

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0014.4
(568 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
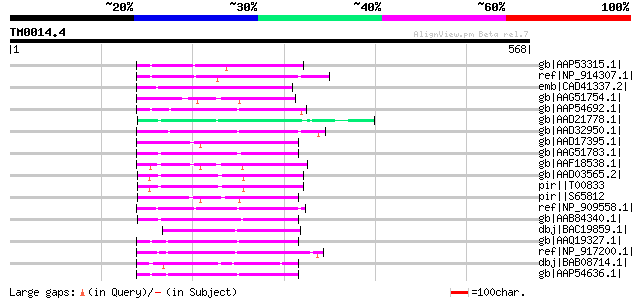
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP53315.1| putative reverse transcriptase [Oryza sativa (jap... 94 9e-18
ref|NP_914307.1| reverse transcriptase -like protein [Oryza sati... 91 1e-16
emb|CAD41337.2| OJ991113_30.21 [Oryza sativa (japonica cultivar-... 80 2e-13
gb|AAG51754.1| reverse transcriptase, putative; 100033-105622 [A... 79 3e-13
gb|AAP54692.1| putative reverse transcriptase [Oryza sativa (jap... 78 9e-13
gb|AAD21778.1| putative non-LTR retroelement reverse transcripta... 77 1e-12
gb|AAD32950.1| putative non-LTR retroelement reverse transcripta... 76 3e-12
gb|AAD17395.1| putative non-LTR retroelement reverse transcripta... 74 1e-11
gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Ara... 73 3e-11
gb|AAF18538.1| Very similar to retrotransposon reverse transcrip... 72 4e-11
gb|AAD03565.2| putative non-LTR retroelement reverse transcripta... 72 5e-11
pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabi... 72 5e-11
pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW1... 69 3e-10
ref|NP_909558.1| putative reverse transcriptase [Oryza sativa] g... 69 5e-10
gb|AAB84340.1| putative non-LTR retroelement reverse transcripta... 67 2e-09
dbj|BAC19859.1| orf258 [Oryza sativa (japonica cultivar-group)] 66 3e-09
gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultiva... 65 4e-09
ref|NP_917200.1| P0707D10.17 [Oryza sativa (japonica cultivar-gr... 65 4e-09
dbj|BAB08714.1| non-LTR retroelement reverse transcriptase-like ... 65 8e-09
gb|AAP54636.1| putative reverse transcriptase [Oryza sativa (jap... 64 1e-08
>gb|AAP53315.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37533452|ref|NP_921028.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|20279456|gb|AAM18736.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1509
Score = 94.4 bits (233), Expect = 9e-18
Identities = 57/187 (30%), Positives = 86/187 (45%), Gaps = 6/187 (3%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+ IW D W+P G K P + +V +L P G W+ L+ TF+ VA I
Sbjct: 1133 INIWADPWIPRGWSRK-PMTPRGANLVTKVEELIDPYTGTWDEDLLSQTFWEEDVAAIKS 1191
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQ----NCPSSSNEVGLEPAQWEFF 254
+ H ++ D L W +TVK+ Y+ ++E + CP SN + W+
Sbjct: 1192 IPV-HVEMEDVLAWHFDARGCFTVKSAYKVQREMERRASRNGCPGVSNWESGDDDFWKKL 1250
Query: 255 WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRG 314
WK +++ WR+C + +R LH RG+DVD +CG +E HLF C +
Sbjct: 1251 WKLGVPGKIKHFLWRMCHNTLALRANLHHRGMDVDTRCVMCGRYNEDAGHLFFKCKPVKK 1310
Query: 315 CWFARNL 321
W A NL
Sbjct: 1311 VWQALNL 1317
>ref|NP_914307.1| reverse transcriptase -like protein [Oryza sativa (japonica
cultivar-group)]
Length = 727
Score = 90.5 bits (223), Expect = 1e-16
