

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.19
(295 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
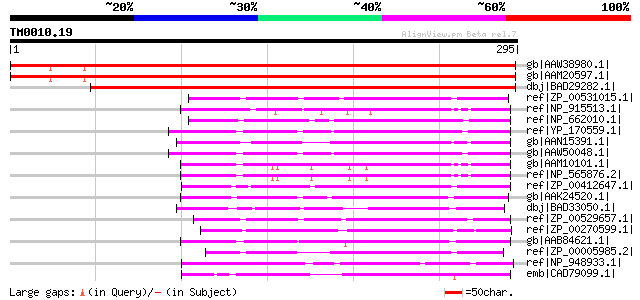
Score E
Sequences producing significant alignments: (bits) Value
gb|AAW38980.1| At3g10130 [Arabidopsis thaliana] gi|6143864|gb|AA... 355 9e-97
gb|AAM20597.1| unknown protein [Arabidopsis thaliana] 353 3e-96
dbj|BAD29282.1| SOUL heme-binding protein-like [Oryza sativa (ja... 333 4e-90
ref|ZP_00531015.1| SOUL heme-binding protein [Chlorobium phaeoba... 114 4e-24
ref|NP_915513.1| P0529H11.19 [Oryza sativa (japonica cultivar-gr... 106 9e-22
ref|NP_662010.1| hypothetical protein CT1119 [Chlorobium tepidum... 105 1e-21
ref|YP_170559.1| hypothetical protein FTT1651 [Francisella tular... 103 8e-21
gb|AAN15391.1| unknown protein [Arabidopsis thaliana] gi|1747381... 103 8e-21
gb|AAW50048.1| hypothetical protein FTT1651 [synthetic construct] 103 8e-21
gb|AAM10101.1| unknown protein [Arabidopsis thaliana] gi|4895187... 100 5e-20
ref|NP_565876.2| SOUL heme-binding family protein [Arabidopsis t... 100 5e-20
ref|ZP_00412647.1| SOUL heme-binding protein [Arthrobacter sp. F... 95 2e-18
gb|AAK24520.1| conserved hypothetical protein [Caulobacter cresc... 94 5e-18
dbj|BAD33050.1| SOUL heme-binding protein-like [Oryza sativa (ja... 94 5e-18
ref|ZP_00529657.1| SOUL heme-binding protein [Chlorobium phaeoba... 92 2e-17
ref|ZP_00270599.1| hypothetical protein Rrub02000642 [Rhodospiri... 89 1e-16
gb|AAB84621.1| unknown [Methanothermobacter thermautotrophicus s... 87 4e-16
ref|ZP_00005985.2| hypothetical protein Rsph03001922 [Rhodobacte... 85 3e-15
ref|NP_948933.1| hypothetical protein RPA3595 [Rhodopseudomonas ... 81 3e-14
emb|CAD79099.1| conserved hypothetical protein [Rhodopirellula b... 77 6e-13
>gb|AAW38980.1| At3g10130 [Arabidopsis thaliana] gi|6143864|gb|AAF04411.1| unknown
protein [Arabidopsis thaliana]
gi|15228209|ref|NP_187624.1| SOUL heme-binding family
protein [Arabidopsis thaliana]
Length = 309
Score = 355 bits (911), Expect = 9e-97
Identities = 182/308 (59%), Positives = 240/308 (77%), Gaps = 14/308 (4%)
Query: 1 MLPLCNASLSSQSSIRTSIPGR-------PNIAITNSASSSERISNRRR------TMSAV 47
M+ + ++S++S S+ +S P + IT S + ++ R +SA
Sbjct: 1 MMMISSSSITSSLSLLSSSPEKLPLINPIQRCPITYSGFRTASVNRAIRRQPQSPAVSAT 60
Query: 48 EARSSLILALASQANSLTQRLVVDVATETARYLFPKRLESRT-LEEALMAVPDLETVKFK 106
E+R SL+LALASQA+S++QRL+ D+A ETA+Y+FPKR +S T LEEA M+VPDLET+ F+
Sbjct: 61 ESRVSLVLALASQASSVSQRLLADLAMETAKYVFPKRFDSSTNLEEAFMSVPDLETMNFR 120
Query: 107 VLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTP 166
VL R D YEIR+VEPYFVAE MPG++GFD GAS+SFNVLAEYLFGKNT+KEKMEMTTP
Sbjct: 121 VLFRTDKYEIRQVEPYFVAETIMPGETGFDSYGASKSFNVLAEYLFGKNTIKEKMEMTTP 180
Query: 167 VYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRK 226
V T K QS G KM+MTTPV T+K +DQ++W MSF++PSKYG+NLPLPKD SV+I+++PRK
Sbjct: 181 VVTRKVQSVGEKMEMTTPVITSKAKDQNQWRMSFVMPSKYGSNLPLPKDPSVKIQQVPRK 240
