

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
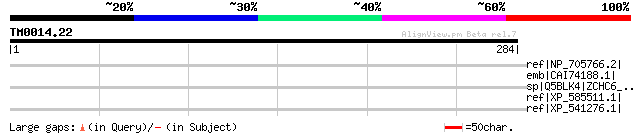
Query= TM0014.22
(284 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_705766.2| zinc finger, CCHC domain containing 6 [Mus musc... 35 2.4
emb|CAI74188.1| hypothetical protein [Theileria annulata] 35 2.4
sp|Q5BLK4|ZCHC6_MOUSE Zinc finger CCHC domain containing protein 6 35 2.4
ref|XP_585511.1| PREDICTED: similar to Protein BAP28, partial [B... 33 7.1
ref|XP_541276.1| PREDICTED: similar to Proprotein convertase sub... 33 9.2
>ref|NP_705766.2| zinc finger, CCHC domain containing 6 [Mus musculus]
gi|60688060|gb|AAH43111.1| Zinc finger, CCHC domain
containing 6 [Mus musculus]
Length = 1474
Score = 35.0 bits (79), Expect = 2.4
Identities = 26/83 (31%), Positives = 43/83 (51%), Gaps = 6/83 (7%)
Query: 175 GKSDYKVVDLHPKLRIWHKILLHCINQRPK-GSSPDYINFCQKAMLFFIQDKRKICLPFF 233
GK + ++V L R W K+ C RP+ G P Y+ F A+ FF+Q +++ LP +
Sbjct: 427 GKLEPRLVPLVIAFRYWAKL---CSIDRPEEGGLPPYV-FALMAV-FFLQQRKEPLLPVY 481
Query: 234 LFSYLKECIRKSITTASIKSAIK 256
L S+++E + S+K K
Sbjct: 482 LGSWIEEFSLNKLGNFSLKDVEK 504
>emb|CAI74188.1| hypothetical protein [Theileria annulata]
Length = 589
Score = 35.0 bits (79), Expect = 2.4
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 1/67 (1%)
Query: 206 SSPDYINFCQKAMLFFIQDKRKICLPFFLF-SYLKECIRKSITTASIKSAIKYIPFGRLL 264
S P Y C K F++Q+K I F S+ K+ I+ SI +SIKS++K ++
Sbjct: 281 SFPGYKRRCSKLFRFWVQEKDLIKFKSIRFLSHNKDVIKSSIVKSSIKSSVKSTAIKSIV 340
Query: 265 SDLFIES 271
IE+
Sbjct: 341 KSPVIEN 347
>sp|Q5BLK4|ZCHC6_MOUSE Zinc finger CCHC domain containing protein 6
Length = 1491
Score = 35.0 bits (79), Expect = 2.4
Identities = 26/83 (31%), Positives = 43/83 (51%), Gaps = 6/83 (7%)
Query: 175 GKSDYKVVDLHPKLRIWHKILLHCINQRPK-GSSPDYINFCQKAMLFFIQDKRKICLPFF 233
GK + ++V L R W K+ C RP+ G P Y+ F A+ FF+Q +++ LP +
Sbjct: 427 GKLEPRLVPLVIAFRYWAKL---CSIDRPEEGGLPPYV-FALMAV-FFLQQRKEPLLPVY 481
Query: 234 LFSYLKECIRKSITTASIKSAIK 256
L S+++E + S+K K
Sbjct: 482 LGSWIEEFSLNKLGNFSLKDVEK 504
>ref|XP_585511.1| PREDICTED: similar to Protein BAP28, partial [Bos taurus]
Length = 1524
Score = 33.5 bits (75), Expect = 7.1
Identities = 26/105 (24%), Positives = 47/105 (44%), Gaps = 7/105 (6%)
Query: 125 GRRIIIIEKSIVNLLGAHTGTGYRFQLTESKVKPRTKDETNKALY------TTYKPGKSD 178
G I + ++ ++ L +G +F L V P+ ++ T+ A Y T ++ +S+
Sbjct: 928 GSPIKEVRRAAIHCLQVLSGVASQFHLVIDHVVPKAEEITSDATYVVQDLATLFEELQSE 987
Query: 179 YKVVDLHPKLRIWHKILLHCINQRPKGSSPDYINFCQKAMLFFIQ 223
K+ H KL K LLHC+ P + D + +L I+
Sbjct: 988 KKLKS-HQKLSETLKNLLHCVYSCPSYVTKDLMKLVLSQLLSMIE 1031
>ref|XP_541276.1| PREDICTED: similar to Proprotein convertase subtilisin/kexin type 5
precursor (Proprotein convertase PC5)
(Subtilisin/kexin-like protease PC5) (PC6)
(Subtilisin-like proprotein convertase 6) (SPC6) [Canis
familiaris]
Length = 2416
Score = 33.1 bits (74), Expect = 9.2
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 153 ESKVKPRTKDETNKALYTTYKPGKS-DYKVVDLHPKLRI-WHKILLHCIN 200
ES KP +E + L TTY G+S D K+ + WH ++LHC N
Sbjct: 543 ESGKKPWYLEECSSTLATTYSSGESYDKKIYSASQSTKTEWHLLVLHCKN 592
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.139 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 487,220,815
Number of Sequences: 2540612
Number of extensions: 20465968
Number of successful extensions: 41011
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 0
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 41008
Number of HSP's gapped (non-prelim): 7
length of query: 284
length of database: 863,360,394
effective HSP length: 126
effective length of query: 158
effective length of database: 543,243,282
effective search space: 85832438556
effective search space used: 85832438556
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0014.22


