

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0017.17
(167 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
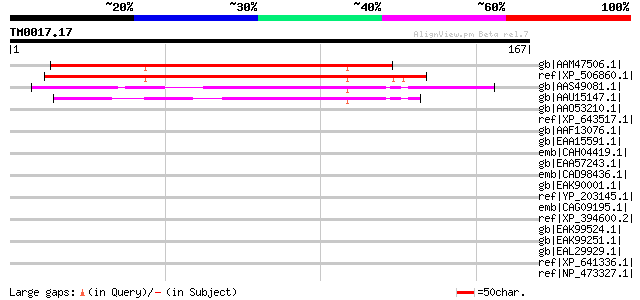
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM47506.1| iron-stress related protein [Citrus junos] 109 2e-23
ref|XP_506860.1| PREDICTED OSJNBa0030C08.45 gene product [Oryza ... 107 2e-22
gb|AAS49081.1| At3g07580 [Arabidopsis thaliana] gi|51970766|dbj|... 86 3e-16
gb|AAU15147.1| At5g48335 [Arabidopsis thaliana] gi|50897196|gb|A... 82 7e-15
gb|AAO53210.1| similar to Plasmodium falciparum (isolate 3D7). H... 42 0.005
ref|XP_643517.1| hypothetical protein DDB0217700 [Dictyostelium ... 42 0.005
gb|AAF13076.1| unknown protein [Arabidopsis thaliana] gi|1523148... 41 0.011
gb|EAA15591.1| hypothetical protein [Plasmodium yoelii yoelii] 38 0.12
emb|CAH04419.1| protein phosphatase 2C [Euplotes vannus] 35 0.78
gb|EAA57243.1| hypothetical protein MG08212.4 [Magnaporthe grise... 35 0.78
emb|CAD98436.1| hypothetical predicted protein, unknown function... 35 1.0
gb|EAK90001.1| uncharacterized protein [Cryptosporidium parvum] ... 35 1.0
ref|YP_203145.1| transcriptional regulator [Xanthomonas oryzae p... 35 1.0
emb|CAG09195.1| unnamed protein product [Tetraodon nigroviridis] 34 1.3
ref|XP_394600.2| PREDICTED: similar to ENSANGP00000022105 [Apis ... 34 1.3
gb|EAK99524.1| hypothetical protein CaO19.10868 [Candida albican... 34 1.7
gb|EAK99251.1| hypothetical protein CaO19.3360 [Candida albicans... 34 1.7
gb|EAL29929.1| GA16728-PA [Drosophila pseudoobscura] 34 1.7
ref|XP_641336.1| hypothetical protein DDB0206474 [Dictyostelium ... 34 1.7
ref|NP_473327.1| hypothetical protein [Plasmodium falciparum 3D7... 33 2.3
>gb|AAM47506.1| iron-stress related protein [Citrus junos]
Length = 166
Score = 109 bits (273), Expect = 2e-23
Identities = 62/112 (55%), Positives = 75/112 (66%), Gaps = 2/112 (1%)
Query: 14 VKRYAPPNQRNRSTNRRKSSDRLDRTNSI-GNDLEKNQFASSRNVQVQDRIDAVSSNLVN 72
VKRYAPPNQRNRS NRRKS DR DR++++ GND EKNQ A+SRN V D D+ SSN N
Sbjct: 19 VKRYAPPNQRNRSLNRRKSGDRFDRSSNLYGNDGEKNQLAASRNAAVTDHGDSGSSNFAN 78
Query: 73 ENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNN-SKDSPEKPVMYSGGTS 123
N R V L+G S S +QLL DR A +++ S D E+PV+YS G S
Sbjct: 79 VNSQPRLVPLEGFSRSEGAQLLKDRGRAIWNLFHDPSIDLSERPVLYSSGAS 130
>ref|XP_506860.1| PREDICTED OSJNBa0030C08.45 gene product [Oryza sativa (japonica
cultivar-group)] gi|50910345|ref|XP_466661.1| putative
iron-stress related protein [Oryza sativa (japonica
cultivar-group)] gi|47497956|dbj|BAD20161.1| putative
iron-stress related protein [Oryza sativa (japonica
cultivar-group)] gi|47496841|dbj|BAD19601.1| putative
iron-stress related protein [Oryza sativa (japonica
cultivar-group)]
Length = 158
Score = 107 bits (266), Expect = 2e-22
Identities = 59/128 (46%), Positives = 81/128 (63%), Gaps = 5/128 (3%)
Query: 12 ESVKRYAPPNQRNRSTNRRKSSDRLDRTNSI-GNDLEKNQFASSRNVQVQDRIDAVSSNL 70