Query: 227 IVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEI 286
IVAVV+FSG+V DEE+++RE +LR AL++D +F+++ G S EVAQYNPPFTLPF RRNE+
Sbjct: 241 IVAVVAFSGYVTDEEIERRERELRRALQNDKKFRVRDGVSFEVAQYNPPFTLPFMRRNEV 300
Query: 287 ALEVERKD 294
+LEVE K+
Sbjct: 301 SLEVENKE 308
>gb|AAM20597.1| unknown protein [Arabidopsis thaliana]
Length = 309
Score = 353 bits (906), Expect = 3e-96
Identities = 181/308 (58%), Positives = 239/308 (76%), Gaps = 14/308 (4%)
Query: 1 MLPLCNASLSSQSSIRTSIPGR-------PNIAITNSASSSERISNRRR------TMSAV 47
M+ + ++S++S S+ +S P + IT S + ++ R +SA
Sbjct: 1 MMMISSSSITSSLSLLSSSPEKLPLINPIQRCPITYSGFRTASVNRAIRRQPQSPAVSAT 60
Query: 48 EARSSLILALASQANSLTQRLVVDVATETARYLFPKRLESRT-LEEALMAVPDLETVKFK 106
E+R SL+LAL SQA+S++QRL+ D+A ETA+Y+FPKR +S T LEEA M+VPDLET+ F+
Sbjct: 61 ESRVSLVLALPSQASSVSQRLLADLAMETAKYVFPKRFDSSTNLEEAFMSVPDLETMNFR 120
Query: 107 VLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTP 166
VL R D YEIR+VEPYFVAE MPG++GFD GAS+SFNVLAEYLFGKNT+KEKMEMTTP
Sbjct: 121 VLFRTDKYEIRQVEPYFVAETIMPGETGFDSYGASKSFNVLAEYLFGKNTIKEKMEMTTP 180
Query: 167 VYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRK 226
V T K QS G KM+MTTPV T+K +DQ++W MSF++PSKYG+NLPLPKD SV+I+++PRK
Sbjct: 181 VVTRKVQSVGEKMEMTTPVITSKAKDQNQWRMSFVMPSKYGSNLPLPKDPSVKIQQVPRK 240
Query: 227 IVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEI 286
IVAVV+FSG+V DEE+++RE +LR AL++D +F+++ G S EVAQYNPPFTLPF RRNE+
Sbjct: 241 IVAVVAFSGYVTDEEIERRERELRRALQNDKKFRVRDGVSFEVAQYNPPFTLPFMRRNEV 300
Query: 287 ALEVERKD 294
+LEVE K+
Sbjct: 301 SLEVENKE 308
>dbj|BAD29282.1| SOUL heme-binding protein-like [Oryza sativa (japonica
cultivar-group)] gi|50251400|dbj|BAD28427.1| SOUL
heme-binding protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 287
Score = 333 bits (854), Expect = 4e-90
Identities = 164/248 (66%), Positives = 199/248 (80%), Gaps = 1/248 (0%)
Query: 48 EARSSLILALASQANSLTQRLVVDVATETARYLFP-KRLESRTLEEALMAVPDLETVKFK 106
EAR+SL+LALA+QA + +QR +A E +Y FP +R E RTLEEALM+VPDLETV F+
Sbjct: 37 EARASLVLALAAQALAASQRRAAGLAAEAVKYAFPPRRFEPRTLEEALMSVPDLETVPFR 96
Query: 107 VLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTP 166
VL R YEIREVE Y+VAE TMPG+SGFDF+G+SQSFNVLA YLFGKNT E+MEMTTP
Sbjct: 97 VLKREAEYEIREVESYYVAETTMPGRSGFDFNGSSQSFNVLASYLFGKNTTSEQMEMTTP 156
Query: 167 VYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRK 226
V+T K + DG KMDMTTPV T K +++KW MSF++PSKYG +LPLPKD SV IKE+P K
Sbjct: 157 VFTRKGEPDGEKMDMTTPVITKKSANENKWKMSFVMPSKYGPDLPLPKDPSVTIKEVPAK 216
Query: 227 IVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEI 286
IVAV +FSG V D+++ QRE +LRE L+ D QF++K + VE+AQYNPPFTLPF RRNEI
Sbjct: 217 IVAVAAFSGLVTDDDISQRESRLRETLQKDSQFRVKDDSVVEIAQYNPPFTLPFTRRNEI 276
Query: 287 ALEVERKD 294
ALEV+R D
Sbjct: 277 ALEVKRLD 284
>ref|ZP_00531015.1| SOUL heme-binding protein [Chlorobium phaeobacteroides BS1]
gi|67915333|gb|EAM64656.1| SOUL heme-binding protein