Identities = 64/217 (29%), Positives = 100/217 (45%), Gaps = 9/217 (4%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+ IWED W+P G + P L +VADL P G W+ PLVE F+ + IL
Sbjct: 375 IDIWEDPWIPAGVTRR-PITPRRGTVLNKVADLIDPATGWWDKPLVEEIFWESDARNILA 433
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGY-----EWH*QLENQNCPSSSNEVGLEPAQWEF 253
+R D D + W +Y+VK+ Y + + Q SSS V +W+
Sbjct: 434 IPVR-LDTNDFVAWHFDTRGMYSVKSAYHVLEDQRERSAKKQEGGSSSGAVNQRNFEWDK 492
Query: 254 FWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSR 313
W P+V++ WR+ ++P+++ + RR D + P+C DE H F+ C +
Sbjct: 493 IWSLQCIPKVKQFIWRLAHNSLPLKLNIKRRVPDAETLCPVCKRFDENGGHCFLQCKPMK 552
Query: 314 GCWFARNLGLRLLSGSVLRLCSVIDSAEVLNQ*VNGR 350
CW R L L + S+ +L S D + + + N R
Sbjct: 553 LCW--RILCLEDIRLSLTQLVSARDVVQTILKLENDR 587
>emb|CAD41337.2| OJ991113_30.21 [Oryza sativa (japonica cultivar-group)]
gi|50925420|ref|XP_472970.1| OJ991113_30.21 [Oryza
sativa (japonica cultivar-group)]
Length = 541
Score = 80.1 bits (196), Expect = 2e-13
Identities = 52/173 (30%), Positives = 79/173 (45%), Gaps = 3/173 (1%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
VRIW+D W+P S +K + G RL V L + W+ L++ P V IL
Sbjct: 83 VRIWKDQWIPRQSSLKLSPQNG-RCRLRWVHQLINQDTNSWDADLIKNVCSPLDVNEILK 141
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEW--H*QLENQNCPSSSNEVGLEPAQWEFFWK 256
+ H+ + D L W V+TV++ Y H QL+ SSS+ E W W
Sbjct: 142 IKLPHRGMEDFLAWHFEKTGVFTVRSAYHLALHNQLKANELGSSSSSTSGERKIWASLWT 201
Query: 257 APSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSC 309
AP +V+ AWR+ + +R +++D + +CGL E H +SC
Sbjct: 202 APVPQKVKIFAWRLARECLATMENRKKRKLEIDSTCRICGLGGEDGFHAVISC 254
>gb|AAG51754.1| reverse transcriptase, putative; 100033-105622 [Arabidopsis thaliana]
gi|25403084|pir||G86419 probable reverse transcriptase,
100033-105622 [imported] - Arabidopsis thaliana
Length = 1557
Score = 79.3 bits (194), Expect = 3e-13
Identities = 52/182 (28%), Positives = 91/182 (49%), Gaps = 15/182 (8%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+ +W DNW+ + P Q+ +RV DL + WN L++ FF ++ +
Sbjct: 1206 ISVWMDNWIYDNGPRIPLQKHFSVSLNLRVKDLINLDGRCWNRDLLQDLFF-----QVDI 1260
Query: 199 TLIRHQ----DVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQ---W 251
LI + D+ D W+ + Y+VK+GY W + N P + +P+
Sbjct: 1261 DLICKKKPVVDMDDFWVWLHSKSGEYSVKSGY-W--LVFQSNKPELLFKACRQPSTNGLK 1317
Query: 252 EFFWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNV 311
E W +FP+++ WR+ + A+PV ++ RRG++VDP +CG E E + H+ +C+V
Sbjct: 1318 EKIWSTSTFPKIKLFLWRILSAALPVADQILRRGMNVDPCCQICGQEGESINHVLFTCSV 1377
Query: 312 SR 313
+R
Sbjct: 1378 AR 1379
>gb|AAP54692.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37536206|ref|NP_922405.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|27311287|gb|AAO00713.1|
retrotransposon protein, putative, unclassified [Oryza
sativa (japonica cultivar-group)]
Length = 1557
Score = 77.8 bits (190), Expect = 9e-13
Identities = 53/191 (27%), Positives = 84/191 (43%), Gaps = 7/191 (3%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
