

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0020.10
(535 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
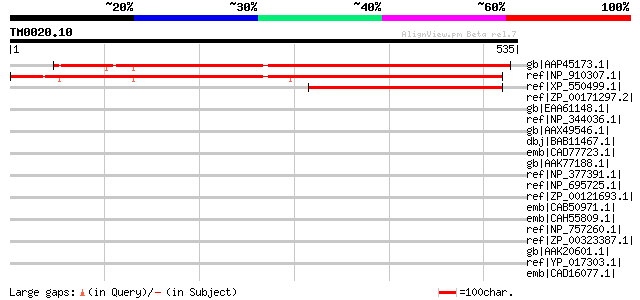
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP45173.1| hypothetical protein 177O13.25 [Solanum bulbocast... 538 e-151
ref|NP_910307.1| unknown protein [Oryza sativa (japonica cultiva... 424 e-117
ref|XP_550499.1| unknown protein [Oryza sativa (japonica cultiva... 221 6e-56
ref|ZP_00171297.2| COG2864: Cytochrome b subunit of formate dehy... 47 0.002
gb|EAA61148.1| hypothetical protein AN7277.2 [Aspergillus nidula... 46 0.003
ref|NP_344036.1| Permease, multidrug resistance protein [Sulfolo... 40 0.24
gb|AAX49546.1| cation/H+ exchanger [Arabidopsis thaliana] 39 0.32
dbj|BAB11467.1| Na+/H+ antiporter-like protein [Arabidopsis thal... 39 0.32
emb|CAD77723.1| cytochrome oxidase subunit I [Rhodopirellula bal... 39 0.41
gb|AAK77188.1| cytochrome oxidase subunit I [Periclistus brandti] 39 0.41
ref|NP_377391.1| hypothetical protein ST1433 [Sulfolobus tokodai... 39 0.41
ref|NP_695725.1| hypothetical transmembrane protein with unknown... 39 0.41
ref|ZP_00121693.1| COG0477: Permeases of the major facilitator s... 39 0.41
emb|CAB50971.1| SPBC1105.08 [Schizosaccharomyces pombe] gi|19113... 39 0.54
emb|CAH55809.1| putative transport protein [Escherichia coli] 39 0.54
ref|NP_757260.1| Hypothetical protein yjiJ [Escherichia coli CFT... 39 0.54
ref|ZP_00323387.1| COG0477: Permeases of the major facilitator s... 39 0.54
gb|AAK20601.1| cytochrome oxidase subunit I [Diadasia martialis] 39 0.54
ref|YP_017303.1| multidrug resistance protein, putative [Bacillu... 38 0.71
emb|CAD16077.1| HYPOTHETICAL TRANSMEMBRANE PROTEIN [Ralstonia so... 38 0.71
>gb|AAP45173.1| hypothetical protein 177O13.25 [Solanum bulbocastanum]
Length = 553
Score = 538 bits (1385), Expect = e-151
Identities = 268/493 (54%), Positives = 348/493 (70%), Gaps = 18/493 (3%)
Query: 47 REAYNGYEEVGEKPSRFEVMGWYLYEFCSYFIQTVLIPVVFPLIISQLQNLPAD----WA 102
R +Y G ++ KP++ EV GWYLY CSYFI TVLIP+VFPLIISQ P
Sbjct: 47 RGSYGG-DDQEMKPTKIEVFGWYLYGMCSYFIHTVLIPIVFPLIISQTLYWPQQPQLGIV 105
Query: 103 KNNRPAGTHCSQKELHLYSKLTNQTIT---SNFSALEWTSIGWAIGLAIAAPILGFLSFH 159
KN + G C Q+EL LY LT + + + +SALEWTSI W IGL +AAP+LGFLS H
Sbjct: 106 KNAKGLG--CRQRELELYEMLTKRRMNLAGTEYSALEWTSISWIIGLILAAPVLGFLSIH 163
Query: 160 LD-GHFPKFITVASTAVGVFFCLPAGFFKVTAIFIPYIAGIIAASTVATAAHTHHLGLMI 218
LD G + + I ASTA+G FCLPAGFFK IF PYIA I+AA+T+ ++ H HLGLM+
Sbjct: 164 LDYGGYQQLIAAASTAIGGLFCLPAGFFKTRWIFPPYIAAIVAANTIVSSCHARHLGLMV 223
Query: 219 RSFTGPTLKRSQFPIRQGISSKLSLHATAAGCLGSALISAFTYHMIRELNVNEPDIMSLW 278
R G +++SQF R+ ++S LSLH+TAAGCLG+AL+SAFTYHM+R+ + SLW
Sbjct: 224 RGLVGSPIRKSQFSDRKAVASWLSLHSTAAGCLGAALMSAFTYHMLRKSD----SFTSLW 279
Query: 279 IVSIFSGLIWLVGIFHVATAI--GRTTD-EISFSSRLHPFSVFKYPHAIGALASVFISSF 335
+VSIFSGLIW +G+ H+AT+ G+ D + + + H S+FKYPHA G+LA VF+SSF
Sbjct: 280 VVSIFSGLIWFIGMVHIATSNRPGQNADLHTNSAPKTHVVSIFKYPHAAGSLAGVFLSSF 339