[Chlorobium phaeobacteroides BS1]
Length = 205
Score = 114 bits (284), Expect = 4e-24
Identities = 66/187 (35%), Positives = 106/187 (56%), Gaps = 12/187 (6%)
Query: 105 FKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMT 164
+ ++ + +EIRE + +AE + G S + + F+ LA+Y+FG N EK+ MT
Sbjct: 29 YSIVKKDGAFEIREYDAMIIAETLLDGSYR---STSGKGFSKLAKYIFGSNVGSEKIAMT 85
Query: 165 TPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGA-NLPLPKDSSVRIKEI 223
PV +++G K+ MT PV K KW M+F++P++Y NLP P D + I+E+
Sbjct: 86 APVL---QEAEGEKISMTAPVIQEKAGT--KWKMAFVMPAEYTLQNLPKPVDPDILIREV 140
Query: 224 PRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRR 283
P + VA V +SG +++ + KL E L+ G +K + A Y+PP+T+PF RR
Sbjct: 141 PARKVASVRYSGLHSEKNIANWSAKLTEWLEKQG---VKAVSVPRSASYDPPWTIPFLRR 197
Query: 284 NEIALEV 290
NEI ++V
Sbjct: 198 NEIHIDV 204
>ref|NP_915513.1| P0529H11.19 [Oryza sativa (japonica cultivar-group)]
gi|20805177|dbj|BAB92846.1| SOUL heme-binding
protein-like [Oryza sativa (japonica cultivar-group)]
Length = 216
Score = 106 bits (264), Expect = 9e-22
Identities = 82/214 (38%), Positives = 111/214 (51%), Gaps = 29/214 (13%)
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATM-PGKSGFDFSGASQSFNVLAEYLFG----K 154
+ET K +VL YE+R+ P VAE T P + D G F VL Y+ +
Sbjct: 10 VETPKHEVLHTGAGYEVRKYPPCVVAEVTYDPAEMKGDRDGG---FTVLGNYIGALGNPQ 66
Query: 155 NTLKEKMEMTTPVYTSKNQSDGVKMD-------------MTTPVYTAKMEDQDK--WTMS 199
NT EK++MT PV TS + + + M MT PV TA+ Q K TM
Sbjct: 67 NTKPEKIDMTAPVITS-GEPESIAMTAPVITSGEPEPVAMTAPVITAEERSQGKGQMTMQ 125
Query: 200 FILPSKYGA--NLPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDG 257
F+LPSKY P P D V ++++ + VV FSG D+ VK++ L+ AL+ DG
Sbjct: 126 FLLPSKYSKVEEAPRPTDERVVLRQVGERKYGVVRFSGLTGDKVVKEKAEWLKAALEKDG 185
Query: 258 QFKIKKGTSVEVAQYNPPFTLPFQRRNEIALEVE 291
F + KG V +A+YNPPFTLP R NE+ + VE
Sbjct: 186 -FTV-KGPFV-LARYNPPFTLPPLRTNEVMVPVE 216
>ref|NP_662010.1| hypothetical protein CT1119 [Chlorobium tepidum TLS]
gi|21647087|gb|AAM72352.1| lipoprotein, putative
[Chlorobium tepidum TLS]
Length = 215
Score = 105 bits (263), Expect = 1e-21
Identities = 63/189 (33%), Positives = 106/189 (55%), Gaps = 13/189 (6%)
Query: 105 FKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGAS-QSFNVLAEYLFGKNTLKEKMEM 163
+++L +E+R P +AE + KS +S AS + FN LA Y+FGKN K + M
Sbjct: 29 YELLKHDGAFEVRRYGPMVIAETILDEKS---YSAASGKGFNRLAGYIFGKNRSKTSISM 85
Query: 164 TTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGA-NLPLPKDSSVRIKE 222
T PV ++ K+ MT PV + + W+M+F+LP + + P P D V+++E
Sbjct: 86 TAPVLQERSSE---KISMTAPVL--QQPQKGGWSMAFVLPEGFTLQSAPEPLDPEVKLRE 140
Query: 223 IPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQR 282
+P +AVV+FSG + +++ +L+ LK G + + ++A Y+PP+T+PF R
Sbjct: 141 LPPSTIAVVTFSGLHSAANLEKYSRQLQAWLKKQGYRALSEP---KLASYDPPWTIPFLR 197
Query: 283 RNEIALEVE 291
RNE+ + +E
Sbjct: 198 RNEVQIRIE 206
>ref|YP_170559.1| hypothetical protein FTT1651 [Francisella tularensis subsp.