VR+W+D WLP + P + R+ VADL L N G W+ + F P V IL
Sbjct: 1301 VRVWQDPWLPRDLSRR-PITPKNNCRIKWVADLMLDN-GMWDANKINQIFLPVDVEIILK 1358
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAP 258
+D D + W ++V+T Y + SSS++V + A WE WK
Sbjct: 1359 LRTSSRDEEDFIAWHPDKLGNFSVRTAYRLAENWAKEEASSSSSDVNIRKA-WELLWKCN 1417
Query: 259 SFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFA 318
+V+ WR + +P +R +++ + +CG+E E H C ++ W A
Sbjct: 1418 VPSKVKIFTWRATSNCLPTWDNKKKRNLEISDTCVICGMEKEDTMHALCRCPQAKHLWLA 1477
Query: 319 ----RNLGLRL 325
+L LR+
Sbjct: 1478 MKESNDLSLRM 1488
>gb|AAD21778.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25410938|pir||G84429 hypothetical protein
At2g01840 [imported] - Arabidopsis thaliana
Length = 1715
Score = 77.4 bits (189), Expect = 1e-12
Identities = 62/262 (23%), Positives = 105/262 (39%), Gaps = 24/262 (9%)
Query: 140 RIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILVT 199
+IWED WLP P + DE ++VADL+ N W+ + E P
Sbjct: 1277 KIWEDPWLPTLPPRPARGPILDED--MKVADLWRENKREWDPVIFEGVLNPEDQQLAKSL 1334
Query: 200 LIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAPS 259
+ + D W T + YTV++GY W N N + + + W+
Sbjct: 1335 YLSNYAARDSYKWAYTRNTQYTVRSGY-WVATHVNLTEEEIINPLEGDVPLKQEIWRLKI 1393
Query: 260 FPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFAR 319
P+++ WR + A+ +L R + DP+ C DE + H+ +C+ ++ W +
Sbjct: 1394 TPKIKHFIWRCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSYAQVVWRSA 1453
Query: 320 NLGLRLLSGSVLRLCSVIDSAEVLNQ*VNGRTNKSLGGSCLPCGRLRIN*LFRRKLGLWL 379
N SGS RLC + E + + G+ N++L + + W+
Sbjct: 1454 N-----FSGS-NRLCFTDNLEENIRLILQGKKNQNLP-------------ILNGLMPFWI 1494
Query: 380 --RFWHGQQRLFAPRLCRSPWR 399
R W + +L R PW+
Sbjct: 1495 MWRLWKSRNEYLFQQLDRFPWK 1516
>gb|AAD32950.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411805|pir||C84554 hypothetical protein
At2g17610 [imported] - Arabidopsis thaliana
Length = 773
Score = 75.9 bits (185), Expect = 3e-12
Identities = 60/212 (28%), Positives = 90/212 (42%), Gaps = 10/212 (4%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+++W+DNWLP P ++V+DL + G WN L+ + I
Sbjct: 387 IQLWKDNWLPLNPPRPPVGTCDSIYSQLKVSDLLI--EGRWNEDLLCKLIHQNDIPHIRA 444
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPS--SSNEVGLEPAQWEFFWK 256
D + W+ T D Y+VK+GY +L Q S S NEV + + WK
Sbjct: 445 IRPSITGANDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASLPSPNEVSAQTV-FTNIWK 503
Query: 257 APSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
+ P+++ WR A+P L RR + D + CG E V HL C VS+ W
Sbjct: 504 QNAPPKIKHFWWRSAHNALPTAGNLKRRRLITDDTCQRCGEASEDVNHLLFQCRVSKEIW 563
Query: 317 FARNLGLRLLSGSVLRLCSV---IDSAEVLNQ 345
++L G L S ++S + LNQ
Sbjct: 564 --EQAHIKLCPGDSLMSNSFNQNLESIQKLNQ 593
>gb|AAD17395.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411640|pir||H84529 hypothetical protein
At2g15510 [imported] - Arabidopsis thaliana
Length = 1138
Score = 73.9 bits (180), Expect = 1e-11