+ +KRY PP RNR+ NRRKS DR ++ + + ND EK+ S +N+ + SN
Sbjct: 7 DGIKRYTPPVHRNRANNRRKSGDRAEKASYLYNNDGEKSHAPSLKNLPPIIPHETFFSNP 66
Query: 71 VNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNN-SKDSPEKPVMYSGGT-SVW--T 126
N+ +R + L+GC S A+QLLNDRW AA+ YN+ S DSP+KPVMYSG + S W
Sbjct: 67 QNDYSQTRLIPLEGCCASEAAQLLNDRWAAAMNLYNDQSYDSPDKPVMYSGSSGSSWGHG 126
Query: 127 HFRLPHQI 134
H +LPHQ+
Sbjct: 127 HMKLPHQM 134
>gb|AAS49081.1| At3g07580 [Arabidopsis thaliana] gi|51970766|dbj|BAD44075.1|
unnamed protein product [Arabidopsis thaliana]
Length = 143
Score = 86.3 bits (212), Expect = 3e-16
Identities = 56/150 (37%), Positives = 78/150 (51%), Gaps = 18/150 (12%)
Query: 8 APPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAVS 67
A S + RY PPNQR S NRRKS DR ++ ND E+NQ A
Sbjct: 2 ADSSSNQNRYTPPNQRKASGNRRKSGDR--TSSQQNNDNERNQ------------PPATG 47
Query: 68 SNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNN-SKDSPEKPVMYSGGTSVWT 126
+ N + R + + CS SAA QL++ RW A + Y++ + D +P+MY GG SVW
Sbjct: 48 LQMENISQTQRIITIKDCSRSAAYQLMSQRWAAEMHQYDDPTVDLSVRPIMYYGG-SVWG 106
Query: 127 HFRLPHQIMSPAASAPPSVSQLDFLGELNR 156
+LPHQI++ A P S D++ E+ R
Sbjct: 107 --KLPHQILAAANKTLPPPSPTDYMTEVRR 134
>gb|AAU15147.1| At5g48335 [Arabidopsis thaliana] gi|50897196|gb|AAT85737.1|
At5g48335 [Arabidopsis thaliana]
gi|42573618|ref|NP_974905.1| expressed protein
[Arabidopsis thaliana]
Length = 114
Score = 81.6 bits (200), Expect = 7e-15
Identities = 51/119 (42%), Positives = 67/119 (55%), Gaps = 23/119 (19%)
Query: 15 KRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAVSSNLVNEN 74
KRY PPNQRNRS+NRR+S N+ SS++V V EN
Sbjct: 18 KRYTPPNQRNRSSNRRRSR----------GSFHSNEGESSQSVAV---------GFQREN 58
Query: 75 PYSRFVALDGCSCSAASQLLNDRWTAAIQSYNN-SKDSPEKPVMYSGGTSVWTHFRLPH 132
+ ++++GCS S A QLL+DRWTAA+ YN+ S D E+P+MY GG VW +LPH
Sbjct: 59 SSPKIISVEGCSRSEAFQLLSDRWTAAMHLYNDPSVDLSERPMMYYGG-DVWG--KLPH 114
>gb|AAO53210.1| similar to Plasmodium falciparum (isolate 3D7). Hypothetical
protein [Dictyostelium discoideum]
Length = 1264
Score = 42.4 bits (98), Expect = 0.005
Identities = 30/118 (25%), Positives = 51/118 (42%), Gaps = 1/118 (0%)
Query: 6 HPAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDA 65
HP P + N N +TN +++ + TN+ N+ N ++ N + +
Sbjct: 402 HPHHPHQRPPPQYGYNNNNNNTNNTNNNNN-NNTNNNNNNNNNNNNNNNNNNNNNNNNNN 460
Query: 66 VSSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNNSKDSPEKPVMYSGGTS 123
++N +N N S A Q ND+ TAAI N +DS P+++ G T+
Sbjct: 461 NNNNNINSNNNSNNAAQQTQPSQQPPQEYNDQNTAAINLNNGMEDSTSLPIIHLGATN 518
>ref|XP_643517.1| hypothetical protein DDB0217700 [Dictyostelium discoideum]
gi|60471731|gb|EAL69687.1| hypothetical protein
DDB0217700 [Dictyostelium discoideum]
Length = 1290
Score = 42.4 bits (98), Expect = 0.005
Identities = 30/118 (25%), Positives = 51/118 (42%), Gaps = 1/118 (0%)
Query: 6 HPAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDA 65
HP P + N N +TN +++ + TN+ N+ N ++ N + +
Sbjct: 429 HPHHPHQRPPPQYGYNNNNNNTNNTNNNNN-NNTNNNNNNNNNNNNNNNNNNNNNNNNNN 487
