

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.7
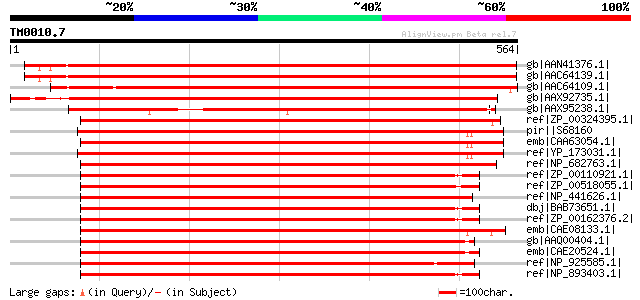
(564 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN41376.1| putative signal recognition particle 54CP protein... 846 0.0
gb|AAC64139.1| signal recognition particle 54 kDa subunit precur... 843 0.0
gb|AAC64109.1| signal recognition particle 54 kDa subunit precur... 835 0.0
gb|AAX92735.1| signal recognition particle protein, putative [Or... 733 0.0
gb|AAX95238.1| signal recognition particle protein, putative [Or... 644 0.0
ref|ZP_00324395.1| COG0541: Signal recognition particle GTPase [... 499 e-140
pir||S68160 probable RNA binding protein ffh - Synechococcus sp.... 493 e-138
emb|CAA63054.1| signal sequence binding protein [Synechococcus s... 491 e-137
ref|YP_173031.1| signal recognition particle receptor [Synechoco... 490 e-137
ref|NP_682763.1| signal recognition particle protein [Thermosyne... 489 e-136
ref|ZP_00110921.1| COG0541: Signal recognition particle GTPase [... 486 e-135
ref|ZP_00518055.1| Signal recognition particle protein [Crocosph... 484 e-135
ref|NP_441626.1| signal recognition particle protein [Synechocys... 484 e-135
dbj|BAB73651.1| signal recognition particle protein [Nostoc sp. ... 483 e-135
ref|ZP_00162376.2| COG0541: Signal recognition particle GTPase [... 483 e-135
emb|CAE08133.1| signal recognition particle protein (SRP54) [Syn... 465 e-129
gb|AAQ00404.1| Signal recognition particle GTPase [Prochlorococc... 456 e-127
emb|CAE20524.1| signal recognition particle protein (SRP54) [Pro... 454 e-126
ref|NP_925585.1| signal recognition particle protein SRP54 [Gloe... 442 e-122
ref|NP_893403.1| signal recognition particle protein (SRP54) [Pr... 435 e-120
>gb|AAN41376.1| putative signal recognition particle 54CP protein precursor
[Arabidopsis thaliana] gi|396701|emb|CAA79981.1| 54CP
[Arabidopsis thaliana] gi|7406404|emb|CAB85514.1| signal
recognition particle 54CP protein precursor [Arabidopsis
thaliana] gi|15293131|gb|AAK93676.1| putative signal
recognition particle 54CP protein precursor [Arabidopsis
thaliana] gi|15237580|ref|NP_196014.1| signal
recognition particle 54 kDa protein, chloroplast / 54
chloroplast protein / SRP54 (FFC) [Arabidopsis thaliana]
gi|480296|pir||S36637 signal recognition particle 54CP
protein precursor - Arabidopsis thaliana
gi|586038|sp|P37107|SR5C_ARATH Signal recognition
particle 54 kDa protein, chloroplast precursor (SRP54)
(54 chloroplast protein) (54CP) (FFC)
Length = 564
Score = 846 bits (2186), Expect = 0.0
Identities = 445/557 (79%), Positives = 496/557 (88%), Gaps = 11/557 (1%)
Query: 17 SSHNRTVCSSSPSPN---SLKLSSPWTNASI------SSRNSFTKEIWGWVQAKSVAVRR 67
SS NR C+ S + N SS +T +I SSRN T+EIW WV++K+V V
Sbjct: 7 SSVNRVPCTLSCTGNRRIKAAFSSAFTGGTINSASLSSSRNLSTREIWSWVKSKTV-VGH 65
Query: 68 RRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVR 127
R VRAEMFGQLT GLEAAW+KLKGEEVLTK+NI EPMRDIRRALLEADVSLPVVR
Sbjct: 66 GRYRRSQVRAEMFGQLTGGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVR 125
Query: 128 RFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQ 187
RFVQSVSDQAVG+GVIRGVKPDQQLVKIVH+ELVKLMGGEVSEL FAKSG TVILLAGLQ
Sbjct: 126 RFVQSVSDQAVGMGVIRGVKPDQQLVKIVHDELVKLMGGEVSELQFAKSGPTVILLAGLQ 185
Query: 188 GVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSE 247
GVGKTTVCAKL+ YLKKQGKSCML+AGD+YRPAAIDQL ILG+QVGVPVYTAGTDVKP++
Sbjct: 186 GVGKTTVCAKLACYLKKQGKSCMLIAGDVYRPAAIDQLVILGEQVGVPVYTAGTDVKPAD 245
Query: 248 IARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAA 307
IA+QGL+EAKK +DVVI+DTAGRLQIDK MMDELK+VKK LNPTEVLLVVDAMTGQEAA
Sbjct: 246 IAKQGLKEAKKNNVDVVIMDTAGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAA 305
Query: 308 ALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRM 367
ALVTTFN+EIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERM+DLEPFYPDRM
Sbjct: 306 ALVTTFNVEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRM 365
Query: 368 AGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVM 427
AGRILGMGDVLSFVEKA EVM+QEDAE LQ+KIMSAKFDFNDFLKQTRAVA+MGS++RV+
Sbjct: 366 AGRILGMGDVLSFVEKATEVMRQEDAEDLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVL 425
Query: 428 GMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTE 487
GMIPGM KV+PAQIREAEKNL MEAMIE MTPEERE+PELLAESP RRKR+A++SGKTE
Sbjct: 426 GMIPGMGKVSPAQIREAEKNLLVMEAMIEVMTPEERERPELLAESPERRKRIAKDSGKTE 485
Query: 488 QQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQRS 547
QQVS +VAQ+FQMRV+MKNLMGVM+GGS+P LS LE+ALK E+KAPPGTARR+++++ R
Sbjct: 486 QQVSALVAQIFQMRVKMKNLMGVMEGGSIPALSGLEDALKAEQKAPPGTARRKRKADSRK 545
Query: 548 LFADSA-ARPTPRGFGS 563
F +SA ++P PRGFGS
Sbjct: 546 KFVESASSKPGPRGFGS 562
