

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0016.18
(931 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
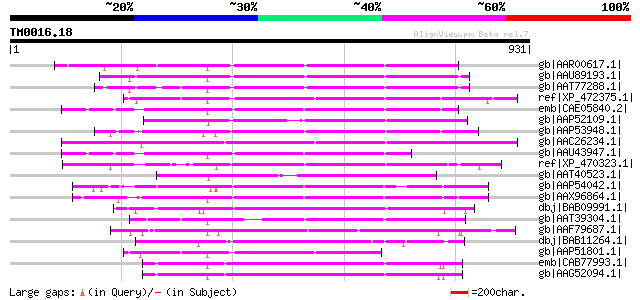
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR00617.1| hypothetical protein [Oryza sativa (japonica cult... 402 e-110
gb|AAU89193.1| transposon protein, putative, mutator sub-class [... 395 e-108
gb|AAT77288.1| putative polyprotein [Oryza sativa (japonica cult... 387 e-105
ref|XP_472375.1| OSJNBb0118P14.3 [Oryza sativa (japonica cultiva... 368 e-100
emb|CAE05840.2| OSJNBa0091C07.2 [Oryza sativa (japonica cultivar... 350 9e-95
gb|AAP52109.1| putative transposon protein [Oryza sativa (japoni... 333 1e-89
gb|AAP53948.1| putative mutator-like transposase [Oryza sativa (... 333 2e-89
gb|AAC26234.1| contains similarity to maize transposon MuDR (GB:... 331 6e-89
gb|AAU43947.1| hypothetical protein [Oryza sativa (japonica cult... 299 3e-79
ref|XP_470323.1| putative transposon protein [Oryza sativa (japo... 292 3e-77
gb|AAT40523.1| putative mutator transposable element [Solanum de... 287 1e-75
gb|AAP54042.1| putative mutator protein [Oryza sativa (japonica ... 286 2e-75
gb|AAX96864.1| transposon protein, putative, mutator sub-class [... 283 1e-74
dbj|BAB09991.1| mutator-like transposase-like [Arabidopsis thali... 276 2e-72
gb|AAT39304.1| putative transposon MuDR mudrA-like protein [Sola... 276 3e-72
gb|AAF79687.1| F9C16.9 [Arabidopsis thaliana] gi|25405857|pir||D... 270 1e-70
dbj|BAB11264.1| unnamed protein product [Arabidopsis thaliana] 259 4e-67
gb|AAP51801.1| putative mutator-like transposase [Oryza sativa (... 255 4e-66
emb|CAB77993.1| putative MuDR-A-like transposon protein [Arabido... 251 6e-65
gb|AAG52094.1| putative Mutator-like transposase; 12516-14947 [A... 250 1e-64
>gb|AAR00617.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50901476|ref|XP_463171.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 981
Score = 402 bits (1034), Expect = e-110
Identities = 251/789 (31%), Positives = 382/789 (47%), Gaps = 78/789 (9%)
Query: 81 IRMLWKLLGMQWILG-GVVDIYLEHTVELPEVVRNEMQKEDAVLDDEDDVFVEVTEVEVI 139
+++ W L G + G +VD +E + + +V N+++ LD D V E V+
Sbjct: 78 LKVYWLLPGKNLVDGLRIVDSEVE--INVMNLVCNKVKNFVIYLDHIDHVSGRNQEDIVV 135
Query: 140 PMTQEAPTVAVPEVEASNIDSEDEICKNL-------TNDGFDDVDEPFDVQQDGDGRDDV 192
E P V P ++ N+ ++ D++++ + DG DD
Sbjct: 136 SPVSELPIVMSPVRGCQQKGDGNQGGSNVGVQACGVVDEVADEIEDDVEYDSDGSEVDDS 195
Query: 193 NRNSGPNNGVQDGDE--SDNNVGFIPSEVAAQHNIDM---------------EYESDDLL 235
+ D D+ DN G + + A + E SD+
Sbjct: 196 DFVDSDYEFQDDDDDLFEDNVDGDVVDQGPAPKKFNQKKVAGGKLKGKRVIREEGSDEES 255
Query: 236 SDYSC---DEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAV 292
SD C E + +R++ F ED + F++GM F S E +K I +S+ N +
Sbjct: 256 SDEECLELPENPDEINLRFKSFNPEDMNNPV-FKVGMVFPSVELLRKTITEYSLKNRVDI 314
Query: 293 IYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIA 352
+ND+ R++ C E C + Y + + +KT P H C + + + TA W+A
Sbjct: 315 KMPRNDRTRIKAHCA-EGCPWNMYASLDSRVHGFIVKTYVPQHKCRKEWVLQRCTANWLA 373
Query: 353 -----------------------------------KNARKIAKEAVEGDALKQFTLITSY 377
AR++A + + GD + Q+ ++ Y
Sbjct: 374 LKYIESFRADSRMTLPSFAKTVQKEWNLTPSRSKLARARRLALKEIYGDEVAQYNMLWDY 433
Query: 378 AAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTK 437
EL R N ++ + L +F C Y L+ CK F CRP I +DGCH+KTK
Sbjct: 434 GNELRRSNPGSSFYLKLD------DGKFSCLYFSLDACKRGFLSGCRPIICLDGCHIKTK 487
Query: 438 YGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQK 497
+GG LL AVG DPND FP+A AVVE E +W WFL L D+G W ++DKQK
Sbjct: 488 FGGQLLTAVGIDPNDCIFPIAMAVVEVESFSTWSWFLQTLKDDVGIVNTYPWTIMTDKQK 547
Query: 498 GLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWA-SKATYKELWEEKMQEI 556
GL+ + FP EHR+CVRHLY NF+Q F G E+ +WA ++++ + W K +E+
Sbjct: 548 GLIPAVQQLFPDSEHRFCVRHLYQNFQQSFKG--EILKNQLWACARSSSVQEWNTKFEEM 605
Query: 557 KKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWI 616
K +NE AY WL + W R FS FPKCD+ +NN E FN IL AR+ P++TM E I
Sbjct: 606 KALNEDAYNWLEQMAPNTWVRAFFSDFPKCDILLNNSCEVFNKYILEAREMPILTMLEKI 665
Query: 617 RKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQ 676
+ LMTRF +++++KW+G + PK RK+L + + + +G+FQVE G +
Sbjct: 666 KGQLMTRFFNKQKEAQKWQGPICPKIRKKLLKIAEQANICYVLPAGKGVFQVEERG--TK 723
Query: 677 FIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPI 736
+IVD+ + C C W+L GIPC HA+AC + L E+++ Y ++ Y I P
Sbjct: 724 YIVDVVTKHCDCRRWDLTGIPCCHAIACIREDHLSEEDFLPHCYSINAFKAVYAENIIPC 783
Query: 737 NGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQK 796
N + W KM+ ILPP ++ GRPKK RR++P E +N KL + G + C CH+
Sbjct: 784 NDKANWEKMNGPQILPPVYEKKVGRPKKSRRKQPQEVQGRNGPKLTKHGVTIHCSYCHEA 843
Query: 797 GHNQRKCPL 805
HN++ C L
Sbjct: 844 NHNKKGCEL 852
>gb|AAU89193.1| transposon protein, putative, mutator sub-class [Oryza sativa
(japonica cultivar-group)]
Length = 1030
Score = 395 bits (1015), Expect = e-108
Identities = 241/712 (33%), Positives = 361/712 (49%), Gaps = 65/712 (9%)
Query: 161 EDEICKNLTNDGFDDVDEPF---DVQQDGDGRDDVNRNSGPNNGVQDGDESDNNV----- 212
ED+ + + DD DE F D + D DG DD+ + N + +G +
Sbjct: 210 EDDCDEEDSGTESDDSDEEFYDSDYELD-DGDDDLFVDYVDENVIDEGVAKGKKILKGKK 268
Query: 213 ---GFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQRFRQEDFDKDYKFRLGM 269
+ +A D E E D + D +D G R++++ F +ED + F++G+
Sbjct: 269 ARGSRLKGNLAIVPRGD-ESEDTDEEELNAADSDDEGVRLKFKSFAEEDLNNP-NFKVGL 326
Query: 270 EFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIK 329
F S E+ ++AI +SV N + +ND+ R+R C E C + Y + + +K
Sbjct: 327 VFPSVEKLRQAITEYSVRNRVEIKMPRNDRRRIRAHCA-EGCPWNLYASEDSRAKAFVVK 385
Query: 330 TLNPDHTCGRTFKNRAATAKWIA-----------------------------------KN 354
T + HTC + + + T+KW+A
Sbjct: 386 TCDERHTCQKEWILKRCTSKWLAGKYIEAFRADEKMSLTSFAKTVQLQWNLTLSRSKLAR 445
Query: 355 ARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEG 414
AR+IA + + GD ++Q+ L+ Y EL R N ++ + L F YM ++
Sbjct: 446 ARRIAMKTIHGDEVEQYKLLWDYGKELRRSNPGSSFFLKLD------GSLFSQCYMSMDA 499
Query: 415 CKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFL 474
CK F CRP I +DGCH+KTKYGG LL AVG DPND +P+AFAVVE E +W+WFL
Sbjct: 500 CKRGFLNGCRPLICLDGCHIKTKYGGQLLTAVGMDPNDCIYPIAFAVVEVESLATWKWFL 559
Query: 475 NHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVR 534
L D+G W ++DKQKGL+ + FP EHR+CVRHLY+NF+ +F G EV
Sbjct: 560 ETLKNDLGIENTYPWTIMTDKQKGLIPAVQQVFPESEHRFCVRHLYSNFQLQFKG--EVL 617
Query: 535 DLLMWA-SKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNL 593
+WA ++++ + W + M ++ +N++AY WL +P W R FS FPKCD+ +NN
Sbjct: 618 KNQLWACARSSSVQEWNKNMDVMRNLNKSAYEWLEKLPPNTWVRAFFSEFPKCDILLNNN 677
Query: 594 AEAFNCTILVARDKPLITMFEWIRKYLMTR-FATLREKSKKWKGNVMPKPRKRLDREIKY 652
E FN IL AR+ P++TM E I+ LMTR F +E + +++G + PK RK++ +
Sbjct: 678 CEVFNKYILEARELPILTMLEKIKGQLMTRHFNKQKELADQFQGLICPKIRKKVLKNADA 737
Query: 653 SGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKP 712
+ A QG+FQV E Q+IVDI+ C C W+L GIPC HA++C + +
Sbjct: 738 ANTCYALPAGQGIFQVHER--EYQYIVDINAMYCDCRRWDLTGIPCNHAISCLRHERINA 795
Query: 713 ENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDE 772