Query: 66 VSSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNNSKDSPEKPVMYSGGTS 123
++N +N N S A Q ND+ TAAI N +DS P+++ G T+
Sbjct: 488 NNNNNINSNNNSNNAAQQTQPSQQPPQEYNDQNTAAINLNNGMEDSTSLPIIHLGATN 545
>gb|AAF13076.1| unknown protein [Arabidopsis thaliana] gi|15231484|ref|NP_187415.1|
expressed protein [Arabidopsis thaliana]
Length = 116
Score = 41.2 bits (95), Expect = 0.011
Identities = 23/60 (38%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query: 98 WTAAIQSYNN-SKDSPEKPVMYSGGTSVWTHFRLPHQIMSPAASAPPSVSQLDFLGELNR 156
W A + Y++ + D +P+MY GG SVW +LPHQI++ A P S D++ E+ R
Sbjct: 51 WAAEMHQYDDPTVDLSVRPIMYYGG-SVWG--KLPHQILAAANKTLPPPSPTDYMTEVRR 107
>gb|EAA15591.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1057
Score = 37.7 bits (86), Expect = 0.12
Identities = 30/113 (26%), Positives = 49/113 (42%), Gaps = 4/113 (3%)
Query: 3 ENLHPAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDR 62
EN P +K ++N S+N + LD + SI + + K +N + +
Sbjct: 101 ENSLNNEPDNKLKNNKEIEKKNNSSNLDTQNGLLDISTSIFSAITKRLTEMKKN---EKK 157
Query: 63 IDAVSSNLVNEN-PYSRFVALDGCSCSAASQLLNDRWTAAIQSYNNSKDSPEK 114
ID S N VNEN P + + DG S S+ + + T +Y + + P K
Sbjct: 158 IDTNSENFVNENMPRKKTILKDGNSASSNNNISEKYNTYETDAYLINDEEPTK 210
>emb|CAH04419.1| protein phosphatase 2C [Euplotes vannus]
Length = 327
Score = 35.0 bits (79), Expect = 0.78
Identities = 28/94 (29%), Positives = 42/94 (43%), Gaps = 4/94 (4%)
Query: 64 DAVSSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNNSKDSPEKPVMYSG--G 121
DAV +NL + S F DG ASQ++ D + ++ ++ KD + +Y G
Sbjct: 41 DAVIANLDFDEDTSLFGVFDGHGGKEASQVVKDNYERILKGDSSYKDGDCQKGLYDSFKG 100
Query: 122 TSVWTHFRLPHQIMSPAASAPPSVSQ--LDFLGE 153
T V+ + Q M A + P V L LGE
Sbjct: 101 TDVFLGSKTGKQEMKAVADSNPEVKNPLLKILGE 134
>gb|EAA57243.1| hypothetical protein MG08212.4 [Magnaporthe grisea 70-15]
gi|39946184|ref|XP_362629.1| hypothetical protein
MG08212.4 [Magnaporthe grisea 70-15]
Length = 526
Score = 35.0 bits (79), Expect = 0.78
Identities = 18/50 (36%), Positives = 27/50 (54%)
Query: 6 HPAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSR 55
HPAPP ++K N N ++ S+ R DRT S+G+D + S+R
Sbjct: 287 HPAPPPPAIKTSPHMNNGNGTSAGPPSATRSDRTGSVGSDDVEQSRPSTR 336
>emb|CAD98436.1| hypothetical predicted protein, unknown function [Cryptosporidium
parvum]
Length = 904
Score = 34.7 bits (78), Expect = 1.0
Identities = 25/80 (31%), Positives = 42/80 (52%), Gaps = 7/80 (8%)
Query: 36 LDRTNSIGNDLEKNQFASSRNVQVQDRIDAVSSNLVNENP--YSRFVALDGCSCSAASQL 93
+D+ N+IG L+KN S + + D + N ++EN F+ + S +SQ
Sbjct: 29 IDKVNTIG--LDKNNSYESNSYHLDDESQFIIDNYLSENGELIKEFLGRNDFSNGNSSQ- 85
Query: 94 LNDRWTAAIQSYNNSKDSPE 113
++ TA+ +S NNSK+S E
Sbjct: 86 --NKLTASNESENNSKESQE 103
>gb|EAK90001.1| uncharacterized protein [Cryptosporidium parvum]
gi|66475832|ref|XP_627732.1| hypothetical protein
cgd6_3950 [Cryptosporidium parvum]
Length = 961
Score = 34.7 bits (78), Expect = 1.0
Identities = 25/80 (31%), Positives = 42/80 (52%), Gaps = 7/80 (8%)