>gb|AAC64139.1| signal recognition particle 54 kDa subunit precursor [Arabidopsis
thaliana]
Length = 564
Score = 843 bits (2179), Expect = 0.0
Identities = 444/557 (79%), Positives = 495/557 (88%), Gaps = 11/557 (1%)
Query: 17 SSHNRTVCSSSPSPN---SLKLSSPWTNASI------SSRNSFTKEIWGWVQAKSVAVRR 67
SS NR C+ S + N SS +T +I SSRN T+EIW WV++K+V V
Sbjct: 7 SSVNRVPCTLSCTGNRRIKAAFSSAFTGGTINSASLSSSRNLSTREIWSWVKSKTV-VGH 65
Query: 68 RRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVR 127
R VRA MFGQLT GLEAAW+KLKGEEVLTK+NI EPMRDIRRALLEADVSLPVVR
Sbjct: 66 GRYRRSQVRAVMFGQLTGGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVR 125
Query: 128 RFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQ 187
RFVQSVSDQAVG+GVIRGVKPDQQLVKIVH+ELVKLMGGEVSEL FAKSG TVILLAGLQ
Sbjct: 126 RFVQSVSDQAVGMGVIRGVKPDQQLVKIVHDELVKLMGGEVSELQFAKSGPTVILLAGLQ 185
Query: 188 GVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSE 247
GVGKTTVCAKL+ YLKKQGKSCML+AGD+YRPAAIDQL ILG+QVGVPVYTAGTDVKP++
Sbjct: 186 GVGKTTVCAKLACYLKKQGKSCMLIAGDVYRPAAIDQLVILGEQVGVPVYTAGTDVKPAD 245
Query: 248 IARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAA 307
IA+QGL+EAKK +DVVI+DTAGRLQIDK MMDELK+VKK LNPTEVLLVVDAMTGQEAA
Sbjct: 246 IAKQGLKEAKKNNVDVVIMDTAGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAA 305
Query: 308 ALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRM 367
ALVTTFN+EIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERM+DLEPFYPDRM
Sbjct: 306 ALVTTFNVEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRM 365
Query: 368 AGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVM 427
AGRILGMGDVLSFVEKA EVM+QEDAE LQ+KIMSAKFDFNDFLKQTRAVA+MGS++RV+
Sbjct: 366 AGRILGMGDVLSFVEKATEVMRQEDAEDLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVL 425
Query: 428 GMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTE 487
GMIPGM KV+PAQIREAEKNL MEAMIE MTPEERE+PELLAESP RRKR+A++SGKTE
Sbjct: 426 GMIPGMGKVSPAQIREAEKNLLVMEAMIEVMTPEERERPELLAESPERRKRIAKDSGKTE 485
Query: 488 QQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQRS 547
QQVS +VAQ+FQMRV+MKNLMGVM+GGS+P LS LE+ALK E+KAPPGTARR+++++ R
Sbjct: 486 QQVSALVAQIFQMRVKMKNLMGVMEGGSIPALSGLEDALKAEQKAPPGTARRKRKADSRK 545
Query: 548 LFADSA-ARPTPRGFGS 563
F +SA ++P PRGFGS
Sbjct: 546 KFVESASSKPGPRGFGS 562
>gb|AAC64109.1| signal recognition particle 54 kDa subunit precursor [Pisum
sativum]
Length = 525
Score = 835 bits (2158), Expect = 0.0
Identities = 443/524 (84%), Positives = 475/524 (90%), Gaps = 8/524 (1%)
Query: 46 SRNSFTKEIWGWVQAKSVAVRRRRDM--PGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKE 103
+RNSF +EIWG VQ+KS+ RR DM P VVRAEMFGQLT+GLE+AWNKLK EEVLTKE
Sbjct: 4 TRNSFAREIWGLVQSKSLT-SRRTDMIRPVVVRAEMFGQLTSGLESAWNKLKREEVLTKE 62
Query: 104 NIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKL 163
NIVEPMRDIRRA ADVSLPVVRRFVQSV+D+AVGVGV RGVKPDQQLVKIVH+ELV+L
Sbjct: 63 NIVEPMRDIRRA--RADVSLPVVRRFVQSVTDKAVGVGVTRGVKPDQQLVKIVHDELVQL 120
Query: 164 MGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAID 223
MGGEVSEL FA SG TVILLAGLQGVGKTTVCAKL+ Y KKQGKSCMLVAGD+YRPAAID
Sbjct: 121 MGGEVSELTFANSGPTVILLAGLQGVGKTTVCAKLATYFKKQGKSCMLVAGDVYRPAAID 180
Query: 224 QLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELK 283
LTILGKQV VPVY AGTDVKPS IA+QG EEAKKKKIDVVIVDTAG LQIDKAMMDELK
Sbjct: 181 XLTILGKQVDVPVYAAGTDVKPSVIAKQGFEEAKKKKIDVVIVDTAGSLQIDKAMMDELK 240
Query: 284 EVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVS 343
EVK+ LNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVS
Sbjct: 241 EVKQVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVS 300
Query: 344 GKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSA 403
GKPIKLVGRGERM+DLEPFYPDRMAGRILGMGDVLSFVEKAQEVM +EDAE +Q+KIMSA
Sbjct: 301 GKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMLEEDAEDMQKKIMSA 360
Query: 404 KFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEER 463
KFDFND LKQTR VA+MGSVSRV+GMIPGM+KVTPAQIREAEKNL+ ME +IEAMTPEER
Sbjct: 361 KFDFNDLLKQTRTVAKMGSVSRVLGMIPGMAKVTPAQIREAEKNLQFMEVIIEAMTPEER 420
Query: 464 EKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLE 523
KPELLAE PVRRK VAQESGKTEQQVSQ+VAQLFQMRVRMK LMGVM+GGSMPTLSNLE
Sbjct: 421 GKPELLAEFPVRRKSVAQESGKTEQQVSQLVAQLFQMRVRMKKLMGVMEGGSMPTLSNLE 480
Query: 524 EALKPEEKAPPGTARRRKRSEQRSLFADSAAR---PTPRGFGSK 564
EALK +KAPPGTARR+K+ + LF +++ P PRGFGSK
Sbjct: 481 EALKANKKAPPGTARRKKKGGLKRLFKGRSSKISLPAPRGFGSK 524
>gb|AAX92735.1| signal recognition particle protein, putative [Oryza sativa
(japonica cultivar-group)]
Length = 557
Score = 733 bits (1892), Expect = 0.0
Identities = 382/541 (70%), Positives = 451/541 (82%), Gaps = 26/541 (4%)