E+ + Y + YG I P N ++KW ++ +I PP ++ GRPKK RR+ P E
Sbjct: 796 ESILPNCYTTDAFSKAYGFNIWPCNDKSKWENVNGPEIKPPVYEKKAGRPKKSRRKAPYE 855
Query: 773 PDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIAT 824
KN KL + G C ++ HN C L KKQ S+ A +AT
Sbjct: 856 VIGKNGPKLTKHGVMMHYKYCGEENHNSGGCKL---KKQGISSEEAKRMVAT 904
>gb|AAT77288.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1006
Score = 387 bits (993), Expect = e-105
Identities = 240/721 (33%), Positives = 359/721 (49%), Gaps = 80/721 (11%)
Query: 152 EVEASNIDSEDEICKNLTNDGFDDVDEPF---DVQQDGDGRDDVNRNSGPNNGVQDGDES 208
E + + D ED K+ DD DE F D + D DG DD+ + N + +G
Sbjct: 192 EQQEDDCDEEDSGTKS------DDSDEEFYDSDYELD-DGDDDLFVDYVDENVIDEGVAK 244
Query: 209 DNNV--------GFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQRFRQEDFD 260
+ + +A D E E D + D +D G R++++ F +ED +
Sbjct: 245 GKKILKGKKARGSRLKGNLAIVPRGD-ESEDTDEEELNAADSDDEGVRLKFKSFAEEDLN 303
Query: 261 KDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANI 320
+ ++ ++AI +SV N + +ND+ R+R C +E C + Y +
Sbjct: 304 NP----------NFKKLRQAITEYSVRNRVEIKMPRNDRRRIRAHC-EEGCPWNLYTSED 352
Query: 321 KGTSTYRIKTLNPDHTCGRTFKNRAATAKWIA---------------------------- 352
+ +KT + HTC + + + T+KW+A
Sbjct: 353 SRAKAFVVKTYDERHTCQKEWILKRCTSKWLAGKYIEAFRADEKMSLTSFAKTVQLQWNL 412
Query: 353 -------KNARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRF 405
AR+IA + + GD ++Q+ L+ Y EL R N + + L F
Sbjct: 413 TPSRSKLARARRIAMKTIHGDEVEQYKLLWDYGKELRRSNPGTSFFLKLD------GSLF 466
Query: 406 GCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAE 465
YM ++ CK F CRP I +DGCH+KTKYGG LL AVG DPND +P+AFAVVE E
Sbjct: 467 SQCYMSMDACKRGFLNGCRPLICLDGCHIKTKYGGQLLTAVGMDPNDCIYPIAFAVVEVE 526
Query: 466 CKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQ 525
+W+WFL L D+G W ++DKQKGL+ + FP EHR+CVRHLY+NF+
Sbjct: 527 SLATWKWFLETLKNDLGIENTYPWTIMTDKQKGLIPAVQQVFPESEHRFCVRHLYSNFQL 586
Query: 526 KFGGGTEVRDLLMWA-SKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFP 584
+F G EV +WA ++++ + W + M ++ +N++AY WL +P W R FS FP
Sbjct: 587 QFKG--EVPKNQLWACARSSSVQEWNKNMDVMRNLNKSAYEWLEKLPPNTWVRAFFSEFP 644
Query: 585 KCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTR-FATLREKSKKWKGNVMPKPR 643
KCD+ +NN E FN IL AR+ P++TM E I+ LMTR F +E + +++G + PK R
Sbjct: 645 KCDILLNNNCEVFNKYILEARELPILTMLEKIKGQLMTRHFNKQKELADQFQGLICPKIR 704
Query: 644 KRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVA 703
K++ + + A QG+FQV E Q+IVDI+ C C W+L GIPC HA++
Sbjct: 705 KKVLKNADAANTCYALPAGQGIFQVHER--EYQYIVDINAMHCDCRRWDLTGIPCNHAIS 762
Query: 704 CFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPK 763
C + + E+ + Y + YG I P N ++KW ++ +I PP ++ GRPK
Sbjct: 763 CLRHERINAESILPNCYTTDAFSKAYGFNIWPCNDKSKWENINGPEIKPPVYEKKAGRPK 822
Query: 764 KLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIA 823
K RR+ P E KN KL + G C C ++ HN C L KKQ S+ A +A
Sbjct: 823 KSRRKAPYEVIGKNGPKLTKHGVMMHCKYCGEENHNSGGCKL---KKQGISSEEAKRMVA 879
Query: 824 T 824
T
Sbjct: 880 T 880
>ref|XP_472375.1| OSJNBb0118P14.3 [Oryza sativa (japonica cultivar-group)]
gi|38346143|emb|CAD40681.2| OSJNBb0118P14.3 [Oryza
sativa (japonica cultivar-group)]
Length = 939
Score = 368 bits (944), Expect = e-100
Identities = 235/764 (30%), Positives = 355/764 (45%), Gaps = 78/764 (10%)
Query: 204 DGDESDNNVGFIPSEVAAQHNID-----MEYESDDLLSDYSCDEEDNGSRVRYQRFRQED 258
D D DN V ++ HN + + E + L+ +EE+ + ++ FR
Sbjct: 194 DDDLFDNCV---EGKIELAHNKEEGSDYVSSEDERLMMPDISEEEEEKIKFSFKSFRANV 250
Query: 259 FDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCA 318
+ +F++ + FA E +KAI +++ N A+ +N K R+ C E C ++ +
Sbjct: 251 DMETPEFKVSIMFADVVELRKAIDQYTIKNQVAIKKTRNTKTRIEAKC-AEGCPWMLSAS 309
Query: 319 NIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAK------------------------- 353
++ HTC + ++ +A TAK++AK
Sbjct: 310 MDNRVKCLVVREYIEKHTCSKQWEIKAVTAKYLAKRYIEEFRDNDKMTLMSFAKKIQKEL 369
Query: 354 ----------NARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQP 403
AR++A A+ GD + Q+ + Y EL N ++ + L
Sbjct: 370 HLTPSRHKLGRARRMAMRAIYGDEISQYNQLWDYGQELRTSNPGSSFYLNL--------- 420
Query: 404 RFGCF---YMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFA 460
FGCF YM + CK F CRP I +DGCH+KTK+GG +L AVG DPND FP+A A
Sbjct: 421 HFGCFHTLYMSFDACKRGFMFGCRPIICLDGCHIKTKFGGHILTAVGMDPNDCIFPIAIA 480
Query: 461 VVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLY 520
VVE E +SW WFL+ L +D+G W ++D+QKGL+ +F E R+CVRHLY
Sbjct: 481 VVEVESLKSWSWFLDTLKKDLGIENTSAWTVMTDRQKGLVPAVRREFSDAEQRFCVRHLY 540
Query: 521 ANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAF 580
NF+ G T L A +T E W M+++K ++ AY +L IP WCR F
Sbjct: 541 QNFQVLHKGETLKNQLWAIARSSTVPE-WNANMEKMKALSSEAYKYLEEIPPNQWCRAFF 599
Query: 581 SYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFAT-LREKSKKWKGNVM 639
S FPKCD+ +NN +E FN IL AR+ P+++M E IR +M R T +E + W +
Sbjct: 600 SDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIRNQIMNRLYTKQKELERNWPCGIC 659
Query: 640 PKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCR 699
PK ++++++ + + G FQV G Q+IV+++ + C C W+L GIPC
Sbjct: 660 PKIKRKVEKNTEMANTCYVFPAGMGAFQVSDIG--SQYIVELNVKRCDCRRWQLTGIPCN 717
Query: 700 HAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGP 759
HA++C + +KPE+ V Y YE Y + I P+ W KM ++ PP ++
Sbjct: 718 HAISCLRHERIKPEDVVSFCYSTRCYEQAYSYNIMPLRDSIHWEKMQGIEVKPPVYEKKV 777
Query: 760 GRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPA--KKQPTPSQP 817
GRPKK RR++P E + TK+ + G C C GHN+ C A K Q P Q
Sbjct: 778 GRPKKTRRKQPQE--LEGGTKISKHGVEMHCSYCKNGGHNKTSCKKRKADLKLQKQPIQQ 835
Query: 818 AAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAA----------ARARHQAPQPPP 867
A + ++ + H P S + A ++HQ P
Sbjct: 836 MATSSIVDEEELVITWEPVITQEMHLLSIPQKNSNYLSSSVVEAFIKEAASKHQERVP-- 893
Query: 868 ASPAAAARARHQAPQPPPPSPAAAARARHEGEPAA-RSASQPKK 910
A A R + QPP + RA AA + + PKK
Sbjct: 894 -ESAFIATNRREGVQPPLTTSTKQGRATLRKRSAAPKKKNSPKK 936
>emb|CAE05840.2| OSJNBa0091C07.2 [Oryza sativa (japonica cultivar-group)]
gi|50923321|ref|XP_472021.1| OSJNBa0091C07.2 [Oryza
sativa (japonica cultivar-group)]
Length = 879
Score = 350 bits (899), Expect = 9e-95
Identities = 224/754 (29%), Positives = 354/754 (46%), Gaps = 72/754 (9%)
Query: 93 ILGGVVDIY---LEHTVELPEVVRNEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEAPTVA 149
I+G V +IY L+H + V+ N + + V DD ++ + ++ E + E +A
Sbjct: 94 IMGDVAEIYAEILKHNEDDNVVLSNGVSDDSDVEDDIEEEYDSHSDAED---SSETDGIA 150
Query: 150 VPEVEASNIDSEDEICKNLTNDGFDDVDEPFDVQQDGDGRDDVNRNSGPNNGVQDGDESD 209
+ E S D E + + N + V D + V + PN G + D
Sbjct: 151 EDDEERSEDDEEAQENREHANKVKKNPCATITVTSD----NVVLVSDSPNKQDDKGLDDD 206
Query: 210 NNVGFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQRFRQEDFDKDY-KFRLG 268
+ F+ S A ++ D ED + R RF + D KF +G
Sbjct: 207 TDTPFLDSSEEASYD----------------DAEDGEAIRRKSRFPKYDSKAHTPKFEIG 250
Query: 269 MEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRI 328
M FA +FK+A++ + ++ + + + K++ ++R C ++C + + +N +++
Sbjct: 251 MTFAGRAKFKEAVIKYGLVTNRHIRFPKDEAKKIRARCSWKDCPWFIFASNGTNCDWFQV 310