Query: 336 ITMTIFTGGVVFIVGQLCIKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVKMQIVG 395
TM IFT GV++ +G LCI PV++L+ WLTYF+FP+ SL L P ++I+ ++VKMQ++G
Sbjct: 340 TTMCIFTAGVLYSIGDLCIPPVNILYLWLTYFIFPLVSLPLTHPFQQLIRADAVKMQLLG 399
Query: 396 FLFSLISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEGAFSI 455
F+ S I SGFGFYY H +W H+ +F+ IQ+T TGILHAF RVL LDC+PSGKEGAFS+
Sbjct: 400 FILSAIVSGFGFYYRHDNWNKVHIFIFTAIQTTATGILHAFGRVLWLDCSPSGKEGAFSV 459
Query: 456 WYAWMRSAGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIVVLLFGNISDVGGAVAAGHVR 515
W++W+R+ G GF + S SP NI SFG AFC AI G ++L+FGNIS+ GGA AAGHV+
Sbjct: 460 WFSWIRALGACAGFALASTSPQNIEMSFGVAFCAAILGKIILIFGNISNFGGAKAAGHVK 519
Query: 516 DDSERSSAVAGLD 528
D S GLD
Sbjct: 520 DSERGSPVPTGLD 532
>ref|NP_910307.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|7248400|dbj|BAA92723.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 585
Score = 424 bits (1089), Expect = e-117
Identities = 225/540 (41%), Positives = 330/540 (60%), Gaps = 26/540 (4%)
Query: 2 KEVTVEAEGGGHHQGGAQGGAPENGKGTTATRPDLHNRAMKILRAREAYN------GYEE 55
KE + GG+ GG GG G G +P +I REAY Y
Sbjct: 29 KEKAAVPKNGGNGGGGKNGGGNGGGNGGAGAQPG-EETTREIQVVREAYRREPAAPAYVM 87
Query: 56 VGEKPSRFEVMGWYLYEFCSYFIQTVLIPVVFPLIISQLQNLPADWAKNNR--PAGTHCS 113
E P+ E++GWYLY FCSYFI +L+PV+FP II+Q+ +D+ + + G CS
Sbjct: 88 PEEPPAMVELVGWYLYGFCSYFITHLLLPVLFPAIITQVAFPASDFTPDTKYIVKGATCS 147
Query: 114 QKELHLYSKLTNQTIT---SNFSALEWTSIGWAIGLAIAAPILGFLSFHLD-GHFPKFIT 169
E+ +Y +LT +I S S L W+ + WAIG+ IAAPIL + HLD G + I
Sbjct: 148 IHEMSMYQRLTKHSIAIDGSRLSPLGWSGLSWAIGILIAAPILTQAAHHLDRGQYQSLIL 207
Query: 170 VASTAVGVFFCLPAGFFKVTAIFIPYIAGIIAASTVATAAHTHHLGLMIRSFTGPTLKRS 229
+A+T+ G FFCL GFFK +F+ YI I A+ VA A HT +LGLMIR +
Sbjct: 208 IAATSFGSFFCLLTGFFKTVWVFLFYILFIGASIIVAEAVHTRNLGLMIRGLAAHDSGKH 267
Query: 230 QFPIRQGISSKLSLHATAAGCLGSALISAFTYHMIRELNVNEPDIMSLWIVSIFSGLIWL 289
R+ +S+L+L+ TA G +G+AL++AF YHM+R + + LW+VSIFSGLIW
Sbjct: 268 LVLRRRAAASQLTLYCTAIGGIGAALMAAFMYHMLRRTD----QLTGLWVVSIFSGLIWF 323
Query: 290 VGIFH------VATAIGRTTDEISFSSRL-HPFSVFKYPHAIGALASVFISSFITMTIFT 342
+GI H +++ T E +F ++L + ++ +YP AIG+L +VF+SSF TM IFT
Sbjct: 324 IGICHGLFTNRPSSSSPTTAFEPNFFTKLSYSMTLVRYPQAIGSLVAVFLSSFATMCIFT 383
Query: 343 GGVVFIVGQLCIKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVKMQIVGFLFSLIS 402
G ++ +G +CIKPV +L W+ YFLFP+ SL LL P+ +I+ ++V+MQ++GF+ L
Sbjct: 384 SGTLYAIGGVCIKPVLVLVLWILYFLFPLISLPLLHPIQIIIRADAVRMQLLGFIICLFV 443
Query: 403 SGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEGAFSIWYAWMRS 462
SG GFY+ W+ H++V +++QST G+L++F R+L+LD +P GKEGAF++WYA++R
Sbjct: 444 SGAGFYFKSHRWRAAHIIVIALVQSTANGVLYSFGRILLLDASPPGKEGAFAVWYAFVRC 503
Query: 463 AGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIVVLLFGNISDVGGAVAAGHVR--DDSER 520
G ++GF S PG SF AAF + GI+VL+FGN+S++G AAGH++ +D +R
Sbjct: 504 IGAMIGFAAASAGPGRAGGSFAAAFLGSFLGIIVLIFGNVSNIGALKAAGHLKGMEDEKR 563