Query: 2 EAVLASRHFSTPTHSSSHNRTVCSSSPSPNSLKLSSPWTNASISSRNSFTKEIWGWVQAK 61
+A++AS H + + H R V P P+ L SP GW
Sbjct: 25 KAIVASHHLRHSSVPALHLRAV----PGPSFRALPSPGFP--------------GW---- 62
Query: 62 SVAVRRRRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADV 121
RR+R VVRAEMFGQLTTGLE+AWNKL+G + LTK+NI EPMRDIRRALLEADV
Sbjct: 63 ----RRKRGSGLVVRAEMFGQLTTGLESAWNKLRGTDQLTKDNIAEPMRDIRRALLEADV 118
Query: 122 SLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVI 181
SLPVVR F++SV+++AVG VIRGVKP+QQLVK+V++ELV+LMGGEVS+L FAK+ TVI
Sbjct: 119 SLPVVRSFIESVTEKAVGTDVIRGVKPEQQLVKVVNDELVQLMGGEVSDLVFAKTAPTVI 178
Query: 182 LLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGT 241
LLAGLQGVGKTTVCAKL+ YLKK GKSCML+A D+YRPAAIDQLTILGKQVGVPVY+ GT
Sbjct: 179 LLAGLQGVGKTTVCAKLAYYLKKMGKSCMLIAADVYRPAAIDQLTILGKQVGVPVYSEGT 238
Query: 242 DVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAM 301
+ KPS+IA+ G++EAK KK DV+IVDTAGRLQ+DKAMM ELKEVK+++NPTEVLLVVDAM
Sbjct: 239 EAKPSQIAKNGIKEAKSKKTDVIIVDTAGRLQVDKAMMSELKEVKRAVNPTEVLLVVDAM 298
Query: 302 TGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEP 361
TGQEAA+LV+TFN+EIGITGAILTKLDGDSRGGAALS+KEVSGKPIK VGRGERM+DLEP
Sbjct: 299 TGQEAASLVSTFNVEIGITGAILTKLDGDSRGGAALSIKEVSGKPIKFVGRGERMEDLEP 358
Query: 362 FYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMG 421
FYPDRMA RILGMGDVLSFVEKAQEVM+QEDAE+LQ+KI+SAKF+FNDFLKQT+A+AQMG
Sbjct: 359 FYPDRMAQRILGMGDVLSFVEKAQEVMRQEDAEELQKKILSAKFNFNDFLKQTQAIAQMG 418
Query: 422 SVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQ 481
S SR++GMIPGM+KVTPAQIREAEKNLK ME+MI MTPEERE+PELLAES RR RVA+
Sbjct: 419 SFSRIIGMIPGMNKVTPAQIREAEKNLKFMESMINVMTPEERERPELLAESRERRIRVAK 478
Query: 482 ESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRK 541
ESGK E+QVSQ+VAQLF+MR +M+ +MG MQG P + L +++K EE+A GT +RR+
Sbjct: 479 ESGKNERQVSQLVAQLFRMRAQMQKMMGAMQGQDTPDMEGLMDSIKAEEQAAAGTGKRRR 538
Query: 542 R 542
+
Sbjct: 539 K 539
>gb|AAX95238.1| signal recognition particle protein, putative [Oryza sativa
(japonica cultivar-group)]
Length = 1072
Score = 644 bits (1660), Expect = 0.0
Identities = 350/493 (70%), Positives = 403/493 (80%), Gaps = 48/493 (9%)
Query: 66 RRRRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPV 125
RRRR VVRAEMFGQLTTGLE+AWNKL+G +VLTKENIVEPMRDIRRALLEADVSLPV
Sbjct: 58 RRRRGSGLVVRAEMFGQLTTGLESAWNKLRGVDVLTKENIVEPMRDIRRALLEADVSLPV 117
Query: 126 VRRFVQSVSDQAVGVGVIRGVKPDQQLVK-----------------IVHEELVKLMGGEV 168
VRRFV S+S++A+G +IRGV+P+QQLVK IVH+ELVKLMGGEV
Sbjct: 118 VRRFVSSISEKALGSDLIRGVRPEQQLVKTNLCAFSTGQITYSVVQIVHDELVKLMGGEV 177
Query: 169 SELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTIL 228
S+L FAKSG TVILLAGLQ D+YRPAAIDQLTIL
Sbjct: 178 SDLVFAKSGPTVILLAGLQ---------------------------DVYRPAAIDQLTIL 210
Query: 229 GKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKS 288
G+QVGVPVY+ GT+ KP++I + +EEAK+K ID +++DTAGRLQIDK+MM ELKEVKK+
Sbjct: 211 GEQVGVPVYSEGTEAKPAQITKNAVEEAKRKNIDAIVMDTAGRLQIDKSMMVELKEVKKA 270
Query: 289 LNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIK 348
+NPTEVLLVVDAMTGQEAAALVTTFN+EIGITGAILTKLDGDSRGGAALSVKEVSGKPIK
Sbjct: 271 VNPTEVLLVVDAMTGQEAAALVTTFNIEIGITGAILTKLDGDSRGGAALSVKEVSGKPIK 330
Query: 349 LVGRGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFN 408
VGRGERM+DLE FYPDRMA R+LGMGDVLSFVEKAQEVM+QEDA +LQ+KIMSAKFDFN
Sbjct: 331 FVGRGERMEDLELFYPDRMAQRVLGMGDVLSFVEKAQEVMRQEDAVELQKKIMSAKFDFN 390
Query: 409 DFLKQTRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPEL 468
DFLKQ++ VA+MGS+SRV+GMIPGM+KVTPAQIREAEK L +E+MI AMT EEREKPEL
Sbjct: 391 DFLKQSQNVAKMGSMSRVVGMIPGMNKVTPAQIREAEKRLAFVESMINAMTAEEREKPEL 450
Query: 469 LAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQG-GSMPTLSNLEEALK 527
LAES RR RVA+ESGK+EQ+VSQ+VAQLFQMR +M+ LMGVM G ++P + NL E+L
Sbjct: 451 LAESRDRRIRVAEESGKSEQEVSQLVAQLFQMRAQMQKLMGVMTGQEALPGMGNLMESLN 510
Query: 528 PEEKAPPGTARRR 540
+EK G R+R
Sbjct: 511 ADEK---GWRRKR 520
Score = 589 bits (1518), Expect = e-167
Identities = 318/482 (65%), Positives = 367/482 (75%), Gaps = 75/482 (15%)
Query: 66 RRRRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPV 125
RR+R VVRAEMFGQLTTGLE+AWNKL+G + LTK+NI EPMRDIRRALLEADVSLPV
Sbjct: 517 RRKRGSGLVVRAEMFGQLTTGLESAWNKLRGTDQLTKDNIAEPMRDIRRALLEADVSLPV 576
Query: 126 VRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAG 185
VR F++SV+++AVG VIRGVKP+QQLVK
Sbjct: 577 VRSFIESVTEKAVGTDVIRGVKPEQQLVK------------------------------- 605
Query: 186 LQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKP 245
L+ QGKSCML+A D+YRPAAIDQLTILGKQVGVPVY+ GT+ KP
Sbjct: 606 --------------TSLRLQGKSCMLIAADVYRPAAIDQLTILGKQVGVPVYSEGTEAKP 651
Query: 246 SEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQE 305