Query: 329 KTLNPDHTCGRTFKNRAATAKWIA-----------------------------------K 353
KT N H C + NR T++ IA K
Sbjct: 311 KTFNDVHNCPKRRDNRLVTSRRIADKYEHIIKSNPSWKLQSLKKTVRLDMFADVSISKVK 370
Query: 354 NARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLE 413
A+ I + +++ + Y AE++R N +T AI L + P F Y+C +
Sbjct: 371 RAKGIVMRRIYDACRGEYSKVFEYQAEILRSNPGSTVAICLDHEYNW--PVFQRMYVCFD 428
Query: 414 GCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWF 473
CK F CR IG+DGC K G LL A+GRDPN+Q +P+A+AVVE E K++W WF
Sbjct: 429 ACKKGFLAGCRKVIGLDGCFFKGACNGELLCALGRDPNNQMYPIAWAVVEKETKDTWSWF 488
Query: 474 LNHLMRDIG-DNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTE 532
+ L +D+ D WV ISD+QKGL+ E P IEHR C RH+YAN+++K+ +
Sbjct: 489 IGLLQKDLNIDPHGAGWVIISDQQKGLVSAVEEFLPQIEHRMCTRHIYANWRKKY--RDQ 546
Query: 533 VRDLLMW-ASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMN 591
W +KA+ + + ++ ++ A + +HW R F CD N
Sbjct: 547 AFQKPFWKCAKASCRPFFNFCRAKLAQLTPAGAKXXXSTEPMHWSRAWFRIGSNCDSVDN 606
Query: 592 NLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIK 651
N+ E+FN I+ R P+I+MFE IR + R R K+K W G + P K+L++ I
Sbjct: 607 NMCESFNNWIIDIRAHPIISMFEGIRTKVYVRIQQNRSKAKGWLGRICPNILKKLNKYID 666
Query: 652 YSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLK 711
SGN A W + F+V + ++ VD++K++C+C YW+L GIPC HA+ +
Sbjct: 667 LSGNCEAIWNGKDGFEVTDK--DKRYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQ 724
Query: 712 PENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPD 771
PE+Y+ Y Y Y H + P+ G +WP PP + PGRP+K RRR+P
Sbjct: 725 PEDYIADCYSVEVYNKIYDHCMMPMEGMMQWPITGHPKPGPPGYVKMPGRPRKERRRDPL 784
Query: 772 EPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPL 805
E K KL + G+ C +C Q HN R CP+
Sbjct: 785 E--AKKGKKLSKTGTKGRCSQCKQTTHNIRTCPM 816
>gb|AAP52109.1| putative transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37531040|ref|NP_919822.1| putative
transposon protein [Oryza sativa (japonica
cultivar-group)] gi|14488302|gb|AAK63883.1| Putative
transposon protein [Oryza sativa]
Length = 841
Score = 333 bits (855), Expect = 1e-89
Identities = 192/609 (31%), Positives = 310/609 (50%), Gaps = 66/609 (10%)
Query: 241 DEEDNGS-RVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDK 299
D +D+G R++++ F ED K+ F++GM F S E +KAI +S+ + +N++
Sbjct: 156 DSDDDGEVRLKFKAFMAEDV-KNLVFKVGMVFPSVEVLRKAITEYSLKARVDIKMPRNEQ 214
Query: 300 NRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAKN----- 354
R+R C E C + Y + + + +KT +H C + + + TAKW+++
Sbjct: 215 KRLRAHCV-EGCPWNLYASFDSRSKSMMVKTYLGEHKCQKEWVLKRCTAKWLSEKYIETF 273
Query: 355 ------------------ARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRY 396
AR++A + + GD ++Q+ + +Y AEL R N + + L
Sbjct: 274 RANDKMTLGGFAKSKLARARRLAFKDIYGDEIQQYNQLWNYGAELRRSNPGSCFFLNL-- 331
Query: 397 IPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFP 456
+ F YM + CK CRP I +DGCH+KTK+GG +L AVG DPND +P
Sbjct: 332 ----VDSCFSTCYMSFDACKRGSLSGCRPLICLDGCHIKTKFGGQILTAVGIDPNDCIYP 387
Query: 457 LAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCV 516
+A VVE E SWRWFL L D+G + W ++DKQK
Sbjct: 388 IAIVVVETESLRSWRWFLQTLKEDLGIDNTYPWTIMTDKQK------------------- 428
Query: 517 RHLYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWC 576
+FK G +++ L ++++ + W M+E+K +N+ AY WL +P W
Sbjct: 429 ----VHFK-----GENLKNQLWACARSSSEVEWNANMEEMKSLNQDAYEWLQKMPPKTWV 479
Query: 577 RHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTR-FATLREKSKKWK 635
+ FS FPKCD+ +NN E FN IL AR+ P+++MFE I+ L++R ++ +E +++W
Sbjct: 480 KAYFSEFPKCDILLNNNCEVFNKYILEARELPILSMFEKIKSQLISRHYSKQKEVAEQWH 539
Query: 636 GNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNG 695
G + PK RK++ + + +G+FQVE F ++IVD+ + C C W+L G
Sbjct: 540 GPICPKIRKKVLKNADMANTCYVLPAGKGIFQVEDRNF--KYIVDLSAKHCDCRRWDLTG 597
Query: 696 IPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEI 755
IPC HA++C + + E+ + Y + Y I P N KW +++ ++ PP
Sbjct: 598 IPCNHAISCLRSERISAESILPPCYSLEAFSRAYAFNIWPYNDMTKWVQVNGPEVKPPIY 657
Query: 756 KRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQP--- 812
++ GRP K RR+ P E KN K+ + G C C + HN++ CPL A+ +P
Sbjct: 658 EKKVGRPPKSRRKAPHEVQGKNGPKMSKHGVEMHCSFCKEPRHNKKGCPLRKARLRPKLK 717
Query: 813 TPSQPAAAA 821
T PAA++
Sbjct: 718 TSRIPAASS 726
>gb|AAP53948.1| putative mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|37534718|ref|NP_921661.1| putative
mutator-like transposase [Oryza sativa (japonica
cultivar-group)]
Length = 1005
Score = 333 bits (854), Expect = 2e-89
Identities = 221/738 (29%), Positives = 354/738 (47%), Gaps = 65/738 (8%)
Query: 152 EVEASNIDSEDEICK--NLTNDGFDDVDE--------PFDVQQDGDGRDDVNRNSGPNNG 201
E A ++++E + C T+D D+V E +V++ GD D N +G
Sbjct: 172 EEAAEDLEAESDGCDATGFTSDEDDEVREIRTKYKEFMSEVKKRGDIPID---NPIEVDG 228
Query: 202 VQDGDESDNNVGFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQ-RFRQED-F 259
+Q GD N + A++ ++SD D S DE+ +G R + RF D F
Sbjct: 229 IQGGDIPLGNNQVLEGGDGAEY-----FDSD---GDASYDEDSDGVFTRRKCRFPIFDSF 280
Query: 260 DKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCAN 319
+F + M F ++ K AI +++ + Y KN++ R+R C+ + C ++ Y ++
Sbjct: 281 ADTPQFAVDMCFRGKDQLKDAIERYALKKKINIRYVKNEQKRIRAVCRWKGCPWLLYASH 340
Query: 320 IKGTSTYRIKTLNPDHTCGRTFKNRA---------------ATAKWIAKNARKIAKEAVE 364
+ ++I T NP+H C KN+ A W A+ ++ +E +
Sbjct: 341 NSRSDWFQIVTYNPNHACCPELKNKRLSTRRICDRYESTIKANPSWKAREMKETVQEDMG 400
Query: 365 GDA--------------------LKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPR 404
D +++ + YA +L R N N+ I L P
Sbjct: 401 VDVSITMIKRAKTHVMKKIMDTQTGEYSKLFDYALKLQRSNPGNSVHIALD--PEEEDHV 458
Query: 405 FGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEA 464
F FY+C + C+ F CR IG+DGC LK G LL A+GRD N+Q +P+A+AVVE
Sbjct: 459 FQRFYVCFDACRRGFLEGCRRIIGLDGCFLKGPLKGELLSAIGRDANNQLYPIAWAVVEY 518
Query: 465 ECKESWRWFLNHLMRDIG-DNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANF 523
E K+SW WFL HL +DI G WVFI+D+QKGLL + + FP EHR C RH+YAN+
Sbjct: 519 ENKDSWNWFLGHLQKDINIPVGAAGWVFITDQQKGLLSIVSTLFPFAEHRMCARHIYANW 578
Query: 524 KQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYF 583
++K E + +KA ++L+ +++ + L IHW R F
Sbjct: 579 RKKHRL-QEYQKRFWKIAKAPNEQLFNHYKRKLAAKTPRGWQDLEKTNPIHWSRAWFRLG 637
Query: 584 PKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPR 643
C+ NN++E+FN I+ +R KP+ITM E IR + R R S++W V P
Sbjct: 638 SNCESVDNNMSESFNSWIIESRFKPIITMLEDIRIQVTRRIQENRSNSERWTMTVCPNII 697
Query: 644 KRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVA 703
++ ++ + W F+V + +F VD+ ++C+C YW+++GIPC+HA A
Sbjct: 698 RKFNKIRHRTQFCHVLWNGDAGFEVRDKKW--RFTVDLTSKTCSCRYWQVSGIPCQHACA 755
Query: 704 CFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPK 763
+P N +H+ + Y+ Y H++ P+ E+ WP LPP +K+ PGRPK
Sbjct: 756 ALFKMAQEPNNCIHECFSLERYKKTYQHVLQPVEHESAWPVSPNPKPLPPRVKKMPGRPK 815
Query: 764 KLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIA 823
K RR++P +P K+ TK + G+ C RC GHN R C + ++ P A +
Sbjct: 816 KNRRKDPSKP-VKSGTKSSKVGTKIRCRRCGNYGHNSRTCCVKMQQEHAESYNPPATNAS 874
Query: 824 TNQAPQPPPSPAAAARAR 841