>ref|XP_550499.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|55295891|dbj|BAD67759.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 230
Score = 221 bits (562), Expect = 6e-56
Identities = 97/207 (46%), Positives = 148/207 (70%), Gaps = 2/207 (0%)
Query: 316 SVFKYPHAIGALASVFISSFITMTIFTGGVVFIVGQLCIKPVHLLFFWLTYFLFPVFSLS 375
++ +YP AIG+L +VF+SSF TM IFT G ++ +G +CIKPV +L W+ YFLFP+ SL
Sbjct: 2 TLVRYPQAIGSLVAVFLSSFATMCIFTSGTLYAIGGVCIKPVLVLVLWILYFLFPLISLP 61
Query: 376 LLQPLLRVIKMNSVKMQIVGFLFSLISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHA 435
LL P+ +I+ ++V+MQ++GF+ L SG GFY+ W+ H++V +++QST G+L++
Sbjct: 62 LLHPIQIIIRADAVRMQLLGFIICLFVSGAGFYFKSHRWRAAHIIVIALVQSTANGVLYS 121
Query: 436 FSRVLVLDCAPSGKEGAFSIWYAWMRSAGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIV 495
F R+L+LD +P GKEGAF++WYA++R G ++GF S PG SF AAF + GI+
Sbjct: 122 FGRILLLDASPPGKEGAFAVWYAFVRCIGAMIGFAAASAGPGRAGGSFAAAFLGSFLGII 181
Query: 496 VLLFGNISDVGGAVAAGHVR--DDSER 520
VL+FGN+S++G AAGH++ +D +R
Sbjct: 182 VLIFGNVSNIGALKAAGHLKGMEDEKR 208
>ref|ZP_00171297.2| COG2864: Cytochrome b subunit of formate dehydrogenase [Ralstonia
eutropha JMP134]
Length = 398
Score = 47.0 bits (110), Expect = 0.002
Identities = 34/116 (29%), Positives = 55/116 (47%), Gaps = 18/116 (15%)
Query: 334 SFITMTIFTGGVVFIVGQLCIKPV--HLLFFWLTYFLF-------PVFSLSLLQPLLRVI 384
SF+ + + G+V + G+ + P+ H LF WL+Y L PVF+LS++ + +
Sbjct: 188 SFVLLGV--SGIVMLFGKHFLLPIMGHTLFGWLSYLLKNIHNLAGPVFTLSIIVAFVIFV 245
Query: 385 KMNSVKMQIVGFLFSLISSGFG-------FYYGHSHWKWGHLVVFSVIQSTGTGIL 433
+ N V ++ SL G F +G W WG LV+F +I S +L
Sbjct: 246 RDNLPSRDDVRWVTSLGGLASGKHVPSGRFNFGEKMWFWGGLVIFGLILSASGWVL 301
>gb|EAA61148.1| hypothetical protein AN7277.2 [Aspergillus nidulans FGSC A4]
gi|67900580|ref|XP_680546.1| hypothetical protein
AN7277_2 [Aspergillus nidulans FGSC A4]
gi|49106303|ref|XP_411414.1| hypothetical protein
AN7277.2 [Aspergillus nidulans FGSC A4]
Length = 2150
Score = 45.8 bits (107), Expect = 0.003
Identities = 49/210 (23%), Positives = 88/210 (41%), Gaps = 18/210 (8%)
Query: 292 IFHVATAIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITMTIFTGGVVFIVGQ 351
+F T + T SF+SR F V I A+ I+S +T T++ + + GQ
Sbjct: 1600 LFCSLTPVSATRPYTSFTSRSTMFIVI-----IAGFAT--ITSPLTATVYFPLLPTLEGQ 1652
Query: 352 LCIKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVKMQIVGFLFSLISSGFGFYYGH 411
+ P + Y +F S ++ PL + + + + +++L + G
Sbjct: 1653 FRVSPQAINMTLTIYVIFQAISPAVFGPLSDTVGRRPIFLLTLA-IYALANLGMAL---- 1707
Query: 412 SHWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEGAFSIWYAWMRSAGLLVGFTV 471
+ +G L++ +QS G A S +V D S + G W + + G +G +
Sbjct: 1708 NKHNYGFLLLLRALQSLGASAAFAISYGIVADVCVSSERGKTMAWVSIALNMGTCLGPII 1767
Query: 472 GSV---SPGNIRTSFGAAFCTAIAGIVVLL 498
G + G+I F A F I G+++LL
Sbjct: 1768 GGLVAYLSGDIEWVFWALF---IVGLLLLL 1794
>ref|NP_344036.1| Permease, multidrug resistance protein [Sulfolobus solfataricus P2]
gi|13816035|gb|AAK42826.1| Permease, multidrug
resistance protein [Sulfolobus solfataricus P2]