S+IA+ G++EAK KK DV+IVDTAGRLQ+DKAMM ELKEVK+++NPTEVLLVVDAMTGQE
Sbjct: 652 SQIAKNGIKEAKSKKTDVIIVDTAGRLQVDKAMMSELKEVKRAVNPTEVLLVVDAMTGQE 711
Query: 306 AA--------------ALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVG 351
AA ALV+TFN+EIGITGAILTKLDGDSRGGAALS+KEVSGKPIK VG
Sbjct: 712 AAFALRLTRISFLIWPALVSTFNVEIGITGAILTKLDGDSRGGAALSIKEVSGKPIKFVG 771
Query: 352 RGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFL 411
RGERM+DLEPFYPDRMA RILGMGDVLSFVEKAQEVM+QEDAE+LQ+KI+SAKF+FNDFL
Sbjct: 772 RGERMEDLEPFYPDRMAQRILGMGDVLSFVEKAQEVMRQEDAEELQKKILSAKFNFNDFL 831
Query: 412 KQTRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAE 471
KQT+A+AQMGS SR++GMIPGM+KVTPAQIREAEKNLK ME+MI MTPEERE+PELLAE
Sbjct: 832 KQTQAIAQMGSFSRIIGMIPGMNKVTPAQIREAEKNLKFMESMINVMTPEERERPELLAE 891
Query: 472 SPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEK 531
S RR RVA+ESGK E+Q +MG MQG P + L +++K EE+
Sbjct: 892 SRERRIRVAKESGKNERQ----------------KMMGAMQGQDTPDMEGLMDSIKAEEQ 935
Query: 532 AP 533
AP
Sbjct: 936 AP 937
>ref|ZP_00324395.1| COG0541: Signal recognition particle GTPase [Trichodesmium
erythraeum IMS101]
Length = 483
Score = 499 bits (1285), Expect = e-140
Identities = 258/475 (54%), Positives = 346/475 (72%), Gaps = 7/475 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE AW KL+G++ +T NI + + ++RRALLEADV+L VV+ F+ VS +A
Sbjct: 1 MFEALADRLEVAWKKLRGQDKITDSNIQQALGEVRRALLEADVNLQVVKGFIAEVSSKAK 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+ GV+PDQQ +KIVH+EL ++MG + LA A T++L+AGLQG GKTT AKL
Sbjct: 61 GADVLTGVRPDQQFIKIVHDELAQIMGESNAPLAKADQPPTIVLMAGLQGTGKTTASAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K+ +S +LVA D+YRPAAIDQL LGKQ+ VPV+ GTD+ P EIA++GLE+AK
Sbjct: 121 ALHLRKENRSALLVATDVYRPAAIDQLVTLGKQIDVPVFELGTDIDPVEIAQKGLEKAKA 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
ID+VI+DTAGRLQID+ MM EL +K ++ P E LLVVDAMTGQEAA L TFN +IG
Sbjct: 181 DGIDIVIIDTAGRLQIDQDMMAELARIKAAVKPQETLLVVDAMTGQEAATLTRTFNDQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTK+DGD+RGGAALSV+ +SG+PIK +G GE+++ L+PFYP+RMA RILGMGDVL
Sbjct: 241 ITGAILTKMDGDTRGGAALSVRRISGQPIKFIGVGEKVEALQPFYPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE DAEK+QEKI+SAKFDF DFLKQTR + MGS+ +M +IPGM+K++
Sbjct: 301 TLVEKAQEEFDIADAEKMQEKILSAKFDFTDFLKQTRLMKNMGSLGGIMKLIPGMNKISD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
++++ E LK E+MI +MT +ER+ PELLA+SP RR+R+A SG E+ VS++V+
Sbjct: 361 DKLQQGETQLKRCESMINSMTRQERQNPELLAKSPSRRRRIAGGSGYAEKDVSKLVSDFQ 420
Query: 499 QMRVRMKNL-MGVMQGGSMPTLSNLEEALKPEEKAPPG------TARRRKRSEQR 546
+MR M+ + G + G MP + + KAPPG T +K+ +Q+
Sbjct: 421 RMRSLMQKMGQGDLSGMGMPGMPGMGMPGMLGGKAPPGWRNQQSTGSGKKKKKQK 475
>pir||S68160 probable RNA binding protein ffh - Synechococcus sp. (strain PCC
7942)
Length = 496
Score = 493 bits (1268), Expect = e-138
Identities = 257/486 (52%), Positives = 353/486 (71%), Gaps = 12/486 (2%)
Query: 76 RAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSD 135
R+ MF L LE AW KL+G++ +++ N+ E ++++RRALLEADV+L VV+ F+ V +
Sbjct: 9 RSFMFDALAERLEQAWTKLRGQDKISESNVQEALKEVRRALLEADVNLGVVKEFIAQVQE 68
Query: 136 QAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVC 195
+AVG VI G++PDQQ +K+V++ELV++MG LA A TVIL+AGLQG GKTT
Sbjct: 69 KAVGTQVISGIRPDQQFIKVVYDELVQVMGETHVPLAQAAKAPTVILMAGLQGAGKTTAT 128
Query: 196 AKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEE 255
AKL+ +L+K+G+S +LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E+
Sbjct: 129 AKLALHLRKEGRSTLLVATDVYRPAAIDQLITLGKQIDVPVFELGSDANPVEIARQGVEK 188
Query: 256 AKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNL 315
A+++ ID VIVDTAGRLQID MM+EL +VK+++ P EVLLVVD+M GQEAA+L +F+
Sbjct: 189 ARQEGIDTVIVDTAGRLQIDTEMMEELAQVKEAIAPHEVLLVVDSMIGQEAASLTRSFHE 248
Query: 316 EIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMG 375
IGITGAILTKLDGDSRGGAALS++ VSG+PIK VG GE+++ L+PF+P+RMA RILGMG
Sbjct: 249 RIGITGAILTKLDGDSRGGAALSIRRVSGQPIKFVGLGEKVEALQPFHPERMASRILGMG 308
Query: 376 DVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM-S 434
DVL+ VEKAQE + D EK+QEKI++AKFDF DFLKQ R + MGS++ + +IPG+
Sbjct: 309 DVLTLVEKAQEEIDLADVEKMQEKILAAKFDFTDFLKQMRLLKNMGSLAGFIKLIPGLGG 368
Query: 435 KVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVV 494
K+ Q+R+ E+ LK +EAMI +MTP+ER P+LL+ SP RR R+A+ G+TE ++ Q++
Sbjct: 369 KINDEQLRQGERQLKRVEAMINSMTPQERRDPDLLSNSPSRRHRIAKGCGQTEAEIRQII 428
Query: 495 AQLFQMRVRMKNL-------MGVMQ----GGSMPTLSNLEEALKPEEKAPPGTARRRKRS 543
Q QMR M+ + MG M GG MP A +P + +++K S
Sbjct: 429 QQFQQMRTMMQQMSQGGFPGMGGMGMPGFGGGMPGFGGGAPAPQPGFRGYGPPKKQKKGS 488
Query: 544 EQRSLF 549
+++ F
Sbjct: 489 KKKKGF 494
>emb|CAA63054.1| signal sequence binding protein [Synechococcus sp.]