+ Q P+ + + +
Sbjct: 875 HSSPLQIEPAQSKQTKGK 892
>gb|AAC26234.1| contains similarity to maize transposon MuDR (GB:M76978)
[Arabidopsis thaliana] gi|7486726|pir||T01854
hypothetical protein F9D12.2 - Arabidopsis thaliana
Length = 940
Score = 331 bits (849), Expect = 6e-89
Identities = 240/834 (28%), Positives = 372/834 (43%), Gaps = 22/834 (2%)
Query: 93 ILGGVVDIYLEHTVELPEVVRNEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEAPTVAVPE 152
++GG VD+Y+E V P E+AV+++ ++ EV + + P
Sbjct: 94 LIGGEVDVYIEQGVSEPTPFPMSQHGEEAVVEEMEESDAEVEGDKTGEEDRGKDDDVDPV 153
Query: 153 VEASNIDSEDEICKNLTNDGFDDVDEPFDVQQDGDGRDDVNRNSGPNNGVQDGDESDNNV 212
E D N+ ++ + + G D +G GV +G +
Sbjct: 154 AECEGDDPRFRAFYEEINEEDNNHQDAGEAAGQGIPEADEGHVTGQEAGVDEGHVTGQEA 213
Query: 213 GFIPSEVAAQHNIDMEYESDDLL------SDYSCDEEDNGSRVRYQRFRQEDFDKDYK-- 264
G + A Q ID E D + + DEE + + + F KD
Sbjct: 214 GQEAGQEAGQEGIDEEEFEDQCMDTERPDAALDTDEEWDALDREEREIARAKFTKDKPPY 273
Query: 265 FRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTS 324
L +F + EEFK +L + + V K + ++ C E C++ YC K
Sbjct: 274 LWLKQQFNNGEEFKDQLLRYVLKTNYDVKLCKWGQKQLMAICSHENCQWKIYCFVDKRVG 333
Query: 325 TYRIKTLNPDHTCGRTFKNR--AATAKWIAKNARKIAKEAVEGDALKQFTLITSYAAELM 382
+ +KT H + K R +KWI + AR++A + V +QF + Y EL
Sbjct: 334 KWTVKTYVDAHKHAISGKARMLKQVSKWICRKARRMAFDMVIETQRQQFAKLWDYEGELQ 393
Query: 383 RVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGIL 442
R N ++T I+ + +F FY+C E ++ +K CRP IG+DG LK + G +
Sbjct: 394 RSN-KHTHTEIVTIPRADGKQQFDKFYICFEKLRTTWKSCCRPIIGLDGAFLKWELKGEI 452
Query: 443 LIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQV 502
L AVGRD +++ +P+A+A+V E E+W WF+ L D+ + ISDKQKGL+
Sbjct: 453 LAAVGRDADNRIYPIAWAIVRVEDNEAWAWFVEKLKEDLDLGVRAGFTVISDKQKGLINA 512
Query: 503 FAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEA 562
A+ P EHR+C RH+YAN+K+ +G A AT + + M ++ +
Sbjct: 513 VADLLPQAEHRHCARHVYANWKKVYGDYCHESYFWAIAYSATEGD-YSYNMDALRSYDPD 571
Query: 563 AYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMT 622
A L+ WCR FS C+ NNL+E+FN TI AR P++ M E +R+ M
Sbjct: 572 ACDDLLKTDPTTWCRAFFSTHSSCEDVSNNLSESFNRTIREARKLPVVNMLEEVRRISMK 631
Query: 623 RFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQV-EHAGFEDQFIVDI 681
R + L +K+ K + PK + L+ K S + F++ E +G+ + VD+
Sbjct: 632 RISKLCDKTAKCETRFPPKIMQILEGNRKSSKYCQVLKSGENKFEILEGSGY---YSVDL 688
Query: 682 DKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENK 741
++C C WEL GIPC HA+ + PE+YV + ++ Y I P+NGE
Sbjct: 689 LTRTCGCRQWELTGIPCPHAIYVITEHNRDPEDYVDRLLETQVWKATYKDNIEPMNGERL 748
Query: 742 WPKMSQDDILPPEIKRGPGRPKKLRR-REPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQ 800
W + + I P+ + GRPKK R +EP E NPTKL R G + TC C Q GHN+
Sbjct: 749 WKRRGKGRIEVPDKRGKRGRPKKFARIKEPGE-SSTNPTKLTREGKTVTCSNCKQIGHNK 807
Query: 801 RKCPLPPAKKQPTPSQPAAAAIATNQAP-QPPPSPAAAARARHQAPPPPPASPAAAARAR 859
C P + P + N P +P A+ Q+ PP S + A+
Sbjct: 808 GSCKNPTVQLAPPRKRGRPRKCVDNDDPWSIHNAPRRWRAAQSQSTPPVAQSQSTPPVAQ 867
Query: 860 HQAPQPP--PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP 911
Q Q P ++PAA R +A PP + + +G S P+ P
Sbjct: 868 SQPTQNPQHSSAPAANPSGRGRARGHPPGVKNGQGKGKGKGRGKVTDES-PRPP 920
>gb|AAU43947.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 792
Score = 299 bits (765), Expect = 3e-79
Identities = 194/669 (28%), Positives = 315/669 (46%), Gaps = 70/669 (10%)
Query: 93 ILGGVVDIY---LEHTVELPEVVRNEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEAPTVA 149
I+G V +IY L+H + V+ N + + V DD ++ + ++ E + E +A
Sbjct: 94 IMGDVAEIYAEILKHNEDDNVVLSNGVSDDSDVEDDIEEEYDSHSDAED---SSETDGIA 150
Query: 150 VPEVEASNIDSEDEICKNLTNDGFDDVDEPFDVQQDGDGRDDVNRNSGPNNGVQDGDESD 209
+ E S D E + + N + V D + V + PN G + D
Sbjct: 151 EDDEERSEDDEEAQENREHANKVKKNPCATITVTSD----NVVLVSDSPNKQDDKGLDDD 206
Query: 210 NNVGFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQRFRQEDFDKDY-KFRLG 268
+ F+ S A ++ D ED + R RF + D KF +G
Sbjct: 207 TDTPFLDSSEEASYD----------------DAEDGEAIRRKSRFPKYDSKAHTPKFEIG 250
Query: 269 MEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRI 328
M FA +FK+A++ + ++ + + + K++ ++R C ++C + + +N +++
Sbjct: 251 MTFAGRAKFKEAVIKYGLVTNRHIRFPKDEAKKIRARCSWKDCPWFIFASNGTNCDWFQV 310
Query: 329 KTLNPDHTCGRTFKNRAATAKWIA-----------------------------------K 353
KT N H C + NR T++ IA K
Sbjct: 311 KTFNDVHNCPKRRDNRLVTSRRIADKYEHIIKSNPSWKLQSLKKTVRLDMFADVSISKVK 370
Query: 354 NARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLE 413
A+ I + +++ + Y AE++R N +T AI L + P F Y+C +
Sbjct: 371 RAKGIVMRRIYDACRGEYSKVFEYQAEILRSNPGSTVAICLDHEYNW--PVFQRMYVCFD 428
Query: 414 GCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWF 473
CK F CR IG+DGC K G LL A+GRDPN+Q +P+A+AVVE E K++W WF
Sbjct: 429 ACKKGFLAGCRKVIGLDGCFFKGACNGELLCALGRDPNNQMYPIAWAVVEKETKDTWSWF 488
Query: 474 LNHLMRDIG-DNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTE 532
+ L +D+ D WV ISD+QKGL+ E P IEHR C RH+YAN+++K+ +
Sbjct: 489 IGLLQKDLNIDPHGAGWVIISDQQKGLVSAVEEFLPQIEHRMCTRHIYANWRKKY--RDQ 546
Query: 533 VRDLLMW-ASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMN 591
W +KA+ + + ++ ++ A +M+ +HW R F CD N
Sbjct: 547 AFQKPFWKCAKASCRPFFNFCRAKLAQLTPAGAKDMMSTEPMHWSRAWFRIGSNCDSVDN 606
Query: 592 NLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIK 651
N+ E+FN I+ R P+I+MFE IR + R R K+K W G + P K+L++ I
Sbjct: 607 NMCESFNNWIIDIRAHPIISMFEGIRTKVYVRIQQNRSKAKGWLGRICPNILKKLNKYID 666
Query: 652 YSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLK 711
SGN A W + F+V + ++ VD++K++C+C YW+L GIPC HA+ +
Sbjct: 667 LSGNCEAIWNGKDGFEVTDK--DKRYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQ 724
Query: 712 PENYVHQYY 720
PE+Y+ Y
Sbjct: 725 PEDYIADCY 733
>ref|XP_470323.1| putative transposon protein [Oryza sativa (japonica
cultivar-group)] gi|40714668|gb|AAR88574.1| putative
transposon protein [Oryza sativa (japonica
cultivar-group)]
Length = 839
Score = 292 bits (748), Expect = 3e-77
Identities = 228/840 (27%), Positives = 363/840 (43%), Gaps = 108/840 (12%)
Query: 96 GVVDIYLEHTVELPEVVRN---EMQKEDAVLD---DEDDVFVEVTEVEVIPMTQEA--PT 147
G VDIY E V+L + E ++AV D D + +F + + + P
Sbjct: 34 GAVDIYTE-IVKLDMGCSSTLVEAHSDEAVFDLFRDNNSMFPDSLVIAKGGTNTQCDNPN 92
Query: 148 VAVPEVEASNIDSED-EICKNLTNDGFDDVDE--------------PFDVQQDGDGRDDV 192
++ ++ +DS+ T D D+ DE ++ D D+
Sbjct: 93 SSIANIQTEGLDSDSGSDATGFTEDEDDEADEIRINYKQFKKKMKRREEIPLDTPVELDL 152
Query: 193 NRNSGPNNGVQDGDESDNNVGFIPSEVAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQ 252
++ N Q DE D + + SE A + ++D+G +
Sbjct: 153 PQSVDANTITQVNDEGDG-IAYFDSEDDASY------------------DDDSGDSAERR 193
Query: 253 RFRQEDFDKDYKF---RLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQE 309
+ R FD + + M F + K AI +++ K V Y N
Sbjct: 194 KCRFPIFDNHAELPQNAVDMCFRGKMQLKDAITRYALKK-KIVTYNGNH----------- 241
Query: 310 ECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAKNARKIAKEAVEGDALK 369
+ C ++K K L+ C + A W A+ ++ +E + D
Sbjct: 242 -----SCCPDLKN------KRLSTSRICDKYESVIKANPSWKARELKETVQEEMGVDVSM 290
Query: 370 --------------------QFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFY 409