tularensis Schu 4] gi|56605155|emb|CAG46284.1| conserved
hypothetical protein [Francisella tularensis subsp.
tularensis SCHU S4] gi|54114407|gb|AAV29837.1|
NT02FT0503 [synthetic construct]
Length = 208
Score = 103 bits (256), Expect = 8e-21
Identities = 60/201 (29%), Positives = 111/201 (54%), Gaps = 12/201 (5%)
Query: 93 ALMAVPDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGA-SQSFNVLAEYL 151
+++ + + K+ + + DN+ IR P A+ T+ D+ A ++ F L Y+
Sbjct: 18 SIVGINNTPQAKYTNIKKDDNFSIRIYAPLTEAQVTVQDS---DYKSAVNKGFGYLFRYI 74
Query: 152 FGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGA-NL 210
G N K+ ++MT PV K + K+ MT PV K + ++WT++F+LP++Y N
Sbjct: 75 TGANIAKQDIQMTAPV---KIEQSSQKIQMTAPVMV-KGDTNNEWTIAFVLPAQYTLENA 130
Query: 211 PLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVA 270
P P ++ V++ E P +AV++FSGF++ + + KL+ +K++ + + E A
Sbjct: 131 PKPTNNKVKLVEKPETKMAVITFSGFLDKDTIDSNTTKLKAWVKANNYQIVGQ---PEAA 187
Query: 271 QYNPPFTLPFQRRNEIALEVE 291
YNPP+T+PF R NE+ + ++
Sbjct: 188 GYNPPWTIPFLRTNEVMIPIK 208
>gb|AAN15391.1| unknown protein [Arabidopsis thaliana] gi|17473811|gb|AAL38336.1|
unknown protein [Arabidopsis thaliana]
gi|22326906|ref|NP_197514.2| SOUL heme-binding family
protein [Arabidopsis thaliana]
Length = 378
Score = 103 bits (256), Expect = 8e-21
Identities = 69/196 (35%), Positives = 103/196 (52%), Gaps = 27/196 (13%)
Query: 98 PDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTL 157
PDLET K+++L R NYE+R EP+ V E SG S FN +A Y+FGKN+
Sbjct: 203 PDLETPKYQILKRTANYEVRNYEPFIVVETIGDKLSG------SSGFNNVAGYIFGKNST 256
Query: 158 KEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPS-KYGANLPLPKDS 216
EK+ MTTPV+T T + ++ ++PS K ++LP+P +
Sbjct: 257 MEKIPMTTPVFTQ----------------TTDTQLSSDVSVQIVIPSGKDLSSLPMPNEE 300
Query: 217 SVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPP- 275
V +K++ A V FSG ++ V+ +E +LR +L DG + KKG +A+YN P
Sbjct: 301 KVNLKKLEGGFAAAVKFSGKPTEDVVQAKENELRSSLSKDG-LRAKKGCM--LARYNDPG 357
Query: 276 FTLPFQRRNEIALEVE 291
T F RNE+ + +E
Sbjct: 358 RTWNFIMRNEVIIWLE 373
>gb|AAW50048.1| hypothetical protein FTT1651 [synthetic construct]
Length = 243
Score = 103 bits (256), Expect = 8e-21
Identities = 60/201 (29%), Positives = 111/201 (54%), Gaps = 12/201 (5%)
Query: 93 ALMAVPDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGA-SQSFNVLAEYL 151
+++ + + K+ + + DN+ IR P A+ T+ D+ A ++ F L Y+
Sbjct: 44 SIVGINNTPQAKYTNIKKDDNFSIRIYAPLTEAQVTVQDS---DYKSAVNKGFGYLFRYI 100
Query: 152 FGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGA-NL 210
G N K+ ++MT PV K + K+ MT PV K + ++WT++F+LP++Y N
Sbjct: 101 TGANIAKQDIQMTAPV---KIEQSSQKIQMTAPVMV-KGDTNNEWTIAFVLPAQYTLENA 156
Query: 211 PLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVA 270
P P ++ V++ E P +AV++FSGF++ + + KL+ +K++ + + E A
Sbjct: 157 PKPTNNKVKLVEKPETKMAVITFSGFLDKDTIDSNTTKLKAWVKANNYQIVGQ---PEAA 213
Query: 271 QYNPPFTLPFQRRNEIALEVE 291