gi|2500884|sp|Q55311|SR54_SYNP7 SIGNAL SEQUENCE BINDING
PROTEIN gi|45513246|ref|ZP_00164812.1| COG0541: Signal
recognition particle GTPase [Synechococcus elongatus PCC
7942]
Length = 485
Score = 491 bits (1265), Expect = e-137
Identities = 256/483 (53%), Positives = 351/483 (72%), Gaps = 12/483 (2%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE AW KL+G++ +++ N+ E ++++RRALLEADV+L VV+ F+ V ++AV
Sbjct: 1 MFDALAERLEQAWTKLRGQDKISESNVQEALKEVRRALLEADVNLGVVKEFIAQVQEKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI G++PDQQ +K+V++ELV++MG LA A TVIL+AGLQG GKTT AKL
Sbjct: 61 GTQVISGIRPDQQFIKVVYDELVQVMGETHVPLAQAAKAPTVILMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K+G+S +LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E+A++
Sbjct: 121 ALHLRKEGRSTLLVATDVYRPAAIDQLITLGKQIDVPVFELGSDANPVEIARQGVEKARQ 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ ID VIVDTAGRLQID MM+EL +VK+++ P EVLLVVD+M GQEAA+L +F+ IG
Sbjct: 181 EGIDTVIVDTAGRLQIDTEMMEELAQVKEAIAPHEVLLVVDSMIGQEAASLTRSFHERIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTKLDGDSRGGAALS++ VSG+PIK VG GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAILTKLDGDSRGGAALSIRRVSGQPIKFVGLGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM-SKVT 437
+ VEKAQE + D EK+QEKI++AKFDF DFLKQ R + MGS++ + +IPG+ K+
Sbjct: 301 TLVEKAQEEIDLADVEKMQEKILAAKFDFTDFLKQMRLLKNMGSLAGFIKLIPGLGGKIN 360
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
Q+R+ E+ LK +EAMI +MTP+ER P+LL+ SP RR R+A+ G+TE ++ Q++ Q
Sbjct: 361 DEQLRQGERQLKRVEAMINSMTPQERRDPDLLSNSPSRRHRIAKGCGQTEAEIRQIIQQF 420
Query: 498 FQMRVRMKNL-------MGVMQ----GGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQR 546
QMR M+ + MG M GG MP A +P + +++K S+++
Sbjct: 421 QQMRTMMQQMSQGGFPGMGGMGMPGFGGGMPGFGGGAPAPQPGFRGYGPPKKQKKGSKKK 480
Query: 547 SLF 549
F
Sbjct: 481 KGF 483
>ref|YP_173031.1| signal recognition particle receptor [Synechococcus elongatus PCC
6301] gi|56687289|dbj|BAD80511.1| signal recognition
particle receptor [Synechococcus elongatus PCC 6301]
Length = 496
Score = 490 bits (1261), Expect = e-137
Identities = 256/486 (52%), Positives = 351/486 (71%), Gaps = 12/486 (2%)
Query: 76 RAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSD 135
R+ MF L LE AW KL+G++ +++ N+ E ++++RRALLEADV+L VV+ F+ V +
Sbjct: 9 RSFMFDALAERLEQAWTKLRGQDKISESNVQEALKEVRRALLEADVNLGVVKEFIAQVQE 68
Query: 136 QAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVC 195
+AVG VI G++PDQQ +K+V++ELV++MG LA A TVIL+AGLQG GKTT
Sbjct: 69 KAVGTQVISGIRPDQQFIKVVYDELVQVMGETHVPLAQAAKAPTVILMAGLQGAGKTTAT 128
Query: 196 AKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEE 255
AKL+ +L+K+G+S +LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E+
Sbjct: 129 AKLALHLRKEGRSTLLVATDVYRPAAIDQLITLGKQIDVPVFELGSDANPVEIARQGVEK 188
Query: 256 AKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNL 315
A+++ ID VIVDTAGRLQID MM+EL +VK+++ P EVLLVVD+M GQEA +L +F+
Sbjct: 189 ARQEGIDTVIVDTAGRLQIDTEMMEELAQVKEAIAPHEVLLVVDSMIGQEAPSLTRSFHE 248
Query: 316 EIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMG 375
IGITGAILTKLDGDSRGGAALS++ VSG PIK VG GE+++ L+PF+P+RMA RILGMG
Sbjct: 249 RIGITGAILTKLDGDSRGGAALSIRRVSGHPIKFVGLGEKVEALQPFHPERMASRILGMG 308
Query: 376 DVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM-S 434
DVL+ VEKAQE + D EK+QEKI++AKFDF DFLKQ R + MGS++ + +IPG+
Sbjct: 309 DVLTLVEKAQEEIDLADVEKMQEKILAAKFDFTDFLKQMRLLKNMGSLAGFIKLIPGLGG 368
Query: 435 KVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVV 494
K+ Q+R+ E+ LK +EAMI +MTP+ER P+LL+ SP RR R+A+ G+TE ++ Q++
Sbjct: 369 KINDEQLRQGERQLKRVEAMINSMTPQERRDPDLLSNSPSRRHRIAKGCGQTEAEIRQII 428
Query: 495 AQLFQMRVRMKNL-------MGVMQ----GGSMPTLSNLEEALKPEEKAPPGTARRRKRS 543
Q QMR M+ + MG M GG MP A +P + +++K S
Sbjct: 429 QQFQQMRTMMQQMSQGGFPGMGGMGMPGFGGGMPGFGGGAPAPQPGFRGYGPPKKQKKGS 488
Query: 544 EQRSLF 549
+++ F
Sbjct: 489 KKKKGF 494
>ref|NP_682763.1| signal recognition particle protein [Thermosynechococcus elongatus
BP-1] gi|22295699|dbj|BAC09525.1| signal recognition
particle protein [Thermosynechococcus elongatus BP-1]
Length = 469
Score = 489 bits (1258), Expect = e-136
Identities = 250/464 (53%), Positives = 341/464 (72%), Gaps = 1/464 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE+AW L+G++ +T+ NI E +R++RRALLEADV+L V + F+++V +A+
Sbjct: 1 MFDALAERLESAWKTLRGQDKITENNIQEALREVRRALLEADVNLQVAKDFIENVRQRAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV+PDQQ +KIV++ LV++MG S LA + T+IL+AGLQG GKTT AKL
Sbjct: 61 GAEVIAGVRPDQQFIKIVYDALVEVMGESHSPLAHVEPPPTIILMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K+G+S +LVA D+YRPAAIDQL LGKQ+GVPV+ GT+V P EIARQG+ +AK+
Sbjct: 121 ALHLRKEGRSTLLVATDVYRPAAIDQLMTLGKQIGVPVFEMGTEVSPVEIARQGVAKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+D VI+DTAGRLQID MM EL +K+ + P E LLVVDAMTGQEAA L F+ ++G
Sbjct: 181 LGVDTVIIDTAGRLQIDAEMMAELAAIKEGVQPHETLLVVDAMTGQEAANLTRAFHEQVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTKLDGD+RGGAALSV++VSG+PIK +G GE+++ L+PFYPDR+A RILGMGDVL
Sbjct: 241 ITGAILTKLDGDTRGGAALSVRQVSGQPIKFIGVGEKVEALQPFYPDRLASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE + DAEK+ KI+ A+FDF+DFLKQ R + MGS++ ++ +IPGM+K++
Sbjct: 301 TLVEKAQEEVDLADAEKMSRKILEAQFDFDDFLKQLRLLKNMGSLAGIIKLIPGMNKIST 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
Q+++ E+ LK EAMI +MT EER PELLA SP RR+RVA+ SG V+++V+
Sbjct: 361 EQLQQGEQQLKRAEAMINSMTREERRNPELLASSPSRRERVAKGSGYQVADVTKLVSDFQ 420
Query: 499 QMRVRMKNL-MGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRK 541
+MR M+ + G G MP ++ + P+ P ++RK
Sbjct: 421 RMRSLMQQMGQGGFPGMGMPGMNAMGRRGHPQTAKKPKKQKKRK 464
>ref|ZP_00110921.1| COG0541: Signal recognition particle GTPase [Nostoc punctiforme PCC
73102]
Length = 488
Score = 486 bits (1250), Expect = e-135