+++ + YA ELMR N ++ + L P + F FY
Sbjct: 291 TMVKRAKSRVIKKVMDAHSGEYSRLFDYALELMRSNPGSSVHVALD--PDENEHVFQRFY 348
Query: 410 MCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKES 469
+CL+ C+ F CR IG DGC LK G LL AVGRD N+Q +P+A+AVVE E S
Sbjct: 349 VCLDACRRGFLDGCRRVIGFDGCFLKGVVKGELLSAVGRDANNQIYPIAWAVVEYENASS 408
Query: 470 WRWFLNHLMRDIG-DNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFG 528
W WFL HL +D+ G WVF++D+QKGLL V FP EHR C RH+YAN++++
Sbjct: 409 WNWFLGHLQKDLNIPYGGDGWVFLTDQQKGLLSVIEHLFPKAEHRMCARHIYANWRKRHR 468
Query: 529 GGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDV 588
E + ++++ L+ ++ + L IHWCR F CD
Sbjct: 469 L-QEYQKRFWKIARSSNAVLFNHYKSKLANKTPMGWEDLEKTNPIHWCRAWFKLGSNCDS 527
Query: 589 TMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDR 648
NN+ E+FN I+ AR KP+ITM E IR + R + S++W + P K++++
Sbjct: 528 VENNICESFNNWIIEARFKPIITMLEDIRMKVTRRIQENKTNSERWTMGICPNILKKINK 587
Query: 649 EIKYSGNWV-ATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFND 707
I+++ + W F+V + +F VD+ +C+C YW+++GIPC+HA A +
Sbjct: 588 -IRHATQFCHVLWNGSSGFEVREKKW--RFTVDLSANTCSCRYWQISGIPCQHACAAYFK 644
Query: 708 KGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRR 767
+P N+V+ + Y Y ++ P+ E+ WP + LPP +K+ PG PK+ RR
Sbjct: 645 MAEEPNNHVNMCFSIDQYRNTYQDVLQPVEHESVWPLSTNPRPLPPRVKKMPGSPKRARR 704
Query: 768 REPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQA 827
++P E + KRGG S CG CH+KGHN R C KK+ S + A
Sbjct: 705 KDPTEAAGSSTKSSKRGG-SVKCGFCHEKGHNSRGC-----KKKMEHSSSHYPSHPDQAA 758
Query: 828 PQPPPSPAAAARAR-----HQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQ 882
P + ++ A R Q+ A R+R + P S + + + P+
Sbjct: 759 PSGEANDSSVAHRRGKTLLSQSQGSAVQLAIGATRSRQSRGKQPAISSNSKTTKKGKMPE 818
>gb|AAT40523.1| putative mutator transposable element [Solanum demissum]
Length = 707
Score = 287 bits (735), Expect = 1e-75
Identities = 176/542 (32%), Positives = 276/542 (50%), Gaps = 65/542 (11%)
Query: 264 KFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRF-IAYCANIKG 322
+ LGM + +EFK+A++A+ GK++ + K+D+ R R C+ C++ I +
Sbjct: 184 ELELGMVLDNKKEFKEAVVANQAKIGKSIRWTKDDRERARAKCRTNACKWRILGSLMQRD 243
Query: 323 TSTYRIKTLNPDHTC-GRTFKNRAATAKWIAKN--------------------------- 354
ST++IKT +HTC G + N+ + WIA+
Sbjct: 244 ISTFQIKTFVSEHTCFGWNYNNKTINSSWIARKYVDRVKSNKNWRTSEFRDTLSRELKFH 303
Query: 355 -----ARKIAKEAV---EGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQP-RF 405
AR+ ++A+ +GD QF ++ +Y E++R N + + L +P RF
Sbjct: 304 VSMHQARRAKEKAIAMIDGDINDQFGILWNYCNEIVRTNPGTSVFMKLTPNEILNKPMRF 363
Query: 406 GCFYMCLEGCKSAFKLACRPFIGIDGCHLKT-KYGGILLIAVGRDPNDQYFPLAFAVVEA 464
FY+C CK+ FK CR IG++GC LK YG LL AV D N+ FP+A+A+VE
Sbjct: 364 QRFYICFAACKAGFKADCRKIIGVNGCWLKNIMYGAQLLSAVTLDGNNNIFPIAYAIVEK 423
Query: 465 ECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFK 524
E KE W+WFL +LM D+ E++++F CVRHL+ NFK
Sbjct: 424 ENKEIWQWFLTYLMNDL--EIEEQYLF-----------------------CVRHLHNNFK 458
Query: 525 QKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFP 584
+ G +++ L A+ AT + ++ M ++ ++++ AY WL A W R FS P
Sbjct: 459 RAGYSGMALKNALWKAASATTIDRFDACMTDLFELDKDAYAWLSAKVPSEWSRSHFSPLP 518
Query: 585 KCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGN-VMPKPR 643
KCD+ +NN E FN IL ARDKP++ + E IR LMTR ++REK++KW N + P +
Sbjct: 519 KCDILLNNQCEVFNKFILDARDKPIVKLLETIRHLLMTRINSIREKAEKWNLNDICPTIK 578
Query: 644 KRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVA 703
K+L + +K + N++ ++V D + VD+ ++C+C WEL+G+PC+HA++
Sbjct: 579 KKLAKTMKKAANYIPQRSNMWNYEVIGPVEGDNWAVDLYNRTCSCRQWELSGVPCKHAIS 638
Query: 704 CFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPK 763
K + NYV Y TY Y I P+NG + WPK PP + K
Sbjct: 639 SIWLKNDEVLNYVDDCYKVDTYRKIYEASILPMNGLDLWPKSLNPPPFPPSYLNNKKKGK 698
Query: 764 KL 765
K+
Sbjct: 699 KM 700
>gb|AAP54042.1| putative mutator protein [Oryza sativa (japonica cultivar-group)]
gi|37534906|ref|NP_921755.1| putative mutator protein
[Oryza sativa (japonica cultivar-group)]
Length = 929
Score = 286 bits (732), Expect = 2e-75
Identities = 220/795 (27%), Positives = 352/795 (43%), Gaps = 103/795 (12%)
Query: 114 NEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEAPTVA------VPEVEASNIDSED----- 162
NE++K A+ + + + V + P+ +AP+ + +P +A + D E+
Sbjct: 157 NEVRKGKAISSEIPENRLVVYNCDNDPLASQAPSESESNSEYIPGDDAGSDDDEEIIEIE 216
Query: 163 ----EICKNLTNDGFDDVDEPFDVQQDGDGRDDVNRNSGPNNGVQDGDESDNNVGFIPSE 218
++ + + D++D DV G G ++N +G G DG
Sbjct: 217 KHYKDMKRKVKAGQLDNLD---DVYLGGQGWQNMNNEAG---GEGDG------------- 257
Query: 219 VAAQHNIDMEYESDDLLSDYSCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFK 278
I E D+ + + D E + ++ R++++ F LGM+F+S ++FK
Sbjct: 258 ------IAAESPEDESMEEIGTDGEVSTKENKFPRYKKKVGTPC--FELGMKFSSKKQFK 309
Query: 279 KAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCG 338
KA+ +++ K + + K+D RVR C C ++ + + +++I TL H C
Sbjct: 310 KAVTKYALAERKVINFIKDDLKRVRAKCDWASCPWVCLLSKNTRSDSWQIVTLESLHACP 369
Query: 339 RTFKNRAATAKWIAKNARKI---------------AKEAVEGDA----LK---------- 369
N+ TA IA+ KI +E + G+A LK
Sbjct: 370 PRRDNKMVTATRIAEKYGKIILANPGWNLAHMKATVQEEMFGEASVPKLKRAKAMVMKKA 429
Query: 370 ------QFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLAC 423
Q+ + +Y EL+R N +T ++ R I T P F Y+CL+ CK F C
Sbjct: 430 MDATKGQYQKLYNYQLELLRSNPGSTV-VVNREIGTD-PPVFKRIYICLDACKRGFVDGC 487
Query: 424 RPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGD 483
R +G+DGC K G LL A+GRD N+Q +P+A+AVV E E W WFL+ L D+
Sbjct: 488 RKVVGLDGCFFKGATNGELLCAIGRDANNQMYPIAWAVVHKENNEEWDWFLDLLCSDLKV 547
Query: 484 NGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKA 543
WVFISD+QKG++ + P EHR C RH+YAN+K+ F E + +KA
Sbjct: 548 LDGTGWVFISDQQKGIINAVEKWAPQAEHRNCARHIYANWKRHF-SDKEFQKKFWRCAKA 606
Query: 544 TYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILV 603
+ L+ ++ ++ A ++ HW R F CD NN+ E+FN IL
Sbjct: 607 PCRMLFNLARAKLAQVTPAGAQAILNTHPEHWSRAWFKLGSNCDSVDNNMCESFNKWILE 666
Query: 604 ARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQ 663
AR P+ITM E IR+ +M R + R+ S +W V P K+L+ I S A
Sbjct: 667 ARFFPIITMLETIRRKVMVRISEQRKVSARWNTVVCPGILKKLNVYITESAYCHAICNGA 726
Query: 664 GLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRA 723
F+V+H ++F V +DK+ C+C L + + Y
Sbjct: 727 DSFEVKH--HTNRFTVQLDKKECSCRTNTL-------------------DEFTADCYKVD 765
Query: 724 TYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKR 783
++ Y H + P+ G N WP+ ++ + P + PGRP+ RRRE EP K P+K +
Sbjct: 766 AFKSTYKHCLLPVEGMNAWPEDDREPLTAPGYIKMPGRPRTERRREAHEPAK--PSKASK 823
Query: 784 GGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQ 843
G+ C C Q GHN+ C + S A + N P A +R R
Sbjct: 824 FGTKVRCRTCKQVGHNKSSCHKHNPAQTAGGSSQQVATPSHNLVLSNTPQSCAQSRKRKA 883
Query: 844 APPPPPASPAAAARA 858
+ A+ ++A
Sbjct: 884 TGTLTTTTSASQSKA 898
>gb|AAX96864.1| transposon protein, putative, mutator sub-class [Oryza sativa
(japonica cultivar-group)]
Length = 868
Score = 283 bits (725), Expect = 1e-74
Identities = 216/790 (27%), Positives = 334/790 (41%), Gaps = 79/790 (10%)