YNPP+T+PF R NE+ + ++
Sbjct: 214 GYNPPWTIPFLRTNEVMIPIK 234
>gb|AAM10101.1| unknown protein [Arabidopsis thaliana] gi|4895187|gb|AAD32774.1|
unknown protein [Arabidopsis thaliana]
gi|15451064|gb|AAK96803.1| Unknown protein [Arabidopsis
thaliana] gi|13877685|gb|AAK43920.1| Unknown protein
[Arabidopsis thaliana] gi|25360399|pir||D84799
hypothetical protein At2g37970 [imported] - Arabidopsis
thaliana
Length = 215
Score = 100 bits (249), Expect = 5e-20
Identities = 79/212 (37%), Positives = 108/212 (50%), Gaps = 26/212 (12%)
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQS-FNVLAEYL--FGK-- 154
+ET K+ V D YEIRE P AE T +F G F +LA+Y+ FGK
Sbjct: 10 VETPKYTVTKSGDGYEIREYPPAVAAEVTYDAS---EFKGDKDGGFQLLAKYIGVFGKPE 66
Query: 155 NTLKEKMEMTTPVYTSKNQS----------DGVKMDMTTPVYTAKMEDQDKW---TMSFI 201
N EK+ MT PV T + + + K++MT+PV T + + + TM F+
Sbjct: 67 NEKPEKIAMTAPVITKEGEKIAMTAPVITKESEKIEMTSPVVTKEGGGEGRKKLVTMQFL 126
Query: 202 LPSKY--GANLPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQF 259
LPS Y P P D V IKE + V+ FSG ++ V ++ KL L+ DG F
Sbjct: 127 LPSMYKKAEEAPRPTDERVVIKEEGGRKYGVIKFSGIASESVVSEKVKKLSSHLEKDG-F 185
Query: 260 KIKKGTSVEVAQYNPPFTLPFQRRNEIALEVE 291
KI G V +A+YNPP+TLP R NE+ + VE
Sbjct: 186 KI-TGDFV-LARYNPPWTLPPFRTNEVMIPVE 215
>ref|NP_565876.2| SOUL heme-binding family protein [Arabidopsis thaliana]
Length = 225
Score = 100 bits (249), Expect = 5e-20
Identities = 79/212 (37%), Positives = 108/212 (50%), Gaps = 26/212 (12%)
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQS-FNVLAEYL--FGK-- 154
+ET K+ V D YEIRE P AE T +F G F +LA+Y+ FGK
Sbjct: 20 VETPKYTVTKSGDGYEIREYPPAVAAEVTYDAS---EFKGDKDGGFQLLAKYIGVFGKPE 76
Query: 155 NTLKEKMEMTTPVYTSKNQS----------DGVKMDMTTPVYTAKMEDQDKW---TMSFI 201
N EK+ MT PV T + + + K++MT+PV T + + + TM F+
Sbjct: 77 NEKPEKIAMTAPVITKEGEKIAMTAPVITKESEKIEMTSPVVTKEGGGEGRKKLVTMQFL 136
Query: 202 LPSKY--GANLPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQF 259
LPS Y P P D V IKE + V+ FSG ++ V ++ KL L+ DG F
Sbjct: 137 LPSMYKKAEEAPRPTDERVVIKEEGGRKYGVIKFSGIASESVVSEKVKKLSSHLEKDG-F 195
Query: 260 KIKKGTSVEVAQYNPPFTLPFQRRNEIALEVE 291
KI G V +A+YNPP+TLP R NE+ + VE
Sbjct: 196 KI-TGDFV-LARYNPPWTLPPFRTNEVMIPVE 225
>ref|ZP_00412647.1| SOUL heme-binding protein [Arthrobacter sp. FB24]
gi|66869123|gb|EAL96491.1| SOUL heme-binding protein
[Arthrobacter sp. FB24]
Length = 201
Score = 95.1 bits (235), Expect = 2e-18
Identities = 62/192 (32%), Positives = 103/192 (53%), Gaps = 9/192 (4%)
Query: 101 ETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEK 160
E F V+ R ++E+R + VAE + K+ FD +G + +F +L Y+ G NT +E
Sbjct: 3 EQQPFDVVQRFPDFEVRRYPGHAVAEVKV--KAPFDSAG-NAAFRLLFGYISGNNTARES 59
Query: 161 MEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKY-GANLPLPKDSSVR 219
+ MT PV S S K+ MTTPV + ++ ++F+LP+ A P+P + V
Sbjct: 60 VSMTAPVLQSPAPSR--KLAMTTPVVQSGALGDSEFVVAFVLPASITAATAPVPNNPQVE 117