gi|25394872|pir||C90446 permease, multidrug resistance
protein [imported] - Sulfolobus solfataricus
Length = 469
Score = 39.7 bits (91), Expect = 0.24
Identities = 34/93 (36%), Positives = 46/93 (48%), Gaps = 11/93 (11%)
Query: 419 LVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEG-AFSIWYAWMRSAGLLVGFTVGSVSPG 477
L+VF +IQ+ G +L A S +V D P + G A+ I S G +G VG V G
Sbjct: 96 LIVFRIIQAIGGSMLVANSSSIVADVFPPNRRGKAYGI-----TSLGWNIGALVGIVLGG 150
Query: 478 NIRTSFGAAFCTAI---AGIVVLLFG--NISDV 505
+ T FG + I GIV +L G NI D+
Sbjct: 151 VLTTFFGWQYIFYINVPIGIVAVLLGVMNIKDI 183
>gb|AAX49546.1| cation/H+ exchanger [Arabidopsis thaliana]
Length = 808
Score = 39.3 bits (90), Expect = 0.32
Identities = 55/240 (22%), Positives = 89/240 (36%), Gaps = 36/240 (15%)
Query: 145 GLAIAAPILGFLSFHLDGHFPK-FITVASTAVGV---FFCLPAGF--------------- 185
G+ + +LG LD FPK +TV T + FF AG
Sbjct: 67 GIMLGPSLLGRSKAFLDAVFPKKSLTVLETLANLGLLFFLFLAGLEIDTKALRRTGKKAL 126
Query: 186 -FKVTAIFIPYIAGIIAASTVATAAHTHHLGLMIRSFTGPTLKRSQFPIRQGISSKLSLH 244
+ I +P+ GI ++ + F G L + FP+ I ++L L
Sbjct: 127 GIALAGITLPFALGIGSSFVLKATISKGVNSTAFLVFMGVALSITAFPVLARILAELKLL 186
Query: 245 ATAAG--CLGSALISAFTYHMIRELNV-----NEPDIMSLWIVSIFSGLIWLVGIFHVAT 297
T G + +A ++ ++ L + N ++SLW+ SG +++G +
Sbjct: 187 TTEIGRLAMSAAAVNDVAAWILLALAIALSGSNTSPLVSLWV--FLSGCAFVIGASFIIP 244
Query: 298 AIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITMTIFTGGV--VFIVGQLCIK 355
I R S R H + + LA V + FIT I + F+VG L K
Sbjct: 245 PIFRW-----ISRRCHEGEPIEETYICATLAVVLVCGFITDAIGIHSMFGAFVVGVLIPK 299
>dbj|BAB11467.1| Na+/H+ antiporter-like protein [Arabidopsis thaliana]
gi|42568248|ref|NP_198976.3| cation/hydrogen exchanger,
putative (CHX18) [Arabidopsis thaliana]
Length = 810
Score = 39.3 bits (90), Expect = 0.32
Identities = 55/240 (22%), Positives = 89/240 (36%), Gaps = 36/240 (15%)
Query: 145 GLAIAAPILGFLSFHLDGHFPK-FITVASTAVGV---FFCLPAGF--------------- 185
G+ + +LG LD FPK +TV T + FF AG
Sbjct: 67 GIMLGPSLLGRSKAFLDAVFPKKSLTVLETLANLGLLFFLFLAGLEIDTKALRRTGKKAL 126
Query: 186 -FKVTAIFIPYIAGIIAASTVATAAHTHHLGLMIRSFTGPTLKRSQFPIRQGISSKLSLH 244
+ I +P+ GI ++ + F G L + FP+ I ++L L
Sbjct: 127 GIALAGITLPFALGIGSSFVLKATISKGVNSTAFLVFMGVALSITAFPVLARILAELKLL 186
Query: 245 ATAAG--CLGSALISAFTYHMIRELNV-----NEPDIMSLWIVSIFSGLIWLVGIFHVAT 297
T G + +A ++ ++ L + N ++SLW+ SG +++G +
Sbjct: 187 TTEIGRLAMSAAAVNDVAAWILLALAIALSGSNTSPLVSLWV--FLSGCAFVIGASFIIP 244
Query: 298 AIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITMTIFTGGV--VFIVGQLCIK 355
I R S R H + + LA V + FIT I + F+VG L K
Sbjct: 245 PIFRW-----ISRRCHEGEPIEETYICATLAVVLVCGFITDAIGIHSMFGAFVVGVLIPK 299
>emb|CAD77723.1| cytochrome oxidase subunit I [Rhodopirellula baltica SH 1]
gi|32477652|ref|NP_870646.1| cytochrome oxidase subunit
I [Rhodopirellula baltica SH 1]
Length = 580
Score = 38.9 bits (89), Expect = 0.41
Identities = 70/299 (23%), Positives = 122/299 (40%), Gaps = 45/299 (15%)
Query: 191 IFIPYIAGIIAASTV----ATAAHTHHL---GLMIRSFTGPTLKRSQFPIRQGISSKLSL 243