Query: 36 LDRTNSIGNDLEKNQFASSRNVQVQDRIDAVSSNLVNENP--YSRFVALDGCSCSAASQL 93
+D+ N+IG L+KN S + + D + N ++EN F+ + S +SQ
Sbjct: 29 IDKVNTIG--LDKNNSYESNSYHLDDESQFIIDNYLSENGELIKEFLGRNDFSNGNSSQ- 85
Query: 94 LNDRWTAAIQSYNNSKDSPE 113
++ TA+ +S NNSK+S E
Sbjct: 86 --NKLTASNESENNSKESQE 103
>ref|YP_203145.1| transcriptional regulator [Xanthomonas oryzae pv. oryzae KACC10331]
gi|58428723|gb|AAW77760.1| transcriptional regulator
[Xanthomonas oryzae pv. oryzae KACC10331]
Length = 206
Score = 34.7 bits (78), Expect = 1.0
Identities = 31/108 (28%), Positives = 47/108 (42%), Gaps = 22/108 (20%)
Query: 11 SESVKRYAPPNQRNRST----------NRRKSSDRLDRT--NSIGNDLEKNQFASSRNVQ 58
S V + P N R T +S+ RL T N+ N++ KNQF+S
Sbjct: 68 SSDVAQIPPANAPERGTVTPIDLDAIIEHNRSNSRLGTTVLNAFNNNIFKNQFSSL---- 123
Query: 59 VQDRIDAVSSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYN 106
DR+ N P+S AL+ C AAS + +R+++ I + N
Sbjct: 124 --DRL----KNADLHRPFSAVDALEQCPAQAASAVALNRYSSMIDAPN 165
>emb|CAG09195.1| unnamed protein product [Tetraodon nigroviridis]
Length = 488
Score = 34.3 bits (77), Expect = 1.3
Identities = 40/163 (24%), Positives = 69/163 (41%), Gaps = 15/163 (9%)
Query: 7 PAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRN--------VQ 58
P+P E + P R + + R + LD SI + +SS + V+
Sbjct: 3 PSPLDERIVVALPRPVRPQELHLRLDTSYLDSITSISSSSSSGSGSSSSSSAVISTTVVK 62
Query: 59 VQDRIDAVSSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNNSKDSPEKP-VM 117
VQ + D V+ + + +R ++ C CS+AS + QS ++S S P +
Sbjct: 63 VQAKADIVTY-MPSSKGSTRSLS---CGCSSASCCSVTTYERDSQSLSHSPVSASSPNLS 118
Query: 118 YSG--GTSVWTHFRLPHQIMSPAASAPPSVSQLDFLGELNRQM 158
Y+G G V H L ++P S PS ++ EL +++
Sbjct: 119 YAGPPGPRVSNHEALSTSSLTPGTSKAPSAVRVIHPNELAKRL 161
>ref|XP_394600.2| PREDICTED: similar to ENSANGP00000022105 [Apis mellifera]
Length = 675
Score = 34.3 bits (77), Expect = 1.3
Identities = 33/127 (25%), Positives = 55/127 (42%), Gaps = 15/127 (11%)
Query: 18 APPNQRNRSTNRRKSSDRLDRT--NSIGNDLEKNQFASSRNVQVQDRIDAVS--SNLVNE 73
APP +R + R ++ D+T N++ +N+ + +++ D ++ + NL E
Sbjct: 195 APPTKRRKC--RENANTNADKTISNALSAKDSENKTSENQSTTGSDPVEIIPLMPNLKLE 252
Query: 74 NPYSRFVALDGCSCSAASQLLNDRWTAAI---QSYNNSKDSPEKP----VMYSGGTSVWT 126
P ++ DG SCS Q L D I + +N+ D +KP YS S
Sbjct: 253 MP--EYLVQDGSSCSYEEQSLGDSSLNKIGIEDTSSNTPDHDQKPDITQTFYSSNQSTSD 310
Query: 127 HFRLPHQ 133
L HQ
Sbjct: 311 SLDLKHQ 317
>gb|EAK99524.1| hypothetical protein CaO19.10868 [Candida albicans SC5314]
Length = 668
Score = 33.9 bits (76), Expect = 1.7
Identities = 36/164 (21%), Positives = 61/164 (36%), Gaps = 18/164 (10%)
Query: 7 PAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAV 66
P PP + ++ T KSS N GN + K+ S + +
Sbjct: 388 PPPPQQQPQQQKSTTTTTTITTSPKSSTNTIELNKKGNRINKSISKSPSGINTPSSSSDI 447
Query: 67 SSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQS-------YNNSKDSPEKPVMY- 118
SS + R ++ S S+ S L +D T+ I S Y ++ +SP +
Sbjct: 448 SSK--SSKSRKRKLSTSSSSSSSNSSLSDDNLTSNIGSTSINSTNYKSNYNSPPSEQEFD 505