Query: 220 IKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLP 279
I+ +P + AV+ FSG + ++R L+EAL G +K + A+++PPF
Sbjct: 118 IRAVPGSVAAVLGFSGRGTEAAFEKRNSVLQEALAQAG---LKPVGAPRFARFDPPFKPW 174
Query: 280 FQRRNEIALEVE 291
F R+NE+ ++E
Sbjct: 175 FLRKNEVVQDIE 186
>gb|AAK24520.1| conserved hypothetical protein [Caulobacter crescentus CB15]
gi|16126788|ref|NP_421352.1| hypothetical protein CC2549
[Caulobacter crescentus CB15] gi|25360398|pir||D87565
conserved hypothetical protein CC2549 [imported] -
Caulobacter crescentus
Length = 208
Score = 94.0 bits (232), Expect = 5e-18
Identities = 60/192 (31%), Positives = 100/192 (51%), Gaps = 12/192 (6%)
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKE 159
+E FKV+ ++++R+ VAE T+ SG A++ F +LA Y+FG N ++
Sbjct: 26 VEEPVFKVVLHEGDFDVRDYPALVVAEVTV---SGDQKQAANRGFRLLAGYIFGGNRTRQ 82
Query: 160 KMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGAN-LPLPKDSSV 218
+ MT PV G + MT PV + + +W + F +PS+Y LP P D V
Sbjct: 83 SIAMTAPV---AQAPAGQTIAMTAPV--TQTQSAGQWVVRFTMPSRYSLEALPEPNDPQV 137
Query: 219 RIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTL 278
+++ IP +AV+ FSG + V+ + L++ L + +++ +AQYN P+T
Sbjct: 138 KLRLIPPSRLAVLRFSGLAGADTVEVKTADLKKRLSA---HQLQATGPATLAQYNTPWTP 194
Query: 279 PFQRRNEIALEV 290
F RRNE+ + V
Sbjct: 195 WFMRRNEVMIPV 206
>dbj|BAD33050.1| SOUL heme-binding protein-like [Oryza sativa (japonica
cultivar-group)] gi|50725456|dbj|BAD32927.1| SOUL
heme-binding protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 381
Score = 94.0 bits (232), Expect = 5e-18
Identities = 65/191 (34%), Positives = 94/191 (49%), Gaps = 25/191 (13%)
Query: 98 PDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTL 157
PD+ET K+ +L R NYEIR P+ + EA G +G+S FN + Y+FGKN
Sbjct: 208 PDIETPKYLILKRTANYEIRSYPPFLIVEA-----KGDKLTGSS-GFNNVTGYIFGKNAS 261
Query: 158 KEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSS 217
E + MTTPV+T SD D++ + +D D +LP P +
Sbjct: 262 SETIAMTTPVFT--QASDDKLSDVSIQIVLPMNKDLD--------------SLPAPNTEA 305
Query: 218 VRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFT 277
V ++++ I AV FSG +E V Q+E +LR L D +K +A+YN P T
Sbjct: 306 VNLRKVEGGIAAVKKFSGRPKEEIVIQKEKELRSQLLKD---VLKPQHGCLLARYNDPRT 362
Query: 278 LPFQRRNEIAL 288
F RNE+ +
Sbjct: 363 QSFIMRNEVLI 373
>ref|ZP_00529657.1| SOUL heme-binding protein [Chlorobium phaeobacteroides DSM 266]
gi|67774427|gb|EAM34113.1| SOUL heme-binding protein
[Chlorobium phaeobacteroides DSM 266]
Length = 198
Score = 92.0 bits (227), Expect = 2e-17
Identities = 60/186 (32%), Positives = 100/186 (53%), Gaps = 13/186 (6%)
Query: 108 LSRRDN-YEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTP 166
L ++D +E+R AE + G + +F+ LA Y+FGKN K+K+ MT P
Sbjct: 24 LQKKDGVFEVRHYGRTVYAETVVDGAYA---KTSGVAFSRLAGYIFGKNRAKQKIPMTAP 80
Query: 167 VYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGAN-LPLPKDSSVRIKEIPR 225