Query: 114 NEMQKEDAVLDDEDDVFVEVTEVEVIPMTQEAPTVAVPEVEASNIDSEDEICKNLTNDGF 173
N + DA+ +E+DV T ++P+ P + V + + S +D+ +
Sbjct: 105 NVNSEADAMNGNEEDV---QTNERLLPVQIPNPAMVVYKAKESPHGYDDDEEGAAIERHY 161
Query: 174 DDVDEPFDVQQDGDGRDDVN-----RNSGPNNGVQDGDESDNNVGFIPSEVAAQHNIDME 228
D+ + + DDV N+ G +GD +D E
Sbjct: 162 KDLKKKLRTSKVAS-LDDVTFGGHKTNANMATGTNEGDGNDT-----------------E 203
Query: 229 YESDDLLSDYSCDEEDNGSRVRYQRFRQEDFDKDY---KFRLGMEFASTEEFKKAILAHS 285
Y D D EE + +R + F K F LGM+F+ +FKKAI A++
Sbjct: 204 YIDSD---DEESVEEGSDGELRVRGSNNARFKKKPGYPTFELGMKFSCKSQFKKAITAYA 260
Query: 286 VMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRA 345
+ K + + K+D RVR C C ++ +N + ++I T H C N+
Sbjct: 261 LAERKVINFVKDDPKRVRAKCDWHSCPWVCLLSNNSRSDGWQIVTFENFHACPPRRDNKL 320
Query: 346 ATAKWIA-----------------------------------KNARKIAKEAVEGDALKQ 370
TA IA K A+ I + Q
Sbjct: 321 VTATRIADKYGKFIIANPSWPLAHMKATIQEEMFANVSVSKLKRAKSIVMKKAMDATKGQ 380
Query: 371 FTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGID 430
+ + +Y EL+R N +T ++ R + P F Y+CL+ CK F CR IG+D
Sbjct: 381 YQKLYNYQKELLRSNPGSTV-VVNREVDMD-PPVFKRIYICLDACKRGFISGCRKVIGLD 438
Query: 431 GCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWV 490
GC K G LL A+GRD N+Q + +A+AV+ E E W WFL+ L D+ WV
Sbjct: 439 GCFFKGATNGKLLCAIGRDANNQMYLVAWAVIHKENNEEWDWFLDLLCGDLKVGDGSGWV 498
Query: 491 FISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMW-ASKATYKELW 549
FISD+QKG++ + P EHR C RH+YA++K+ F ++ W +KA L+
Sbjct: 499 FISDQQKGIINAVEKWAPEAEHRNCARHIYADWKRHF--NEKILQKKFWRCAKAPCILLF 556
Query: 550 EEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPL 609
++ ++ +M HW R F CD NNL E+FN IL AR P+
Sbjct: 557 NLARAKLAQLTPPGAQAIMNTHPQHWSRAWFRLGSNCDSVDNNLCESFNKWILEARFFPI 616
Query: 610 ITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVE 669
ITM E IR+ +M R S +W + P K+L+ I S A ++V+
Sbjct: 617 ITMLETIRRKVMVRIHDQITTSARWNTAICPGILKKLNAYITKSAFSHAICNGASSYEVK 676
Query: 670 HAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICY 729
H +++F V +DK +C+ YW+L+G+PC HA++C K + Y+ Y ++ Y
Sbjct: 677 H--HDNRFTVQLDKMACSYRYWQLSGLPCPHAISCIFFKTNSLDGYISDCYSVTEFKKTY 734
Query: 730 GHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYT 789
H + P G N WP + + P + PGRP+ RRREP E K T++ + G+
Sbjct: 735 SHCLEPFEGMNNWPYDDRQPLNAPGYIKMPGRPRTERRREPTEAPK--ATRMSKMGTKIR 792
Query: 790 CGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPP 849
G H P +P+ + T + S + + A H P P
Sbjct: 793 WGPSTAASHPTGS---PAGTNAVSPAGGLVLSNYTQGRKRKSISSTSTSSAIHSVRPAPS 849
Query: 850 ASPAAAARAR 859
A A+
Sbjct: 850 KRKAQMQEAQ 859
>dbj|BAB09991.1| mutator-like transposase-like [Arabidopsis thaliana]
Length = 825
Score = 276 bits (707), Expect = 2e-72
Identities = 201/708 (28%), Positives = 318/708 (44%), Gaps = 77/708 (10%)
Query: 186 GDGRDDVNRNSGPNNGVQDGDESDNNVGFIPSEVAAQHN----IDMEYESDDLLSDYSCD 241
GDG + + + V DG E D FI E + N +D E +D + + D
Sbjct: 126 GDG--SIGDGAAGDGAVGDGSECDRV--FIDMEARVEQNLAGFVDEEEHDNDESTPLNSD 181
Query: 242 EEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKK--NDK 299
E+ G R RY R ++ D R+ F S EFK+A++ +++ G + + + ++K
Sbjct: 182 CEE-GER-RYVRCKKGSGD----IRIEQVFDSIAEFKEAVIDYALKKGVNIKFTRWGSEK 235
Query: 300 NRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTCGR-------------------- 339
+ VR + C+F YCA + Y +KT +H C +
Sbjct: 236 SEVRCSIGGN-CKFRIYCAYDEKIGVYMVKTFIDEHACTKDGFCKVLKSRIIVDMFLNDI 294
Query: 340 ----TFKNRAA-----------TAKWIAKNARKIAKEAVEGDALKQFTLITSYAAELMRV 384
TFK +A + + AR A + ++ + +QF I Y +L+
Sbjct: 295 RQDPTFKVKAMQKAIEERYSLIASSDQCRKARGKALKMIQDEYDEQFARIHDYKEQLLET 354
Query: 385 NHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLI 444
N +T + I + +F FY+C + + + CRP IGIDGC LK G LL
Sbjct: 355 NLGSTVEVETITIDGIV--KFDKFYVCFDALRKTWLAYCRPIIGIDGCFLKNNMKGQLLA 412
Query: 445 AVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFA 504
AVGRD N+Q++P+A+AVVE E +SW WF+ L D+ + ISD+QKGLL
Sbjct: 413 AVGRDANNQFYPVAWAVVETENIDSWLWFIRKLKSDLKLQDGDGFTLISDRQKGLLNTVD 472
Query: 505 EDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAY 564
++ P +EHR C RH+Y N ++ + G ++L +K+ + + E+K+ ++ Y
Sbjct: 473 QELPKVEHRMCARHIYGNLRRVYPGKDLSKNLFCAVAKSFNDYEYNRAIDELKQFDQGVY 532
Query: 565 WWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRF 624
+M + R F C+ NN +E++N I AR+ PL+ M E IR+ M R
Sbjct: 533 DAVMMRNPRNCSRAFFGCKSSCEDVSNNFSESYNNAINKAREMPLVEMLETIRRQTMIRI 592
Query: 625 ATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQV--EHAGFEDQFIVDID 682
K+ K +G K + E Y G F+V + GF VD++
Sbjct: 593 DMRLRKAIKHQGKFTLKVANTIKEETIYRKYCQVVPCGNGQFEVLENNHGFR----VDMN 648
Query: 683 KQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVH-QYYLRATYEICYGHMISPINGENK 741
++C C W + GIPCRH + +K + PE V+ +YL + Y IS +NG
Sbjct: 649 AKTCACRRWNMTGIPCRHVLRIILNKKVNPEELVNSDWYLASKNLKIYSESISGVNGIGF 708
Query: 742 WPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNP-----------TKLKRGGSSYTC 790
W + + I PP G + +K ++ + + P KK TK+ + G S C
Sbjct: 709 WIRSGEPRIQPPPRPTGTKKKEKRKKGKNESPKKKKKGKGVQEPPEKRTKVSKEGRSIRC 768
Query: 791 GRCHQKGHNQRKCPLPPAKKQPTPSQ-----PAAAAIATNQAPQPPPS 833
GRC +GHN R CP + P + + +I TN+ PS
Sbjct: 769 GRCSTEGHNTRSCPSVGVVNKRWPKKTPDGVSQSMSIGTNEGTSQRPS 816
>gb|AAT39304.1| putative transposon MuDR mudrA-like protein [Solanum demissum]
Length = 873
Score = 276 bits (705), Expect = 3e-72
Identities = 192/644 (29%), Positives = 299/644 (45%), Gaps = 76/644 (11%)
Query: 215 IPSEVAA--QHNIDMEYESDDLLSDYSCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFA 272
+P EVA + N + E +++ + S Y+ DE + R R + D+ + GM F
Sbjct: 119 VPVEVATDCESNEEDEDQNEPIPSYYNSDELEV-FRKEKNREISDKLDRFLELEKGMCFK 177
Query: 273 STEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLN 332
+E K+ + +S++ A+ +K+D R+R+ C C F + + ++IKTLN
Sbjct: 178 DLKEAKRIVSFYSIVRKVALKVEKSDSTRLRYLC-DIGCPFECLISEDRKNQGFKIKTLN 236
Query: 333 PDHTCGRT-FKNRAATAKWIA-----------------------------------KNAR 356
H+CG FKNR AT + +A K +
Sbjct: 237 TKHSCGENVFKNRRATQEALAYYFKKKLQNNPKYSVNDMRQDLDDNFNLNVSYSKMKRVK 296
Query: 357 KIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAI-ILRYIPTTLQPRFGCFYMCLEGC 415
++ E +EG + +F + YA EL N I I R + RF Y+C++
Sbjct: 297 RLVLEKLEGSYIDEFNKLEGYAQELRDSNPGTDVIINISRDALEQGKRRFLRMYVCIQAL 356
Query: 416 KSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLN 475
K+ +K G+ ++PLA+AVV+ E +W+WF+
Sbjct: 357 KNGWK-------------------GV----------KHFYPLAWAVVDRETSRTWKWFIE 387
Query: 476 HLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRD 535
L + + F+SD KGLL ++ FP HR+C RH+ AN+ + + G ++R
Sbjct: 388 LLRNSLDLANGEGVTFMSDMHKGLLDAVSQVFPKAHHRWCARHIEANWSKAWKG-VQMRK 446
Query: 536 LLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAE 595
LL W++ +TY+E + ++++ + +++ A L+ P +WCR F K NN E
Sbjct: 447 LLWWSAWSTYEEEFHDQLKVMGAVSKQAAKDLVWYPAQNWCRAYFDTVCKNHSCENNFTE 506
Query: 596 AFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRL-DREIKYSG 654
+FN IL AR KP+I M E IR +M R L E+ K WKG+ P + D I
Sbjct: 507 SFNKWILEARAKPIIKMLENIRIKIMNRLQKLEEEGKNWKGDFSPYAMELYNDFNIIAQC 566