I + Y+ G+++A V A A H L G M + F + I L +
Sbjct: 41 IGLMYLIGVMSAFAVGGLLALAIRLHLLNPNGWMFSGAEANNIYNQVFTLHGAIMVFLFI 100
Query: 244 HATAAGCLGSALISAFTYHMIRELNVNEPDIMSLWIVSIFSGLIWLVGIFHVATAIGRTT 303
+ LG+ L+ M+ +V P + ++ S +W+ G TA+
Sbjct: 101 IPSIPAALGNFLVPV----MLGAKDVGFPRL------NLSSFYLWVFGALFFVTAL---- 146
Query: 304 DEISFSSRLHPFSVFKYPHAIGALASVFISS---FIT--MTIFTGGVVFIVGQLCIKPVH 358
FSS L F P++ SV +++ FI +IFTG + FIV ++P
Sbjct: 147 ----FSSGLDTGWTFYTPYSTTTDTSVILATLGAFILGFSSIFTG-LNFIVTINTMRPPG 201
Query: 359 LLFFWLTYFLFPVFSLSLLQPL------LRVIKMNSVKMQIVGFLFSLISSGFGFYYGHS 412
+ +F + FL+ ++ S++Q L + ++ + + + +G +F +G Y H
Sbjct: 202 MTWFKMPLFLWATYATSIIQVLATPVLGITLLLLIAERTMHIG-IFDPEFNGDPVTYQHF 260
Query: 413 HWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEGAFSIWYAWMRSAGLLVGFTV 471
W + H V+ + IL AF + L S K + A+ A L+GF V
Sbjct: 261 FWFYSHPAVYIM-------ILPAFGIISELISVHSHKHIFGYRFIAYSSIAIALLGFLV 312
>gb|AAK77188.1| cytochrome oxidase subunit I [Periclistus brandti]
Length = 359
Score = 38.9 bits (89), Expect = 0.41
Identities = 36/134 (26%), Positives = 60/134 (43%), Gaps = 11/134 (8%)
Query: 343 GGVVFIVGQLCIKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVKMQIVG-----FL 397
GG+ FI L ++P + +T F++ +F +L + + + M +
Sbjct: 144 GGINFITTILNMRPNLMSMDKITLFVWSIFLTVILLVVSLPVLAGGITMLLFDRNMNTSF 203
Query: 398 FSLISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEGAFSIWY 457
F + G Y H W +GH V+ +I G G++ S ++ +C G G+ + Y
Sbjct: 204 FDPLGGGDPILYQHLFWFFGHPEVYILI-LPGFGMV---SHMIYTECGKKGTFGSLGMMY 259
Query: 458 AWMRSAGLLVGFTV 471
A M S GLL GF V
Sbjct: 260 A-MISIGLL-GFVV 271
>ref|NP_377391.1| hypothetical protein ST1433 [Sulfolobus tokodaii str. 7]
gi|15622509|dbj|BAB66500.1| 701aa long conserved
hypothetical protein [Sulfolobus tokodaii str. 7]
Length = 701
Score = 38.9 bits (89), Expect = 0.41
Identities = 70/298 (23%), Positives = 122/298 (40%), Gaps = 55/298 (18%)
Query: 215 GLMIRSFTGPTLKRSQFPIRQGISSKLSLHATAAGCLGSAL-ISAFTYHMIRELNVNEPD 273
GL+ R F GP P+ G+ L+ + G L A I Y I ++ +
Sbjct: 404 GLLAREFFGP------LPVG-GLQELLNNPSAPVGPLYYAWPIPVSFYEKISDIIPTGAN 456
Query: 274 -IMSLWIVSIFSG--LIWLVGIFHVATAIGRTTDEISFSSRLHPFSVFKYPHAIGALASV 330
I++ ++S+ G L+++ + V AI + E F RL F ++ P V
Sbjct: 457 AIINTILLSLLLGSILLFVSTLLGVINAIKKKDKEFLFLDRLPLFIIYIVP------LIV 510
Query: 331 FISSFITMTIFTGGVVFIVGQLCIKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVK 390
F+ FI ++ + G I+G + +YFLF S + P ++++ V
Sbjct: 511 FLYGFINISNYQGEEAMILGGI------------SYFLF--HSGTPAPPSVQILADILVI 556
Query: 391 MQIVGFLFSLISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILH------AFSRVLVLDC 444
+G +++ ++ + H H G ++F I+ L +F RVLV
Sbjct: 557 WVEIGLIYNWVAKAY-LLKKHEHASTGSAIIFGFIEGGFEAALLLLSNTISFIRVLVFAI 615
Query: 445 APSGKEGAFSIWYAWMRSAGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIVVLLFGNI 502
A + I YA+ A L G +P ++ + F AGIV+L+ GN+
Sbjct: 616 AH------YYILYAFSYMAYLAAG------NPSSLLSVF-----INPAGIVILIIGNL 656
>ref|NP_695725.1| hypothetical transmembrane protein with unknown function