V + +K+ MT PV K D W MSF++P LP P D +V+++E
Sbjct: 81 VL---QEPVSLKIPMTAPVLQEKKGDG--WLMSFVMPDGSRLETLPEPLDPAVKLREAEG 135
Query: 226 KIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNE 285
+ VAV+ ++G +++ +++ L+E + G I + + A Y+PP+T+PF RRNE
Sbjct: 136 RSVAVIGYAGLHSEKNIRKYAGLLKEWIGKKGYRAISEPRA---ASYDPPWTIPFLRRNE 192
Query: 286 IALEVE 291
+ ++VE
Sbjct: 193 VQIDVE 198
>ref|ZP_00270599.1| hypothetical protein Rrub02000642 [Rhodospirillum rubrum]
Length = 179
Score = 89.4 bits (220), Expect = 1e-16
Identities = 62/182 (34%), Positives = 92/182 (50%), Gaps = 11/182 (6%)
Query: 112 DNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSK 171
D EIR P AE G + +F +L Y+ G NT ++ + MT PV
Sbjct: 7 DGVEIRHYGPRVAAEVAARHSGGA--GERTHAFRLLFAYITGANTARQNLPMTKPVGVGA 64
Query: 172 NQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANL-PLPKDSSVRIKEIPRKIVAV 230
++ MT PV T + F LP+ A P+P D V +++I + +AV
Sbjct: 65 VGGASQRLAMTIPVATGAGA-----ALQFFLPAGLTAQTAPVPSDPRVTLRDIAAQDMAV 119
Query: 231 VSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEIALEV 290
+ FSGF + EV +R+ +LR++L + G G +V Y+PPF+LPF RRNE+A+ V
Sbjct: 120 LGFSGFRHGIEVDRRKAQLRQSLTASGW--TASGEAVAYF-YDPPFSLPFLRRNEVAVPV 176
Query: 291 ER 292
ER
Sbjct: 177 ER 178
>gb|AAB84621.1| unknown [Methanothermobacter thermautotrophicus str. Delta H]
gi|15678143|ref|NP_275258.1| hypothetical protein MTH115
[Methanothermobacter thermautotrophicus str. Delta H]
gi|7482353|pir||B69020 hypothetical protein MTH115 -
Methanobacterium thermoautotrophicum (strain Delta H)
Length = 189
Score = 87.4 bits (215), Expect = 4e-16
Identities = 62/196 (31%), Positives = 101/196 (50%), Gaps = 12/196 (6%)
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQ-SFNVLAEYLFGKNTLK 158
+E+ ++ V + +EIR Y +A+ + F A F++LA Y+FG N K
Sbjct: 2 VESPEYTVELKDGKFEIRRYPGYILAQVDVEAS----FRDAMVIGFSILANYIFGGNRRK 57
Query: 159 EKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQD--KWTMSFILPSKYG-ANLPLPKD 215
E++ MT+PV T N ++ M PV +D D K+ +SF +PS Y LP P D
Sbjct: 58 EELPMTSPV-TGVNLGSSERIPMKVPVTEEVPDDADSGKYRISFTMPSSYTLETLPEPLD 116
Query: 216 SSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPP 275
+R +E + A FSG VN + QR +L+E L+ + I+ ++ +AQYN P
Sbjct: 117 DRIRFREEKDQRFAAYRFSGRVNSDMAAQRIAELKEWLERN---SIEPRSNFIIAQYNHP 173
Query: 276 FTLPFQRRNEIALEVE 291
F R+NE+ ++++
Sbjct: 174 AVPGFLRKNEVLVKID 189
>ref|ZP_00005985.2| hypothetical protein Rsph03001922 [Rhodobacter sphaeroides 2.4.1]
Length = 179
Score = 84.7 bits (208), Expect = 3e-15
Identities = 58/174 (33%), Positives = 82/174 (46%), Gaps = 26/174 (14%)
Query: 115 EIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSKNQS 174
EIR PY VAE TM G + ++ F VLA Y+FG N ++EMT PV
Sbjct: 25 EIRRYGPYLVAEVTMAGDRS---TAITRGFRVLARYIFGGNAESRRIEMTVPV------- 74
Query: 175 DGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGAN-LPLPKDSSVRIKEIPRKIVAVVSF 233
+ +D WT+ F +P+ A+ LP PKDS +R +P AV F