Query: 655 NWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPEN 714
V + QG VE ED+ +V+++++ CTC W+L GI C HA+ + +P +
Sbjct: 567 CQVQSNGDQGYEVVEG---EDRHVVNLNRKKCTCRTWDLTGILCPHAIKAYLHDKQEPLD 623
Query: 715 YVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPD 774
+ +Y R Y + Y H I P+ GE W + PPEI + GRPK R+RE DE
Sbjct: 624 QLSWWYSREAYMLVYMHKIQPVRGEKFWKVDPSHAMEPPEIHKLVGRPKLKRKREKDEAR 683
Query: 775 KKNPT-KLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQP 817
K+ R G TCG C GHNQR+CP+ KQP P
Sbjct: 684 KREGVWSASRKGLKMTCGHCSATGHNQRRCPMLQRSKQPAQDVP 727
>gb|AAF79687.1| F9C16.9 [Arabidopsis thaliana] gi|25405857|pir||D96503 protein
F9C16.9 [imported] - Arabidopsis thaliana
Length = 946
Score = 270 bits (691), Expect = 1e-70
Identities = 232/814 (28%), Positives = 347/814 (42%), Gaps = 105/814 (12%)
Query: 181 DVQQDGDGRDDVNRNSGPNNGVQDGD--ESDNNVGF-------------IPSEVAAQHNI 225
+V G+ +VN G V +G+ E + N G +E + N
Sbjct: 134 EVNNGGNAEAEVNDGGGVEAEVNNGESVEEEGNGGLDMEEEDCEDFEAEFGAEAREEENE 193
Query: 226 DMEYESDDLLSD--------YSCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEF 277
DD+ D +S DE DN Q D D + RL F + E+F
Sbjct: 194 SDGDSGDDIWDDERIPDPLVHSDDEGDNDDE---DGAPQTD-DPEESLRLAKTFNNPEDF 249
Query: 278 KKAILAHSVMNGKAVIYKKNDKNRVRFACKQEE--CRFIAYCANIKGTSTYRIKTLNPDH 335
K A+L +S+ + ++ RV C + C++ YC+ K ++K H
Sbjct: 250 KIAVLRYSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQKMQVKVYKDGH 309
Query: 336 TCGRTFKNRAATAKWIA-------KNARKIAKEAVEGDALKQFTL--------------- 373
+C R+ ++ + IA + KI K+ + + +++ L
Sbjct: 310 SCVRSGYSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYKLTVTEDQCSKAKTMVM 369
Query: 374 -------------ITSYAAELMRVNHENTAAIILRYIPT--TLQPRFGCFYMCLEGCKSA 418
I Y A++ R N T I PT +LQ RF ++C + K +
Sbjct: 370 RERQATHQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQ-RFLRLFICFKSQKES 428
Query: 419 FKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLM 478
+K CRP IGIDG LK G LL A GRD +++ P+A+AVVE E ++W WF+ L
Sbjct: 429 WKQTCRPIIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVEIENDDNWDWFVRMLS 488
Query: 479 RDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLM 538
R + + ISDKQ GL++ P EHR C RH+ N+K+ R L
Sbjct: 489 RTLDLQDGRNVAIISDKQSGLVKAIHSVIPQAEHRQCARHIMENWKRNSHDMELQR--LF 546
Query: 539 WASKATYKE-LWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAF 597
W +Y E + M+ +K N +A+ L+ + W R F C+ +NNL+E+F
Sbjct: 547 WKIVRSYTEGEFGAHMRALKSYNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESF 606
Query: 598 NCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIK--YSGN 655
N TI AR KPL+ M E IR+ M R EK + + KR EI+ +G+
Sbjct: 607 NRTIREARRKPLLDMLEDIRRQCMVR----NEKRYVIADRLRTRFTKRAHAEIEKMIAGS 662
Query: 656 WVA-TWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPEN 714
V W+A+ G D F VD++ +C C W++ GIPC HA + K K E+
Sbjct: 663 QVCQRWMARHNKHEIKVGPVDSFTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVED 722
Query: 715 YVHQYYLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRG-PGRPKKLRRR----E 769
YV +Y + ++ Y I P+ G+ WP +++ +LPP +RG PGRPK RR E
Sbjct: 723 YVSDWYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFE 782
Query: 770 PDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCP----LPPAK-------KQPTPSQPA 818
+ T+L R TC C +GHN++ C PPAK K P
Sbjct: 783 SSTASSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKRPRGRPRKDKDPRMVF 842
Query: 819 AAAIATNQAPQ---PPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAAR 875
T Q PQ P S A ++ H AP P+ A + A Q Q + ++
Sbjct: 843 FGQGQTWQEPQAQGAPSSHGAPSQVFHGAPLSQPSQEAPSQGAPSQGAQ-------SLSQ 895
Query: 876 ARHQAPQPPPPSPAAAARA--RHEGEPAARSASQ 907
+AP P+ R+ R E + R SQ
Sbjct: 896 PSQEAPSQAAPTETRGTRSVRRGESQRGGRGGSQ 929
>dbj|BAB11264.1| unnamed protein product [Arabidopsis thaliana]
Length = 733
Score = 259 bits (661), Expect = 4e-67
Identities = 175/638 (27%), Positives = 306/638 (47%), Gaps = 63/638 (9%)
Query: 226 DMEYESDDLLSDYSCDEEDNGSRVRYQR---FRQEDFDKD---YKFRLGMEFASTEEFKK 279
D Y D L+ + +EE ++ + R +D D D F +G EF S E+ K
Sbjct: 60 DDPYLDDVLIGEREEEEELESEQLDFYRDDYVESDDSDDDGARIDFYVGQEFVSKEKCKD 119
Query: 280 AILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKTLNPDHTC-- 337
I +++ + +K++++N++ C Q+ C++ Y + + +++ H+C
Sbjct: 120 TIEKYAIREKVNIHFKRSERNKIEGVCVQDCCKWRIYASITSRSDKMVVQSYKGIHSCYP 179
Query: 338 --------------------------------GRTFKNRAATAKWIAKNARKIAKEAVEG 365
R + N K ++AR+I K +
Sbjct: 180 IGVVDLYSAPKIAADFINEFRTNSNLSAGQIMQRLYLNGLRVTKTKCQSARQIMKHIISD 239
Query: 366 DALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRP 425
+ ++QFT + Y EL + N +T IIL T F FY C E K+ +K ACR
Sbjct: 240 EYVEQFTRMYDYVEELRKTNPGST--IILG----TKDRIFEKFYTCFEAQKTGWKTACRR 293
Query: 426 FIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNG 485
I + G LK + G LL AVGRDPND + +A+A+V E K W+WF+ L D+
Sbjct: 294 VIHLGGTFLKGRMKGQLLTAVGRDPNDAMYIIAWAIVPVENKVYWQWFMELLGEDLRLEL 353
Query: 486 EKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATY 545
SD+QKGL+ P EHR C RH++AN ++++ + + ++A
Sbjct: 354 GNGLALSSDQQKGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYN 413
Query: 546 KELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVAR 605
+ ++ ++++++K I AY + +W R FS K NN++E++N + AR
Sbjct: 414 ETVFWKQLEKMKTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAR 473
Query: 606 DKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWV-AQG 664
+KP++ + E IR+++M ++ + G + PK +++ K S W + +G
Sbjct: 474 EKPVVALLEDIRRHIMASNLVKLKEMQNVTGLITPKAIAIMEKR-KKSLKWCYPFSNGRG 532
Query: 665 LFQVEHAGFEDQFIVDI-DKQSCTCNYWELNGIPCRHAVAC----FNDKGLKPENYVHQY 719
+++V+H +++++V + DK SCTC ++++GIPC H ++ + + L PE + +
Sbjct: 533 IYEVDHG--KNKYVVHVRDKTSCTCREYDVSGIPCCHIMSAMWAEYKETKL-PETAILDW 589
Query: 720 YLRATYEICYGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRR-REP-DEPDKKN 777
Y +++CY ++ P+NG W S ++PP + PGRPKK R ++P +E +N
Sbjct: 590 YSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSEN 649
Query: 778 PTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPS 815
K + TC C Q GHN+R C + K P PS
Sbjct: 650 SQK-----ALVTCSNCGQTGHNKRTCQIELVPKPPNPS 682
>gb|AAP51801.1| putative mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|37530424|ref|NP_919514.1| putative
mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|18542920|gb|AAL75755.1| Putative
mutator-like transposase [Oryza sativa]
Length = 812
Score = 255 bits (652), Expect = 4e-66
Identities = 160/510 (31%), Positives = 247/510 (48%), Gaps = 60/510 (11%)
Query: 204 DGDESDNNVGFIPSEVAAQHNID--MEYESDD----LLSDYSCDEEDNGS-RVRYQRFRQ 256
D D DN V ++ HN + +Y S + ++ D S +EE+ + ++ FR
Sbjct: 194 DDDLFDNCV---EGQIELAHNKEEGSDYGSSEDERLMMPDISEEEEEEEEIKFSFKSFRA 250
Query: 257 EDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAY 316
+ + +F++ M FA E +KAI +++ N A+ +N K R+ C E C ++
Sbjct: 251 DVDMETPEFKVSMMFADAVELRKAIDQYTIKNRVAIKKTRNTKTRIEAKC-AEGCPWMLS 309
Query: 317 CANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAK----------------------- 353
+ ++ HT + ++ +A TAK++ K
Sbjct: 310 ASMDNRVKCLVVREYIEKHTRSKQWEIKAVTAKYLTKRYIEEFRDNDKMTLMSFAKKIQK 369