Identities = 49/181 (27%), Positives = 81/181 (44%), Gaps = 7/181 (3%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+ +W D W+ + +P Q ++V DL W L+E F+PT V I
Sbjct: 708 LNVWMDPWIFDIAPRLPLQRHFSVNLDLKVNDLINFEDRCWKRDLLEELFYPTDVELIT- 766
Query: 199 TLIRHQDVV---DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFW 255
+ VV D W+ Y+VK+GY Q + +N + E W
Sbjct: 767 ---KRNPVVNMDDFWVWLHAKTGEYSVKSGYWLAFQSNKLDLIQQANLLPSTNGLKEQVW 823
Query: 256 KAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGC 315
S P+++ WR+ + A+PV ++ RRG+ +DP +CG E E H+ +C+++R
Sbjct: 824 STKSSPKIKMFLWRILSAALPVADQIIRRGMSIDPRCQICGDEGESTNHVLFTCSMARQV 883
Query: 316 W 316
W
Sbjct: 884 W 884
>gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
gi|25405244|pir||E96519 probable reverse transcriptase,
16838-20266 [imported] - Arabidopsis thaliana
Length = 1142
Score = 72.8 bits (177), Expect = 3e-11
Identities = 48/180 (26%), Positives = 76/180 (41%), Gaps = 7/180 (3%)
Query: 139 VRIWEDNWLPNG--SPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARI 196
+ +W D W+P P K+ + D ++V L WN L++ F P V I
Sbjct: 716 ISVWNDPWIPAQFPRPAKYGGSIVDPS--LKVKSLIDSRSNFWNIDLLKELFDPEDVPLI 773
Query: 197 LVTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWK 256
I + ++ D L W T YTVK+GY N+ ++ A + WK
Sbjct: 774 SALPIGNPNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKA---YIWK 830
Query: 257 APSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
P++R W++ + VPV L +RG+ D CG +E + H C+ +R W
Sbjct: 831 VQCPPKLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHPARQIW 890
>gb|AAF18538.1| Very similar to retrotransposon reverse transcriptase [Arabidopsis
thaliana] gi|25518314|pir||A86359 hypothetical protein
F12K8.9 - Arabidopsis thaliana
Length = 1231
Score = 72.4 bits (176), Expect = 4e-11
Identities = 58/198 (29%), Positives = 88/198 (44%), Gaps = 21/198 (10%)
Query: 139 VRIWEDNWLPNGSP----IKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVA 194
+R+W D WL + IK P D + V+DL W +E FFP V
Sbjct: 780 IRVWIDYWLDDNGLRAPWIKNPIINVD----LLVSDLIDYEKRDWRLDKLEEQFFPDDVV 835
Query: 195 RILVTLIRHQDVV---DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQW 251
+I ++ VV D W Y+VK GY W NQN + E ++P+
Sbjct: 836 KIR----ENRPVVSLDDFWIWKHNKSGDYSVKLGY-W--LASNQNLGQVAIEAMMQPSLN 888
Query: 252 EF---FWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMS 308
+ WK + P+++ W+V + A+PV L RG+ +D CG E E ++H+ S
Sbjct: 889 DLKTQVWKLQTEPKIKVFLWKVLSGAIPVVDLLSYRGMKLDSRCQTCGCEGESIQHVLFS 948
Query: 309 CNVSRGCWFARNLGLRLL 326
C+ R W N+ + LL
Sbjct: 949 CSFPRQVWAMSNIHVPLL 966
>gb|AAD03565.2| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411819|pir||H84557 hypothetical protein
At2g17910 [imported] - Arabidopsis thaliana
Length = 1344
Score = 72.0 bits (175), Expect = 5e-11
Identities = 48/187 (25%), Positives = 84/187 (44%), Gaps = 13/187 (6%)
Query: 141 IWEDNWLPNGS---PIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARIL 197
+W D WL +GS P+ + + + ++V+ L P WN ++ FP I+