Identities = 251/444 (56%), Positives = 331/444 (74%), Gaps = 4/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L+ LEAAW KL+G++ +++ NI + +R++RRALLEADV+L VV+ F+ V +A
Sbjct: 1 MFDALSDRLEAAWKKLRGQDKISQSNIQDALREVRRALLEADVNLQVVKDFISEVEAKAQ 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+ GV+PDQQ +KIVH+ELV++MG E +A A+ T++L+AGLQG GKTT AKL
Sbjct: 61 GAEVVTGVRPDQQFIKIVHDELVEVMGEENVPIAEAQEKPTIVLMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K +SC+LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E A+
Sbjct: 121 ALHLRKLDRSCLLVATDVYRPAAIDQLLTLGKQIDVPVFELGSDADPVEIARQGVERARA 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ ++ VI+DTAGRLQID+ MM EL +K ++ P E LLVVDAMTGQEAA L TF+ +IG
Sbjct: 181 EGVNTVIIDTAGRLQIDEDMMAELARIKATVQPHETLLVVDAMTGQEAANLTRTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTK+DGDSRGGAALSV+++SG PIK VG GE+++ L+PFYPDRMA RILGMGDVL
Sbjct: 241 ITGAILTKMDGDSRGGAALSVRKISGAPIKFVGVGEKVEALQPFYPDRMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE DAEK+QEKI+SAKFDF DFLKQ R + MGS+ + MIPGM+K++
Sbjct: 301 TLVEKAQEEFDLADAEKMQEKILSAKFDFTDFLKQLRMLKNMGSLGGFIKMIPGMNKLSD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
Q+++ E LK EAMI +MT +ER P+LLA SP RR+R+A SG E V+++V F
Sbjct: 361 DQLKQGETQLKRCEAMINSMTRQERHDPDLLASSPSRRRRIAAGSGYRESDVTKLVGD-F 419
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
Q +M++LM M G P + +
Sbjct: 420 Q---KMRSLMQQMGQGGFPGMPGM 440
>ref|ZP_00518055.1| Signal recognition particle protein [Crocosphaera watsonii WH 8501]
gi|67853517|gb|EAM48865.1| Signal recognition particle
protein [Crocosphaera watsonii WH 8501]
Length = 488
Score = 484 bits (1247), Expect = e-135
Identities = 249/444 (56%), Positives = 327/444 (73%), Gaps = 4/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE AW KL+G++ +T+ N+ + ++++RRALLEADV+L VV+ F+ V A+
Sbjct: 1 MFDALADRLEDAWKKLRGQDKITEANMQDALKEVRRALLEADVNLQVVKTFIAEVEKAAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV P QQL+KIV++ELVK+MG LA A+ TVIL+AGLQG GKTT AKL
Sbjct: 61 GAEVIAGVNPGQQLIKIVYDELVKVMGESNVPLAQAEKSPTVILMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
S YL+KQ ++C++VA DIYRPAAIDQL LG+Q+ VPV+ GT P +IA QG+ +AK+
Sbjct: 121 SLYLRKQERTCLMVATDIYRPAAIDQLKTLGEQIEVPVFELGTQANPVDIATQGVAKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
ID VI+DTAGRLQID+ MM EL VKK++NP + LLVVDAMTGQEAA L TF+ +IG
Sbjct: 181 MGIDTVIIDTAGRLQIDEEMMLELARVKKAVNPDDTLLVVDAMTGQEAANLTRTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
+TGAILTK+DGD+RGGAALSV+++SG+PIK VG GE+++ L+PFYPDRMA RIL MGDVL
Sbjct: 241 VTGAILTKMDGDTRGGAALSVRQISGQPIKFVGVGEKVEALQPFYPDRMASRILSMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE + D E++Q KI+ AKFDFNDFLKQ R + MGS ++ MIPGM+K++
Sbjct: 301 TLVEKAQEEIDLSDVEQMQSKILEAKFDFNDFLKQMRLMKNMGSFGSILKMIPGMNKLSS 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
I + E LK EAMI +MT EER+ P+LLA+SP RR+R+A+ SG E +VS+++
Sbjct: 361 TDIEKGEGELKRTEAMISSMTLEERKNPDLLAKSPNRRRRIAKGSGHPEAEVSKLIKNF- 419
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
RM+ +M M G MP + +
Sbjct: 420 ---TRMRGMMQQMGQGGMPAMPGM 440
>ref|NP_441626.1| signal recognition particle protein [Synechocystis sp. PCC 6803]
gi|1653392|dbj|BAA18306.1| signal recognition particle
protein [Synechocystis sp. PCC 6803]
gi|2500885|sp|P74214|SRP54_SYNY3 Signal recognition
particle protein (Fifty-four homolog)
Length = 482
Score = 484 bits (1246), Expect = e-135
Identities = 248/437 (56%), Positives = 327/437 (74%), Gaps = 1/437 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE AW KL+G++ +++ NI E ++++RRALL ADV+L VV+ F++ V +A+
Sbjct: 1 MFDALADRLEDAWKKLRGQDKISESNIKEALQEVRRALLAADVNLQVVKGFIKDVEQKAL 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV P QQ +KIV++ELV LMG LA A+ TVIL+AGLQG GKTT AKL
Sbjct: 61 GADVISGVNPGQQFIKIVYDELVNLMGESNVPLAQAEQAPTVILMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ YL+KQ +S ++VA D+YRPAAIDQL LG+Q+ VPV+ G+D P EIARQG+E+AK+
Sbjct: 121 ALYLRKQKRSALMVATDVYRPAAIDQLKTLGQQIDVPVFDLGSDANPVEIARQGVEKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+D V++DTAGRLQID MM EL E+K+ + P + LLVVDAMTGQEAA L TF+ +IG
Sbjct: 181 LGVDTVLIDTAGRLQIDPQMMAELAEIKQVVKPDDTLLVVDAMTGQEAANLTHTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTKLDGD+RGGAALSV+++SG+PIK VG GE+++ L+PFYPDR+A RIL MGDVL
Sbjct: 241 ITGAILTKLDGDTRGGAALSVRQISGQPIKFVGVGEKVEALQPFYPDRLASRILNMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE + D EKLQ KI+ A FDF+DF+KQ R + MGS+ ++ MIPGM+K++
Sbjct: 301 TLVEKAQEAIDVGDVEKLQNKILEATFDFDDFIKQMRFMKNMGSLGGLLKMIPGMNKLSS 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
I + EK LK EAMI +MT EER+ P+LLA+SP RR+R+AQ SG +E VS+++
Sbjct: 361 GDIEKGEKELKRTEAMISSMTTEERKNPDLLAKSPSRRRRIAQGSGHSETDVSKLITNFT 420
Query: 499 QMRVRMKNL-MGVMQGG 514
+MR M+ + MG M GG
Sbjct: 421 KMRTMMQQMGMGGMPGG 437
>dbj|BAB73651.1| signal recognition particle protein [Nostoc sp. PCC 7120]
gi|17229444|ref|NP_485992.1| signal recognition particle
protein [Nostoc sp. PCC 7120] gi|25294426|pir||AB2050
signal recognition particle protein [imported] - Nostoc
sp. (strain PCC 7120)
Length = 490
Score = 483 bits (1244), Expect = e-135
Identities = 250/444 (56%), Positives = 332/444 (74%), Gaps = 4/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE+AW KL+G++ +++ NI + +R++RRALLEADV+L VV+ F+ V +A
Sbjct: 1 MFDALADRLESAWKKLRGQDKISESNIQDALREVRRALLEADVNLQVVKDFISEVEAKAQ 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV+PDQQ +KIV++ELV++MG E LA + T++L+AGLQG GKTT AKL
Sbjct: 61 GAEVISGVRPDQQFIKIVYDELVQVMGEENVPLAEVEQKPTIVLMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K +SC+LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E AK
Sbjct: 121 ALHLRKLERSCLLVATDVYRPAAIDQLLTLGKQIDVPVFELGSDADPVEIARQGVERAKA 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ ++ VI+DTAGRLQID+ MM EL +K+++ P E LLVVD+MTGQEAA L TF+ +IG