Query: 354 ------------NARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTL 401
AR++A A+ GD + Q+ + Y EL N ++ + L
Sbjct: 370 ELHLTPSRHKLGRARRMAMRAIYGDEISQYNQLWDYGQELRTSNPRSSFYLNL------- 422
Query: 402 QPRFGCF---YMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLA 458
FGCF YM + CK F CRP I +DGCH+KTK+GG +L AVG DPND FP+A
Sbjct: 423 --LFGCFHTLYMSFDACKRGFLSGCRPIICLDGCHIKTKFGGHILTAVGMDPNDCIFPIA 480
Query: 459 FAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRH 518
VVE E +SW WFL+ L +D+G W ++D+QKGL+ +F E R+CVRH
Sbjct: 481 IVVVEVESLKSWSWFLDTLKKDLGIENTSAWTVMTDRQKGLVPAVRREFSHAEQRFCVRH 540
Query: 519 LYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRH 578
LY NF+ G T L A +T E W M+++K ++ AY +L IP WCR
Sbjct: 541 LYQNFQVLHKGETLKNQLWAIARSSTVPE-WNANMEKMKALSSEAYKYLEEIPPNQWCRA 599
Query: 579 AFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFAT-LREKSKKWKGN 637
FS FPKCD+ +NN +E FN IL AR+ P+++M E IR +M R T +E + W
Sbjct: 600 FFSDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIRNQIMNRLYTKQKELERNWPCG 659
Query: 638 VMPKPRKRLDREIKYSGNWVATWVAQGLFQ 667
+ PK ++++++ + + G FQ
Sbjct: 660 LCPKIKRKVEKNTEMANTCYVLPAGMGAFQ 689
>emb|CAB77993.1| putative MuDR-A-like transposon protein [Arabidopsis thaliana]
gi|2565011|gb|AAB81881.1| putative MuDR-A-like
transposon protein [Arabidopsis thaliana]
gi|7487609|pir||T00940 hypothetical protein T3F12.2 -
Arabidopsis thaliana
Length = 761
Score = 251 bits (642), Expect = 6e-65
Identities = 177/624 (28%), Positives = 282/624 (44%), Gaps = 57/624 (9%)
Query: 239 SCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKND 298
S DE+++ VR +R R+ DK F +G F + EFK+AIL ++ +G V+ + +
Sbjct: 86 SSDEDEDPILVRDRRIRRAMGDK---FVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWE 142
Query: 299 KNRVRFACKQE-ECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAK---- 353
K ++ F C +C++ YC+ + +KT H+C K + + IA+
Sbjct: 143 KEKISFKCGMGGKCKWRVYCSYDPDRQLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLD 202
Query: 354 -------------------------------NARKIAKEAVEGDALKQFTLITSYAAELM 382
R +A + ++ + +QF I Y E+
Sbjct: 203 KLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIH 262
Query: 383 RVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGIL 442
N + A I Y + F FY+C ++ + +CRP IG+DG LK G+L
Sbjct: 263 SQN-PGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVL 321
Query: 443 LIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQV 502
L AVG DPN+Q +P+A+AVV++E E+W WF+ + +D+ ++V +SD+ KGLL
Sbjct: 322 LTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSA 381
Query: 503 FAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKEL-WEEKMQEIKKINE 561
++ P EHR CV+H+ N K K ++ L+W +Y E + + + ++ +E
Sbjct: 382 VKQELPNAEHRMCVKHIVENLK-KNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDE 440
Query: 562 AAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLM 621
A Y ++ W R + C+ NN E+FN TI AR K LI M E IR+ M
Sbjct: 441 ALYNDVLNEEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGM 500
Query: 622 TRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDI 681
TR +KS + +G K L E + G+F+V E+ VDI
Sbjct: 501 TRIVKRNKKSLRHEGRFTKYALKMLALEKTDADRSKVYRCTHGVFEVYID--ENGHRVDI 558
Query: 682 DKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENK 741
K C+C W+++GIPC H+ + GL ENY+ +++ + Y P+ G
Sbjct: 559 PKTQCSCGKWQISGIPCEHSYGAMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKY 618
Query: 742 WPKMSQDDILPPEIKRGPGRPKKLRRREP--------DEPDKK-----NPTKLKRGGSSY 788
W S + P PGR K+ ++E + P KK KL + G
Sbjct: 619 WLNSSYGLVTAPPEPILPGRKKEKSKKEKFARIKGKNESPKKKKRKKNEVEKLGKKGKII 678
Query: 789 TCGRCHQKGHNQRKCPLPPAKKQP 812
C C + GHN +C P +K P
Sbjct: 679 HCKSCGEAGHNALRCKKFPKEKVP 702
>gb|AAG52094.1| putative Mutator-like transposase; 12516-14947 [Arabidopsis
thaliana] gi|25406536|pir||A96810 probable Mutator-like
transposase, 12516-14947 [imported] - Arabidopsis
thaliana
Length = 761
Score = 250 bits (639), Expect = 1e-64
Identities = 177/624 (28%), Positives = 281/624 (44%), Gaps = 57/624 (9%)
Query: 239 SCDEEDNGSRVRYQRFRQEDFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKND 298
S DE+++ VR +R R+ DK F +G F + EFK+AIL ++ +G V+ + +
Sbjct: 86 SSDEDEDPILVRDRRIRRAMGDK---FVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWE 142
Query: 299 KNRVRFACKQE-ECRFIAYCANIKGTSTYRIKTLNPDHTCGRTFKNRAATAKWIAK---- 353
K ++ F C +C++ YC+ + +KT H+C K + + IA+
Sbjct: 143 KEKISFKCGMSGKCKWRVYCSYDPDRQLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLD 202
Query: 354 -------------------------------NARKIAKEAVEGDALKQFTLITSYAAELM 382
R +A + ++ + +QF I Y E+
Sbjct: 203 KLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIH 262
Query: 383 RVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGIL 442
N + A I Y + F FY+C ++ + +CRP IG+DG LK G+L
Sbjct: 263 SQN-PGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVL 321
Query: 443 LIAVGRDPNDQYFPLAFAVVEAECKESWRWFLNHLMRDIGDNGEKKWVFISDKQKGLLQV 502
L AVG DPN+Q +P+A+AVV++E E+W WF+ + +D+ ++V +SD+ KGLL
Sbjct: 322 LTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSA 381
Query: 503 FAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMWASKATYKEL-WEEKMQEIKKINE 561
++ P EHR CV+H+ N K K ++ L+W +Y E + + + ++ +E
Sbjct: 382 VKQELPNAEHRMCVKHIVENLK-KNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDE 440
Query: 562 AAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNCTILVARDKPLITMFEWIRKYLM 621
A Y ++ W R + C+ NN E+FN TI AR K LI M E IR+ M
Sbjct: 441 ALYNDVLNEEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGM 500
Query: 622 TRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDI 681
TR +KS + +G K L E + G+F+V G + VDI
Sbjct: 501 TRIVKRNKKSLRHEGRFTKYALKMLALEKTDADRSKVYRCTHGVFEVYIDG--NGHRVDI 558
Query: 682 DKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENK 741
K C+C W+++GIPC HA + GL ENY+ +++ + Y P+ G
Sbjct: 559 PKTQCSCGKWQISGIPCEHAYGAMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKY 618
Query: 742 WPKMSQDDILPPEIKRGPGRPKKLRRR--------EPDEPDKK-----NPTKLKRGGSSY 788
W S + P PGR K+ ++ + + P KK KL + G
Sbjct: 619 WLNSSYGLVTAPPEPILPGRKKEKSKKGKFARIKGKNESPKKKKRKKNEVEKLGKKGKII 678
Query: 789 TCGRCHQKGHNQRKCPLPPAKKQP 812
C C + GHN C P +K P
Sbjct: 679 HCKSCGEAGHNALGCKKFPKEKVP 702
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,767,110,589
Number of Sequences: 2540612
Number of extensions: 87622261
Number of successful extensions: 1248832
Number of sequences better than 10.0: 20974
Number of HSP's better than 10.0 without gapping: 5600
Number of HSP's successfully gapped in prelim test: 17013
Number of HSP's that attempted gapping in prelim test: 700119
Number of HSP's gapped (non-prelim): 175740
length of query: 931
length of database: 863,360,394
effective HSP length: 138
effective length of query: 793
effective length of database: 512,755,938
effective search space: 406615458834
effective search space used: 406615458834
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0016.18