Sbjct: 912 VWTDKWLHDGSNRRPLNRRRFINVD---LKVSQLIDPTSRNWNLNMLR-DLFPWKDVEII 967
Query: 198 VTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFF--- 254
+ D W+ + + +Y+VKTGYE+ L Q E ++P+ F
Sbjct: 968 LKQRPLFFKEDSFCWLHSHNGLYSVKTGYEF---LSKQVHHRLYQEAKVKPSVNSLFDKI 1024
Query: 255 WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRG 314
W + P++R W+ A+PV +L RG+ D +C E+E + H+ C ++R
Sbjct: 1025 WNLHTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQ 1084
Query: 315 CWFARNL 321
W +L
Sbjct: 1085 VWAITHL 1091
>pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabidopsis thaliana
(fragment)
Length = 1365
Score = 72.0 bits (175), Expect = 5e-11
Identities = 48/187 (25%), Positives = 84/187 (44%), Gaps = 13/187 (6%)
Query: 141 IWEDNWLPNGS---PIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARIL 197
+W D WL +GS P+ + + + ++V+ L P WN ++ FP I+
Sbjct: 933 VWTDKWLHDGSNRRPLNRRRFINVD---LKVSQLIDPTSRNWNLNMLR-DLFPWKDVEII 988
Query: 198 VTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFF--- 254
+ D W+ + + +Y+VKTGYE+ L Q E ++P+ F
Sbjct: 989 LKQRPLFFKEDSFCWLHSHNGLYSVKTGYEF---LSKQVHHRLYQEAKVKPSVNSLFDKI 1045
Query: 255 WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRG 314
W + P++R W+ A+PV +L RG+ D +C E+E + H+ C ++R
Sbjct: 1046 WNLHTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQ 1105
Query: 315 CWFARNL 321
W +L
Sbjct: 1106 VWAITHL 1112
>pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW15) - Arabidopsis
thaliana retrotransposon Ta11-1 gi|976278|gb|AAA75254.1|
reverse transcriptase
Length = 1333
Score = 69.3 bits (168), Expect = 3e-10
Identities = 48/182 (26%), Positives = 83/182 (45%), Gaps = 13/182 (7%)
Query: 141 IWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILVTL 200
+W D W+ + P Q+ +RV DL +E F+P + I
Sbjct: 936 VWMDPWIYDNDPRLPLQKHFSVNLDLRVHDLINVEDRCRRRDRLEELFYPADIEII---- 991
Query: 201 IRHQDVV---DCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQ---WEFF 254
++ VV D W+ + Y+VK+GY Q N P E ++P+ E
Sbjct: 992 VKRNPVVSMDDFWVWLHSKSGEYSVKSGYWLAFQT---NKPELIREARVQPSTNGLKEKI 1048
Query: 255 WKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRG 314
W + P+++ WR+ + A+PV ++ RRG+ +DP +CG E E + H+ +C+++R
Sbjct: 1049 WSTLTSPKIKLFLWRILSSALPVAYQIIRRGMPIDPRCQVCGEEGESINHVLFTCSLARQ 1108
Query: 315 CW 316
W
Sbjct: 1109 VW 1110
>ref|NP_909558.1| putative reverse transcriptase [Oryza sativa]
gi|14018103|gb|AAK52166.1| putative reverse
transcriptase [Oryza sativa]
Length = 1185
Score = 68.6 bits (166), Expect = 5e-10
Identities = 47/187 (25%), Positives = 86/187 (45%), Gaps = 7/187 (3%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
V IWED+ +P+ S K G + +V +L P GGW+ L++ F+P RIL
Sbjct: 749 VNIWEDSLIPSSSNGKVLTPRG-RNIITKVEELINPVSGGWDEDLIDTLFWPIDANRILQ 807
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGY--EWH*QLENQNCPSSSNEVGLEPAQWEFFWK 256
I D + W T +++V++ Y +W+ + ++ P W+ WK
Sbjct: 808 IPIS-AGREDFVAWHHTRSGIFSVRSAYHCQWNAKFGSKERLPDGASSSSRPV-WDNLWK 865