Sbjct: 75 ------------SQLPAGEDLWTVRFTMPAVRSASLLPAPKDSRIRFVTVPPSRQAVRRF 122
Query: 234 SGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEIA 287
SG+ D ++++ L + G + K Y+ P TLP+QRRNE+A
Sbjct: 123 SGWPTDHALRRQAEGLAHWIAERG---LPKREGPYFYFYDSPMTLPWQRRNEVA 173
>ref|NP_948933.1| hypothetical protein RPA3595 [Rhodopseudomonas palustris CGA009]
gi|39650513|emb|CAE29036.1| conserved hypothetical
protein [Rhodopseudomonas palustris CGA009]
Length = 209
Score = 81.3 bits (199), Expect = 3e-14
Identities = 66/196 (33%), Positives = 97/196 (48%), Gaps = 15/196 (7%)
Query: 101 ETVKFKVLSR-RDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKE 159
E + VL R D EIR P AE + + D + FN +A G + E
Sbjct: 26 EEPAYTVLDRPSDTIEIRRYAPRVAAEVDLERRGNADGQAFTLLFNYIAGANRGGSGTSE 85
Query: 160 KMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWT-MSFILPSKYGANL-PLPKDSS 217
++ MT PV ++ K+ MT PV TA QD+ T M F LP+ + A+ P P D
Sbjct: 86 RVAMTVPVDVARP----AKIAMTAPVETAT---QDRMTRMRFFLPATFTADTAPKPSDER 138
Query: 218 VRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFT 277
V+I +P + +A + FSG D +++RE +L AL + + + Y+ PFT
Sbjct: 139 VQIVTVPEQTIATLRFSGTGRD--LREREQQLIAALANTPWQPVGAPYGLF---YDAPFT 193
Query: 278 LPFQRRNEIALEVERK 293
LPF RRNE A+EV ++
Sbjct: 194 LPFVRRNEAAVEVAKR 209
>emb|CAD79099.1| conserved hypothetical protein [Rhodopirellula baltica SH 1]
gi|32476962|ref|NP_869956.1| hypothetical protein
RB11397 [Rhodopirellula baltica SH 1]
Length = 207
Score = 77.0 bits (188), Expect = 6e-13
Identities = 56/194 (28%), Positives = 94/194 (47%), Gaps = 25/194 (12%)
Query: 101 ETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEK 160
E+ ++KV+ N+E+RE P + AT + D G SF L Y+ G N ++K
Sbjct: 36 ESAEYKVIESDGNFEVREY-PDLMLVAT---STKIDAQGRDGSFMKLFRYISGANESEQK 91
Query: 161 MEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGA-NLPLPKDSSVR 219
+ MTTPV+ +++D + M F++P + +P P + V
Sbjct: 92 ISMTTPVFMENDKAD------------------SEVQMGFVMPKEVAVEGVPSPTGADVD 133
Query: 220 IKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDG--QFKIKKGTSVEVAQYNPPFT 277
+++ AV+ FSG +N + K+ E KLR ++S G + + VE A Y+PPFT
Sbjct: 134 VRKRSGGRFAVLRFSGRLNKKLAKESETKLRTWMESKGLAADDSPEASGVESASYDPPFT 193
Query: 278 LPFQRRNEIALEVE 291
RRNE+ + ++
Sbjct: 194 PGPLRRNEVLIRLK 207
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.129 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 428,309,608
Number of Sequences: 2540612
Number of extensions: 16421841
Number of successful extensions: 59995
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 59846
Number of HSP's gapped (non-prelim): 94
length of query: 295
length of database: 863,360,394
effective HSP length: 127
effective length of query: 168
effective length of database: 540,702,670
effective search space: 90838048560
effective search space used: 90838048560
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0010.19