Sbjct: 181 EGVNTVIIDTAGRLQIDEDMMAELARIKETVQPHETLLVVDSMTGQEAANLTRTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTK+DGDSRGGAALSV+++SG PIK VG GE+++ L+PFYPDRMA RILGMGDVL
Sbjct: 241 ITGAILTKMDGDSRGGAALSVRQISGAPIKFVGVGEKVEALQPFYPDRMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE DAEK+QEKI+SAKFDF DF+KQ R + MGS+ ++ MIPGM K++
Sbjct: 301 TLVEKAQEEFDLADAEKMQEKILSAKFDFTDFVKQLRMLKNMGSLGGLLKMIPGMGKLSD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
Q+++ E LK EAMI +MT +ER P+LLA SP RR+R+A SG E V+++VA F
Sbjct: 361 DQLKQGETQLKRCEAMINSMTMQERRDPDLLASSPNRRRRIASGSGYKEADVNKLVAD-F 419
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
Q +M++LM M G+ P + +
Sbjct: 420 Q---KMRSLMQQMGQGNFPGMPGM 440
>ref|ZP_00162376.2| COG0541: Signal recognition particle GTPase [Anabaena variabilis
ATCC 29413]
Length = 490
Score = 483 bits (1243), Expect = e-135
Identities = 250/444 (56%), Positives = 332/444 (74%), Gaps = 4/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE+AW KL+G++ +++ NI + +R++RRALLEADV+L VV+ F+ V +A
Sbjct: 1 MFDALADRLESAWKKLRGQDKISESNIQDALREVRRALLEADVNLQVVKDFISEVEAKAQ 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV+PDQQ +KIV++ELV++MG E LA + T++L+AGLQG GKTT AKL
Sbjct: 61 GAEVISGVRPDQQFIKIVYDELVQVMGEENVPLAEVEQKPTIVLMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K +SC+LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E AK
Sbjct: 121 ALHLRKLERSCLLVATDVYRPAAIDQLLTLGKQIDVPVFELGSDADPVEIARQGVERAKA 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ ++ VI+DTAGRLQID+ MM EL +K+++ P E LLVVD+MTGQEAA L TF+ +IG
Sbjct: 181 EGVNTVIIDTAGRLQIDEDMMAELARIKETVQPHETLLVVDSMTGQEAANLTRTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTK+DGDSRGGAALSV+++SG PIK VG GE+++ L+PFYPDRMA RILGMGDVL
Sbjct: 241 ITGAILTKMDGDSRGGAALSVRQISGAPIKFVGVGEKVEALQPFYPDRMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE DAEK+QEKI+SAKFDF DF+KQ R + MGS+ ++ MIPGM K++
Sbjct: 301 TLVEKAQEEFDLADAEKMQEKILSAKFDFTDFVKQLRMLKNMGSLGGLLKMIPGMGKLSD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
Q+++ E LK EAMI +MT +ER P+LLA SP RR+R+A SG E V+++VA F
Sbjct: 361 DQLKQGETQLKRCEAMINSMTIQERRDPDLLASSPNRRRRIASGSGYKEADVNKLVAD-F 419
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
Q +M++LM M G+ P + +
Sbjct: 420 Q---KMRSLMQQMGQGNFPGMPGM 440
>emb|CAE08133.1| signal recognition particle protein (SRP54) [Synechococcus sp. WH
8102] gi|33866152|ref|NP_897711.1| signal recognition
particle protein (SRP54) [Synechococcus sp. WH 8102]
Length = 487
Score = 465 bits (1196), Expect = e-129
Identities = 242/486 (49%), Positives = 344/486 (69%), Gaps = 13/486 (2%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A LKG++ ++ N+ ++D+RRALLEADVSLPVV+ FV V ++AV
Sbjct: 1 MFDELSARFEDAVKGLKGQDKISDTNVEVALKDVRRALLEADVSLPVVKDFVADVREKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV PDQ+ +++VHE+LV++MGG+ + LA A TV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVSPDQKFIQVVHEQLVEVMGGDNAPLAKAAEAPTVVLMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+LK QG+ ++V D+YRPAAI+QL LG Q+GV V++ GT+ KP EIA GL +AK+
Sbjct: 121 GLHLKDQGRRALMVGADVYRPAAIEQLKTLGGQIGVEVFSLGTEAKPEEIAAAGLAKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D ++VDTAGRLQID MM+E+ ++ ++ P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRTAVQPDEVLLVVDSMIGQEAAELTRAFHDQVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ D EK+Q+K+ A FDF+DF++Q R + +MGS+ +M MIPGM+K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEATFDFSDFVQQMRLIKRMGSLGGLMKMIPGMNKLDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
+++ E+ LK +EAMI +MTP+ERE P+LLA P RR+R+A SG V +V+A
Sbjct: 361 GILKQGEQQLKRIEAMIGSMTPKERENPDLLAGQPSRRRRIAAGSGHQPADVDKVLADFQ 420
Query: 499 QMRVRMKNL-------MGVMQG-GSMPTLSNLE--EALKPEEKAPP---GTARRRKRSEQ 545
+MR M+ + MG M G G MP + + + PP G+ +R+K +++
Sbjct: 421 KMRGFMQQMSRGGMPGMGGMPGMGGMPGMGGMPGMGGMPGGGGRPPGRGGSPKRQKPAKK 480
Query: 546 RSLFAD 551
R F D
Sbjct: 481 RRGFGD 486
>gb|AAQ00404.1| Signal recognition particle GTPase [Prochlorococcus marinus subsp.
marinus str. CCMP1375] gi|33240809|ref|NP_875751.1|
Signal recognition particle GTPase [Prochlorococcus
marinus subsp. marinus str. CCMP1375]
Length = 485
Score = 456 bits (1174), Expect = e-127
Identities = 232/439 (52%), Positives = 323/439 (72%), Gaps = 3/439 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A L+GE ++++N+ ++ +RRALLEADVSL VV++FVQ V D+A+
Sbjct: 1 MFDELSARFEDAVKGLRGENKISEDNVDIALKQVRRALLEADVSLSVVKQFVQEVRDKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV P Q+ +++VH+ELV++MGG S LA + +VIL+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVNPGQKFIEVVHQELVEVMGGANSPLADSTKKPSVILMAGLQGAGKTTAAAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
YLK +G ++VA D+YRPAAIDQL LGKQ+ V V++ G + KP +IA GL++A++
Sbjct: 121 GLYLKDKGLKPLMVAADVYRPAAIDQLNTLGKQINVEVFSLGKESKPEDIAASGLKKAQE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D +IVDTAGRLQID+ MM+E+ ++ +++P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EDFDTLIVDTAGRLQIDEEMMNEMVRIRSAVDPDEVLLVVDSMIGQEAADLTRAFHEKVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMAGRILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMAGRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ DAE++Q K A FDF+DF+KQ R + +MGS+ +M MIPGM+K+
Sbjct: 301 TLVEKAQKEVEIADAEQMQRKFQEATFDFSDFVKQMRLIKRMGSLGGLMKMIPGMNKIDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
++ E LK +EAMI +MT E+ +PELLA P RRKRVA SG T +V +V+A
Sbjct: 361 GMLKSGEDQLKRIEAMIGSMTNAEQTQPELLAAQPSRRKRVAFGSGHTPVEVDKVLADFQ 420
Query: 499 QMRVRMKNLMGVMQGGSMP 517
+MR MK + GG +P
Sbjct: 421 KMRGLMKQM---SSGGGLP 436
>emb|CAE20524.1| signal recognition particle protein (SRP54) [Prochlorococcus
marinus str. MIT 9313] gi|33862622|ref|NP_894182.1|
signal recognition particle protein (SRP54)
[Prochlorococcus marinus str. MIT 9313]
Length = 497
Score = 454 bits (1168), Expect = e-126