Query: 257 APSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
P+++ AWR+ +P + L + + V P+C E ++H+ +C ++ W
Sbjct: 866 LEIPPKIKIFAWRMLHGVIPCKGILADKHIAVQGGCPVCSSGVEDIKHMVFTCRRAKLIW 925
Query: 317 FARNLGL 323
+ LG+
Sbjct: 926 --KQLGI 930
>gb|AAB84340.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|7488322|pir||T00814 RNA-directed DNA
polymerase homolog At2g41580 - Arabidopsis thaliana
Length = 1094
Score = 66.6 bits (161), Expect = 2e-09
Identities = 47/179 (26%), Positives = 80/179 (44%), Gaps = 7/179 (3%)
Query: 141 IWEDNWLPNGSPIKFP--QEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+W DNW+ + P + Q + D Q ++V+ L P WN ++ FP +I+
Sbjct: 663 VWMDNWIFDDKPRRPESLQIMVDIQ--LKVSQLIDPFSRNWNLNMLR-DLFPWKEIQIIC 719
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEW-H*QLENQNCPSSSNEVGLEPAQWEFFWKA 257
D W GT +YTVK+ Y+ Q+ Q + + L P + W
Sbjct: 720 QQRPMASRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKI-WNL 778
Query: 258 PSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
S P+++ W+V AV V +L RGV ++ +C ++E + H+ C ++R W
Sbjct: 779 NSAPKIKVFLWKVLKGAVAVEDRLRTRGVLIEDGCSMCPEKNETLNHILFQCPLARQVW 837
>dbj|BAC19859.1| orf258 [Oryza sativa (japonica cultivar-group)]
Length = 258
Score = 65.9 bits (159), Expect = 3e-09
Identities = 43/151 (28%), Positives = 67/151 (43%), Gaps = 1/151 (0%)
Query: 168 VADLFLPNHGGWNTPLVE*TFFPTMVARILVTLIRHQDVVDCLFWMGTVDDVYTVKTGYE 227
VADL L N W+ V+ FFP A IL + D W ++TV++ Y
Sbjct: 15 VADLMLQNTNQWDVEKVQRIFFPLDAAAILNMPRPRTEQDDFWAWAWDRTGIFTVRSAYR 74
Query: 228 WH*QLENQNCPSSSNEVGLEPAQWEFFWKAPSFPRVREVAWRVCTWAVPVRVKLHRRGVD 287
Q + P++ + G E A W+ W+ P++R WRV +P +L R +
Sbjct: 75 ELIQRCGLSEPATGSSTGDE-ATWKALWRLRIMPKIRVFWWRVVKNLLPCAAELRRSHMK 133
Query: 288 VDPSYPLCGLEDEIVEHLFMSCNVSRGCWFA 318
+ PLCG ++E H + C ++ W A
Sbjct: 134 EISNCPLCGNDNETKFHALVECEHAKLFWSA 164
>gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultivar-group)]
Length = 2367
Score = 65.5 bits (158), Expect = 4e-09
Identities = 47/179 (26%), Positives = 79/179 (43%), Gaps = 5/179 (2%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+RIW D+WLP + P RL V+DL + G W+ P + +F + A +++
Sbjct: 1666 IRIWRDSWLPRDHSRR-PITGKANCRLKWVSDL-ITEDGSWDVPKIH-QYFHNLDAEVIL 1722
Query: 199 TL-IRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKA 257
+ I + D + W + +++V++ Y QL N SSS + A WE WK
Sbjct: 1723 NICISSRSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKA-WEMIWKC 1781
Query: 258 PSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
+V+ AWRV + + V +R ++ +C E+E H C + W
Sbjct: 1782 KVPQKVKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHALCRCIQASQLW 1840
>ref|NP_917200.1| P0707D10.17 [Oryza sativa (japonica cultivar-group)]
Length = 892
Score = 65.5 bits (158), Expect = 4e-09
Identities = 56/210 (26%), Positives = 86/210 (40%), Gaps = 10/210 (4%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
VRIW D WLP + G+ RL V++L L +G W+ V F P I
Sbjct: 443 VRIWRDPWLPRDFSRRPITRKGN-CRLKWVSEL-LSENGAWDETRVNQVFLPVDAEAICS 500