[Bifidobacterium longum NCC2705]
gi|23325739|gb|AAN24361.1| hypothetical transmembrane
protein with unknown function [Bifidobacterium longum
NCC2705]
Length = 545
Score = 38.9 bits (89), Expect = 0.41
Identities = 30/128 (23%), Positives = 55/128 (42%), Gaps = 6/128 (4%)
Query: 382 RVIKMNSVKMQIVGFLFSLISSGFGFYYGHSHWKWGH-----LVVFSVIQSTGTGILHAF 436
R+ + N ++V L ++G +G W G +++F++I G+ +
Sbjct: 411 RIEEKNIPVARVVAGACLLYAAGLALAWGLGVWSTGTARENGMLLFALISGFAFGVYDSL 470
Query: 437 SRVLVLDCAPSGKEGAFSIW-YAWMRSAGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIV 495
LV+D P + + YA SAGL + +G++ SFG A ++
Sbjct: 471 GLELVMDSLPDPRRAGHDLGIYALANSAGLALAAIIGALLVNAFEHSFGYYLLFPSAIVM 530
Query: 496 VLLFGNIS 503
VLL G ++
Sbjct: 531 VLLAGIVT 538
>ref|ZP_00121693.1| COG0477: Permeases of the major facilitator superfamily
[Bifidobacterium longum DJO10A]
Length = 545
Score = 38.9 bits (89), Expect = 0.41
Identities = 30/128 (23%), Positives = 55/128 (42%), Gaps = 6/128 (4%)
Query: 382 RVIKMNSVKMQIVGFLFSLISSGFGFYYGHSHWKWGH-----LVVFSVIQSTGTGILHAF 436
R+ + N ++V L ++G +G W G +++F++I G+ +
Sbjct: 411 RIEEKNIPVARVVAGACLLYAAGLALAWGLGVWSTGTARENGMLLFALISGFAFGVYDSL 470
Query: 437 SRVLVLDCAPSGKEGAFSIW-YAWMRSAGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIV 495
LV+D P + + YA SAGL + +G++ SFG A ++
Sbjct: 471 GLELVMDSLPDPRRAGHDLGIYALANSAGLALAAIIGALLVNAFEHSFGYYLLFPSAIVM 530
Query: 496 VLLFGNIS 503
VLL G ++
Sbjct: 531 VLLAGIVT 538
>emb|CAB50971.1| SPBC1105.08 [Schizosaccharomyces pombe]
gi|19113256|ref|NP_596464.1| hypothetical protein
SPBC1105.08 [Schizosaccharomyces pombe 972h-]
gi|7493096|pir||T39285 probable transmembrane protein -
fission yeast (Schizosaccharomyces pombe)
Length = 629
Score = 38.5 bits (88), Expect = 0.54
Identities = 51/226 (22%), Positives = 95/226 (41%), Gaps = 14/226 (6%)
Query: 186 FKVTAIFIPYIAGIIAASTVATAAHTHHLGLMIRSFTGPTLKRSQFPIRQGISSKLSLHA 245
F + I + I GI+A S + A +I F + + + QG+ K +L
Sbjct: 343 FMSSGIVLFAIFGIVAPSRRGSLATATVALFIISGFVSGYVSALSYKLMQGMLRKRNLLL 402
Query: 246 TAAGCLGSALISAFTYHMIRELNVNEPDI-MSLWIVSIFSGLIWLVGIFHVATAIGRTTD 304
T G L +A ++M+ + + S W++ IF L++ V + V + IG +
Sbjct: 403 TPFVVPGFMLAAALFFNMVFWSKSSSSTVPFSSWLLLIFLYLLFTVPLSFVGSLIGFRSR 462
Query: 305 EISFSSRLHPFSVFKYPHAIGALASVFISSFITMTIFTGGVVFIVGQL-CIKPVHLLFFW 363
E + P + P I + S+++SSF I G + F+V + + L+F
Sbjct: 463 EF-----VPPVRTNQIPRQIPS-HSIWLSSF-PSAIIGGSIPFLVILIELFSILDSLWFH 515
Query: 364 LTYFLFPVFSLSLLQPLLRVIKMNSVKMQIVGFLFSLISSGFGFYY 409
YF+F FS ++ + + I+ F L S + +++
Sbjct: 516 PLYFMFG-FSFF----CFGILVTTCIMVSIITVYFQLCSENYNWWW 556
>emb|CAH55809.1| putative transport protein [Escherichia coli]
Length = 392
Score = 38.5 bits (88), Expect = 0.54
Identities = 40/174 (22%), Positives = 68/174 (38%), Gaps = 18/174 (10%)
Query: 131 NFSALEWTSIGWAIGLAIAAPILGFLSFHLDGHFPKFITVASTAVGVFFCLPAGFFK-VT 189
+FS L W + G G + + F +FH F+ ++ A G+ A +
Sbjct: 48 SFSQLSWIASGNYAGYLAGSLLFSFGAFHQPSRLRPFLLASALASGLLILAMAWLLPFIL 107