Identities = 226/444 (50%), Positives = 327/444 (72%), Gaps = 3/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A L+G+ ++ N+ + ++ +RRALL ADVSL VVR FV+ V +AV
Sbjct: 1 MFEELSARFEDAVKGLRGQAKISDTNVEDALKQVRRALLGADVSLEVVREFVEEVRQKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV PDQ+ V++VH++LV++MGG+ + +A A+ TV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVTPDQKFVQVVHQQLVEVMGGDNAPMAEAEDSPTVVLMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+LK QG+ ++VA D+YRPAAIDQL LG+Q+GV V++ G DVKP EIA GL +A++
Sbjct: 121 GLHLKDQGQRPLMVAADVYRPAAIDQLRTLGEQIGVDVFSLGDDVKPEEIAAAGLAKARE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D ++VDTAGRLQID MM+E+ ++ ++ P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRSAVEPDEVLLVVDSMIGQEAAELTRAFHDKVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITG++LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGSVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ D EK+Q+K+ A FDF+DFL+Q R + +MGS+ ++ M+PGM+K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEASFDFSDFLQQMRLIKRMGSLGGLIKMMPGMNKLDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
+++ E+ LK +EAMI +MT +ER +PELLA P RR+R+A SG + V +V+A
Sbjct: 361 GMLKQGEQQLKRIEAMIGSMTADERNQPELLASQPSRRRRIAGGSGHSPADVDKVLADFQ 420
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
+MR M+ + +GG MP + +
Sbjct: 421 KMRGFMQQM---SKGGGMPGMPGM 441
>ref|NP_925585.1| signal recognition particle protein SRP54 [Gloeobacter violaceus
PCC 7421] gi|35213208|dbj|BAC90580.1| signal recognition
particle protein SRP54 [Gloeobacter violaceus PCC 7421]
Length = 497
Score = 442 bits (1136), Expect = e-122
Identities = 234/442 (52%), Positives = 316/442 (70%), Gaps = 5/442 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L+ LE AW KL+G++ +T+ NI E +R++RRALLEADV+ V + FV V D A+
Sbjct: 1 MFESLSEKLEGAWRKLRGQDRITEGNIDEALREVRRALLEADVNFQVAKEFVADVRDDAL 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAF--AKSGTTVILLAGLQGVGKTTVCA 196
G V+ GV PDQQ +KIVH++LV LMG + LA AK V+L+AGLQG GKTT A
Sbjct: 61 GAEVVSGVTPDQQFIKIVHDQLVALMGEQNVPLAEPRAKGRPAVVLMAGLQGTGKTTASA 120
Query: 197 KLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEA 256
KL+ YL+K G+ +L A D+YRPAAIDQL LG Q+ VPV+T G + P +IAR LE A
Sbjct: 121 KLALYLQKNGQKPLLAAADVYRPAAIDQLQTLGGQIQVPVFTLGKEADPVDIARASLERA 180
Query: 257 KKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLE 316
+ DV+IVDTAGRL ID AMM EL+ +K +++P E+LLVVDAMTGQEAA L F+
Sbjct: 181 IADRHDVLIVDTAGRLSIDDAMMAELERIKAAIDPEEILLVVDAMTGQEAANLTRAFHDR 240
Query: 317 IGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGD 376
+GITGAILTKLDGD+RGGAALSV+++SG+PIK VG GE+++ L+PFYP+RMA RILGMGD
Sbjct: 241 LGITGAILTKLDGDTRGGAALSVRKISGQPIKFVGVGEKVEALQPFYPERMASRILGMGD 300
Query: 377 VLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKV 436
VL+ VEKAQE + DA K+++++++ +FDF F+KQ R + MG + V+ MIPGM+K+
Sbjct: 301 VLTLVEKAQEEIDFADAAKMEQQLLTGQFDFESFIKQMRMIKNMGPLGGVLKMIPGMNKI 360
Query: 437 TPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQ 496
+ QIR+ E LK EAMI +MT +ER P+L+ S R++R+++ SG + + V +++
Sbjct: 361 SDDQIRQGEVQLKKAEAMIGSMTTQERRNPDLINLS--RKRRISKGSGVSLEDVGKLLND 418
Query: 497 LFQMRVRMKNLMGVMQ-GGSMP 517
+MR MK + G GG MP
Sbjct: 419 FGKMRQMMKMMGGGGPFGGGMP 440
>ref|NP_893403.1| signal recognition particle protein (SRP54) [Prochlorococcus
marinus subsp. pastoris str. CCMP1986]
gi|33640210|emb|CAE19745.1| signal recognition particle
protein (SRP54) [Prochlorococcus marinus subsp. pastoris
str. CCMP1986]
Length = 498
Score = 435 bits (1118), Expect = e-120
Identities = 225/445 (50%), Positives = 319/445 (71%), Gaps = 5/445 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L++ E A L+GE +++ NI + ++ +++ALL+ADVSL VV+ FV V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNIDDALKQVKKALLDADVSLSVVKEFVSDVKDKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV P Q+ +++V++EL+ +MG E S L ++ TVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNKKETTPTVILMAGLQGAGKTTATGKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGT-DVKPSEIARQGLEEAK 257
YLK++ + +LVA DIYRPAA++QL +G Q + V++A + KP EIAR L A
Sbjct: 121 GLYLKEKEEKVLLVAADIYRPAAVEQLKTIGNQYDLEVFSAKEKNSKPEEIARDALSYAN 180
Query: 258 KKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEI 317
ID +IVDTAGRLQID++MM E+ +K+ P EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ANNIDTLIVDTAGRLQIDESMMGEMVRIKQVTKPDEVLLVVDSMIGQEAADLTKSFHEKV 240
Query: 318 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDV 377
GITGAILTKLDGDSRGGAALS++++SGKPIK +G GE+++ L+PF+P+RMA RILGMGDV
Sbjct: 241 GITGAILTKLDGDSRGGAALSIRKISGKPIKFIGTGEKIEALQPFHPERMASRILGMGDV 300
Query: 378 LSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVT 437
L+ VEKAQ+ ++ DAE +Q+K+ A FDFNDF+KQ R + +MGS+ ++ +IPGM+K+
Sbjct: 301 LTLVEKAQKEVELADAEVMQKKLQEATFDFNDFVKQMRLMKRMGSLGGLIKLIPGMNKID 360
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
I+ E+ LK +E+MI +MT EE++KPE+LA P RR+R+AQ SG + V +V+A
Sbjct: 361 DGMIKNGEEQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYQAKDVDKVLAD- 419
Query: 498 FQMRVRMKNLMGVMQGGSMPTLSNL 522
FQ RM+ M M G MP + +
Sbjct: 420 FQ---RMRGFMKQMSNGGMPGMGGM 441
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 874,842,839
Number of Sequences: 2540612
Number of extensions: 35524783
Number of successful extensions: 127680
Number of sequences better than 10.0: 1310
Number of HSP's better than 10.0 without gapping: 880
Number of HSP's successfully gapped in prelim test: 433
Number of HSP's that attempted gapping in prelim test: 124542
Number of HSP's gapped (non-prelim): 1679
length of query: 564
length of database: 863,360,394
effective HSP length: 133
effective length of query: 431
effective length of database: 525,458,998
effective search space: 226472828138
effective search space used: 226472828138
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0010.7