Query: 199 TLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKAP 258
+ + D + W ++V++ Y L N SSS+ + W W+
Sbjct: 501 IRVSSRQEDDFVAWHPDKHGKFSVRSAYGLACNLVNMETSSSSSSARCK-RMWNLIWQTN 559
Query: 259 SFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCWFA 318
+ +VR AWR T + +R ++ +CGLE E +H+ C +R W A
Sbjct: 560 APQKVRIFAWRAATNCLATMENKAKRKLEQSDICLVCGLEKEDTKHVLCRCPDARNLWGA 619
Query: 319 R-NLGLRLLSGSVLRLCS----VIDSAEVL 343
G L+G LC+ + D E+L
Sbjct: 620 MCEAGNISLNGG--NLCNDPGWIFDQLEIL 647
>dbj|BAB08714.1| non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1197
Score = 64.7 bits (156), Expect = 8e-09
Identities = 54/181 (29%), Positives = 78/181 (42%), Gaps = 11/181 (6%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVR---VADLFLPNHGGWNTPLVE*TFFPTMVAR 195
V +W+D WL + I Q VG L + V DLFLPN W+ +E P +
Sbjct: 762 VLVWQDAWLAHDKRI---QPVGPAPLLSKDWKVKDLFLPNSVDWDEGKIE-AAIPFVKDL 817
Query: 196 ILVTLIRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFW 255
+L D W+G D +YT KTGY+ L + + SS + V L ++ W
Sbjct: 818 VLAIRPSRAGGADKRIWLGAKDGLYTTKTGYKA--TLCDVDSDSSRDPV-LSFNWFDEVW 874
Query: 256 KAPSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGC 315
P++R W+ + A+PV +L R DP CG E E H +C +
Sbjct: 875 NVQCSPKIRLFLWKAVSGALPVGSQLAIRIPSFDPVCCRCG-ELETSSHALFNCMFVQRV 933
Query: 316 W 316
W
Sbjct: 934 W 934
>gb|AAP54636.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37536094|ref|NP_922349.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|13786450|gb|AAK39575.1| putative
reverse transcriptase [Oryza sativa]
Length = 791
Score = 64.3 bits (155), Expect = 1e-08
Identities = 47/179 (26%), Positives = 78/179 (43%), Gaps = 5/179 (2%)
Query: 139 VRIWEDNWLPNGSPIKFPQEVGDEQRLVRVADLFLPNHGGWNTPLVE*TFFPTMVARILV 198
+RIW D WLP + P RL V+DL + G W+ P + +F + A +++
Sbjct: 342 IRIWRDPWLPRDHSRR-PITGKANCRLKWVSDL-ITEDGSWDVPKIH-QYFHNLDAEVIL 398
Query: 199 TL-IRHQDVVDCLFWMGTVDDVYTVKTGYEWH*QLENQNCPSSSNEVGLEPAQWEFFWKA 257
+ I + D + W + +++V++ Y QL N SSS + A WE WK
Sbjct: 399 NICISSRSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKA-WEMIWKC 457
Query: 258 PSFPRVREVAWRVCTWAVPVRVKLHRRGVDVDPSYPLCGLEDEIVEHLFMSCNVSRGCW 316
+V+ AWRV + + V +R ++ +C E+E H C + W
Sbjct: 458 KVPQKVKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHALCRCIQASQLW 516
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.345 0.155 0.589
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,004,405,387
Number of Sequences: 2540612
Number of extensions: 41550173
Number of successful extensions: 149183
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 149002
Number of HSP's gapped (non-prelim): 129
length of query: 568
length of database: 863,360,394
effective HSP length: 133
effective length of query: 435
effective length of database: 525,458,998
effective search space: 228574664130
effective search space used: 228574664130
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0014.4