Query: 190 AIFIPYIAGIIAASTVATAA-----HTHHLGLMIRSFTGP--TLKRSQFPIRQGISSKLS 242
+ I +AG+ +A + + HT H ++ F+G + + G+ LS
Sbjct: 108 VLLIRVLAGVASAGMLIFGSTLIMQHTRHPFVLAALFSGVGIGIALGNEYVLAGLHFDLS 167
Query: 243 LHA--TAAGCLGSALISAFTYHMIRE--------LNVNEPDIMSLWIVSIFSGL 286
AG L ++ A T M + L E IMS W+++I GL
Sbjct: 168 SQTLWQGAGALSGMILIALTLLMPSKKHAIVPMPLAKTEQQIMSWWLLAILYGL 221
>ref|NP_757260.1| Hypothetical protein yjiJ [Escherichia coli CFT073]
gi|26111653|gb|AAN83834.1| Hypothetical protein yjiJ
[Escherichia coli CFT073]
Length = 392
Score = 38.5 bits (88), Expect = 0.54
Identities = 41/174 (23%), Positives = 69/174 (39%), Gaps = 18/174 (10%)
Query: 131 NFSALEWTSIGWAIGLAIAAPILGFLSFHLDGHFPKFITVASTAVGVFFCLPAGFFK-VT 189
+FS L W + G G + + F +FH F+ ++ A G+ A +
Sbjct: 48 SFSQLSWIASGNYAGYLAGSLLFSFGAFHQPSRLRPFLLASALASGLLILAMAWLPPFIL 107
Query: 190 AIFIPYIAGIIAASTVATAA-----HTHHLGLMIRSFTGP--TLKRSQFPIRQGISSKLS 242
+ I +AG+ +A + + HT H ++ F+G + S + G+ LS
Sbjct: 108 VLLIRVLAGVASAGMLIFGSTLIMQHTRHPFVLAALFSGVGIGIALSNEYVLAGLHFDLS 167
Query: 243 LHA--TAAGCLGSALISAFTYHMIRE--------LNVNEPDIMSLWIVSIFSGL 286
AG L ++ A T M + L E IMS W+++I GL
Sbjct: 168 SQTLWQGAGALSGMMLIALTLLMPSKKHAIAPMPLAKTEQQIMSWWLLAILYGL 221
>ref|ZP_00323387.1| COG0477: Permeases of the major facilitator superfamily
[Pediococcus pentosaceus ATCC 25745]
Length = 478
Score = 38.5 bits (88), Expect = 0.54
Identities = 43/175 (24%), Positives = 76/175 (42%), Gaps = 24/175 (13%)
Query: 313 HP---FSVFKYPH-AIGALAS--VFISSFITMTIFTGGVVFIVGQLCIKPVHLLFFWLTY 366
HP FS+FKY G +A+ VFI+ F + +V G L L+
Sbjct: 257 HPLLDFSIFKYSDFTFGLIAALLVFINGFFYNVLMPYYLVNARG---------LSSGLSG 307
Query: 367 FLFPVFSLSLL--QPLLRVI--KMNSVKMQIVGFLFSLISSGF--GFYYGHSHWKWGHLV 420
L + L+++ P+ V+ K NS K+ I+G L++ GF W + +
Sbjct: 308 ILLSIIPLTMVIFGPIGGVLADKFNSTKITIIGVTLVLLAQFLIMGFQLDTPFW---YFI 364
Query: 421 VFSVIQSTGTGILHAFSRVLVLDCAPSGKEGAFSIWYAWMRSAGLLVGFTVGSVS 475
S++ G G+ + + V+ PS + G A R+ G+++G ++ + S
Sbjct: 365 FTSIVTGIGIGMFQSPNNASVMSAVPSNRLGIAGSVNALARNLGMILGVSISTSS 419
>gb|AAK20601.1| cytochrome oxidase subunit I [Diadasia martialis]
Length = 413
Score = 38.5 bits (88), Expect = 0.54
Identities = 40/184 (21%), Positives = 73/184 (38%), Gaps = 20/184 (10%)
Query: 275 MSLWIVSIFSGLIWLVGIFHVATAIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISS 334
+S W++ I L+ L +F+V+ G T P S F Y + +F
Sbjct: 5 ISFWLLPISLTLLILSNLFNVSPGTGWTV--------YPPLSTFLYHSSPSVDMLIFSLH 56
Query: 335 FITMTIFTGGVVFIVGQLCIKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVKMQIV 394
M+ G + F+V + +K + + + ++ F + VF ++L L + ++ M +
Sbjct: 57 ISGMSSILGAMNFMVTIMMMKNLSMNYDQISLFSWSVFITAILLILSLPVLAGAITMLLF 116
Query: 395 G-----FLFSLISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGK 449
F + G Y H W +GH V+ + IL F + + SGK
Sbjct: 117 DRNFNTSFFDPMGGGDPILYQHLFWFFGHPEVYIL-------ILPGFGLISQIIMNESGK 169
Query: 450 EGAF 453
+ F
Sbjct: 170 KEIF 173
>ref|YP_017303.1| multidrug resistance protein, putative [Bacillus anthracis str.
