIEEAKKVE 1390
Query: 121 GLLPHLKGV-----EIDKACAQLKLEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAGV 175
+ E KA A K EE + K AE KR E AKK E+
Sbjct: 1391 EKRKKDEAAKKAEEERKKAEAAKKAEEERKRIEEAKK-AEEERKRIEAAKKVEEE----- 1444
Query: 176 RKQLQEADEVLAKDKAARESAEASDEEKEEEA 207
RK+++EA +V + K E+ +A +E K EA
Sbjct: 1445 RKRIEEAKKVEEERKRIEEAKKAEEERKRIEA 1476
Score = 41.2 bits (95), Expect = 0.024
Identities = 47/167 (28%), Positives = 75/167 (44%), Gaps = 30/167 (17%)
Query: 69 AKGHRKNEKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCATGLLPHLKG 128
A+ +K E+KE KK+ E KR+K +A+K + K
Sbjct: 1332 AEAAKKAERKEKDEAAKKA------EEKRKKDEAAKKVEEERK---------------KA 1370
Query: 129 VEIDKACAQLK-LEEAKAAYSRLKSDAENAAKRAEIAKKQAEDTLAG--VRKQLQEADEV 185
E KA + K +EEAK + K D AAK+AE +K+AE RK+++EA
Sbjct: 1371 EEAKKAEEERKRIEEAKKVEEKRKKD--EAAKKAEEERKKAEAAKKAEEERKRIEEA--- 1425
Query: 186 LAKDKAARESAEASDEEKEEEAKGEGVKKADVKVGGSGEVNQEEQAK 232
K + R+ EA+ + +EE + E KK + + E + E+ +
Sbjct: 1426 -KKAEEERKRIEAAKKVEEERKRIEEAKKVEEERKRIEEAKKAEEER 1471
Score = 37.4 bits (85), Expect = 0.35
Identities = 29/85 (34%), Positives = 44/85 (51%), Gaps = 4/85 (4%)
Query: 155 ENAAKRAEIAKKQAEDTLAGVRKQLQEADEVLAKDKAARESA---EASDEEKEEEAKGEG 211
E A K A+I +K E +A + K+ +EA E + K A E EA+ + +EE K E
Sbjct: 1093 EEAEKNAKIIRKFEELRIADMIKKRKEAGENAEEVKKAEEERKRIEAAKKVEEERKKAEA 1152
Query: 212 VKKADVKVGGSGEVNQ-EEQAKNDQ 235
KKA+ + E + EE+ K D+
Sbjct: 1153 AKKAEEERKRIEEAKKVEEKRKKDE 1177
Score = 34.3 bits (77), Expect = 3.0
Identities = 42/166 (25%), Positives = 68/166 (40%), Gaps = 20/166 (12%)
Query: 63 ASQVETAKGHRKN--EKKETKGVRKKSPLVDEREAKRQKSDASKGTLAVNQALAAASCAT 120
A + A+ RK E K+ + RK+ + E +R++ + +K + + A A
Sbjct: 1409 AEAAKKAEEERKRIEEAKKAEEERKRIEAAKKVEEERKRIEEAKKVEEERKRIEEAKKAE 1468
Query: 121 GLLPHLKGVEIDKACAQ--LKLEEAKAAYSRLK-----SDAENAAKRAEIAKKQAEDTLA 173
K +E K + ++EEAK + K AE KR E AKK E+
Sbjct: 1469 ---EERKRIEAAKKVEEERKRIEEAKKSEEERKRIEEAKKAEEERKRIEAAKKAEEE--- 1522
Query: 174 GVRKQLQE---ADEVLAKDKAARESAEASDEEKEEEAKGEGVKKAD 216
RK+++E A+E + KD E +S + KK D
Sbjct: 1523 --RKRIEEAKKAEEEIKKDSNLAEKKISSSNYETRHIDDNSFKKLD 1566
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.307 0.123 0.319
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 341,900,736
Number of Sequences: 2540612
Number of extensions: 12572800
Number of successful extensions: 137732
Number of sequences better than 10.0: 3983
Number of HSP's better than 10.0 without gapping: 650
Number of HSP's successfully gapped in prelim test: 3512
Number of HSP's that attempted gapping in prelim test: 113070
Number of HSP's gapped (non-prelim): 15499
length of query: 236
length of database: 863,360,394
effective HSP length: 124
effective length of query: 112
effective length of database: 548,324,506
effective search space: 61412344672
effective search space used: 61412344672
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0023.4