Query: 119 --------SGGTSVWTHFRLPHQIMSPAASAPPSVSQLDFLGEL 154
S T+ + F L +Q +S+ S S++D+ L
Sbjct: 506 DHYYSSSSSSSTTTNSTFALSNQDQHNLSSSSSSTSKMDYYTNL 549
>gb|EAK99251.1| hypothetical protein CaO19.3360 [Candida albicans SC5314]
Length = 667
Score = 33.9 bits (76), Expect = 1.7
Identities = 36/164 (21%), Positives = 61/164 (36%), Gaps = 18/164 (10%)
Query: 7 PAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAV 66
P PP + ++ T KSS N GN + K+ S + +
Sbjct: 388 PPPPQQQPQQQKSTTTTTTITTSPKSSTNTIELNKKGNRINKSISKSPSGINTPSSSSDI 447
Query: 67 SSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQS-------YNNSKDSPEKPVMY- 118
SS + R ++ S S+ S L +D T+ I S Y ++ +SP +
Sbjct: 448 SSK--SSKSRKRKLSTSSSSSSSNSSLSDDNLTSNIGSTSINSTNYKSNYNSPPSEQEFD 505
Query: 119 --------SGGTSVWTHFRLPHQIMSPAASAPPSVSQLDFLGEL 154
S T+ + F L +Q +S+ S S++D+ L
Sbjct: 506 DHYYSSSSSSSTTTNSTFALSNQDQHNLSSSSSSTSKMDYYTNL 549
>gb|EAL29929.1| GA16728-PA [Drosophila pseudoobscura]
Length = 478
Score = 33.9 bits (76), Expect = 1.7
Identities = 25/74 (33%), Positives = 36/74 (47%), Gaps = 1/74 (1%)
Query: 11 SESVKR-YAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAVSSN 69
S SV+R PP RNR+ +S R R S ++ E N ++S ++R + +SN
Sbjct: 307 SRSVERTLTPPPHRNRTGGGNGASGRKGRNRSPRHERENNSNSNSSGSNRENRAERNNSN 366
Query: 70 LVNENPYSRFVALD 83
N N SR V D
Sbjct: 367 SNNGNNNSRRVDRD 380
>ref|XP_641336.1| hypothetical protein DDB0206474 [Dictyostelium discoideum]
gi|60469362|gb|EAL67356.1| hypothetical protein
DDB0206474 [Dictyostelium discoideum]
Length = 509
Score = 33.9 bits (76), Expect = 1.7
Identities = 16/63 (25%), Positives = 30/63 (47%)
Query: 21 NQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAVSSNLVNENPYSRFV 80
N+ N T ++ + +N+ N+ N S N + I+ ++N N N ++RF+
Sbjct: 246 NKENNPTTTTTTTTNTESSNNNNNNNNNNPTTSPTNKTNEPSIEKTNTNKANTNEFTRFL 305
Query: 81 ALD 83
LD
Sbjct: 306 GLD 308
>ref|NP_473327.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|4493979|emb|CAB39038.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 2706
Score = 33.5 bits (75), Expect = 2.3
Identities = 20/55 (36%), Positives = 26/55 (46%), Gaps = 1/55 (1%)
Query: 21 NQRNRSTNRRKSSDRLDRTNSIGND-LEKNQFASSRNVQVQDRIDAVSSNLVNEN 74
N N + N RKS R RTN N+ +K F + NV+ DR+ N EN
Sbjct: 2612 NNNNNNNNNRKSRKRSTRTNRKKNERTKKKNFENFLNVRKSDRLRNKEQNKKEEN 2666
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.125 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 277,673,790
Number of Sequences: 2540612
Number of extensions: 10117250
Number of successful extensions: 27830
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 27759
Number of HSP's gapped (non-prelim): 108
length of query: 167
length of database: 863,360,394
effective HSP length: 118
effective length of query: 49
effective length of database: 563,568,178
effective search space: 27614840722
effective search space used: 27614840722
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0017.17