'Ames Ancestor'] gi|30260824|ref|NP_843201.1| multidrug
resistance protein, putative [Bacillus anthracis str.
Ames] gi|49183665|ref|YP_026917.1| multidrug resistance
protein, putative [Bacillus anthracis str. Sterne]
gi|30254273|gb|AAP24687.1| multidrug resistance protein,
putative [Bacillus anthracis str. Ames]
gi|47501102|gb|AAT29778.1| multidrug resistance protein,
putative [Bacillus anthracis str. 'Ames Ancestor']
gi|65318101|ref|ZP_00391060.1| COG0477: Permeases of the
major facilitator superfamily [Bacillus anthracis str.
A2012] gi|49177592|gb|AAT52968.1| multidrug resistance
protein, putative [Bacillus anthracis str. Sterne]
Length = 390
Score = 38.1 bits (87), Expect = 0.71
Identities = 56/226 (24%), Positives = 95/226 (41%), Gaps = 23/226 (10%)
Query: 280 VSIFSGLIWLVGIFHVATAIGRT-TDEISFSSRLHPFSVFKYPHAIGALASVFISSFITM 338
+SI L +V +F + + R T ++ F + K + A F F +
Sbjct: 160 ISILMVLGTIVSLFFLPNNVSRKDTSRTQMMNKEDMFELLKSEPLLQAYIGAFTLMF-SQ 218
Query: 339 TIFTGGVVFIVGQLCIKP----VHLLFFWLTYFLFPVFSLSLLQPLLRVI-KMNSVKMQI 393
I T + V L +K + L F +T LF L P R+ + N K+ +
Sbjct: 219 GIVTYMLPVKVEALALKASTTGMMLSAFGITAILF------FLLPTNRIYDRFNRSKLML 272
Query: 394 VGF-LFSLISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHAFSRVLVLDCAPSGKEG- 451
+G + +L S G + G L + +I G IL L+++ + K G
Sbjct: 273 IGIAVMALALSLLGLFATK-----GMLFIVMMIYGIGFAILFPSINALLVENTTNDKRGK 327
Query: 452 AFSIWYAWMRSAGLLVG-FTVGSVSPG-NIRTSFGAAFCTAIAGIV 495
AF ++YA+ S G++ G FTVG++ ++ G AF AG++
Sbjct: 328 AFGLFYAFF-SLGVVAGSFTVGAIGASPSVSFVIGTAFLLTFAGMI 372
>emb|CAD16077.1| HYPOTHETICAL TRANSMEMBRANE PROTEIN [Ralstonia solanacearum]
gi|17547089|ref|NP_520491.1| HYPOTHETICAL TRANSMEMBRANE
PROTEIN [Ralstonia solanacearum GMI1000]
Length = 384
Score = 38.1 bits (87), Expect = 0.71
Identities = 34/118 (28%), Positives = 54/118 (44%), Gaps = 20/118 (16%)
Query: 334 SFITMTIFTGGVVFIVGQLCIKPV--HLLFFWLTYFLF-------PVFSLSLLQPLLRVI 384
SF+T+ + G+ + G+ + P+ H LF WLTY L P+F++S++ +
Sbjct: 173 SFVTLGV--SGIAMLWGKHFLLPLMGHQLFGWLTYVLKNLHNLVGPLFTVSIVVAFVMWA 230
Query: 385 KMNSVKMQIVGFLFSL--ISSGFG-------FYYGHSHWKWGHLVVFSVIQSTGTGIL 433
N + + +L L + G G F G W WG LVVF +I S +L
Sbjct: 231 HDNLPRRGDLQWLLRLGGMFGGHGAEVPSHRFNAGEKLWFWGGLVVFGLILSASGWVL 288
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 917,691,427
Number of Sequences: 2540612
Number of extensions: 39032480
Number of successful extensions: 146912
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 112
Number of HSP's that attempted gapping in prelim test: 146859
Number of HSP's gapped (non-prelim): 140
length of query: 535
length of database: 863,360,394
effective HSP length: 133
effective length of query: 402
effective length of database: 525,458,998
effective search space: 211234517196
effective search space used: 211234517196
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0020.10


