

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
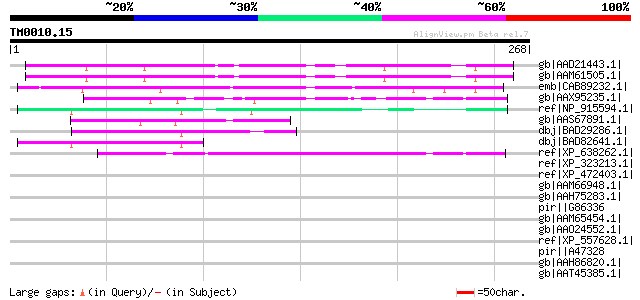
Query= TM0010.15
(268 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD21443.1| expressed protein [Arabidopsis thaliana] gi|18491... 166 5e-40
gb|AAM61505.1| unknown [Arabidopsis thaliana] 165 1e-39
emb|CAB89232.1| putative protein [Arabidopsis thaliana] gi|20465... 164 3e-39
gb|AAX95235.1| conserved hypothetical protein [Oryza sativa (jap... 82 2e-14
ref|NP_915594.1| P0489B03.23 [Oryza sativa (japonica cultivar-gr... 74 4e-12
gb|AAS67891.1| calcium/calmodulin protein kinase [Nicotiana taba... 74 4e-12
dbj|BAD29286.1| unknown protein [Oryza sativa (japonica cultivar... 56 1e-06
dbj|BAD82641.1| unknown protein [Oryza sativa (japonica cultivar... 53 8e-06
ref|XP_638262.1| hypothetical protein DDB0218709 [Dictyostelium ... 48 3e-04
ref|XP_323213.1| hypothetical protein [Neurospora crassa] gi|289... 46 0.001
ref|XP_472403.1| OSJNBa0073L04.10 [Oryza sativa (japonica cultiv... 43 0.008
gb|AAM66948.1| unknown [Arabidopsis thaliana] gi|18394848|ref|NP... 43 0.008
gb|AAH75283.1| Pinin, desmosome associated protein [Xenopus trop... 43 0.008
pir||G86336 hypothetical protein F14O10.9 - Arabidopsis thaliana... 43 0.008
gb|AAM65454.1| unknown [Arabidopsis thaliana] 42 0.014
gb|AAO24552.1| At1g76070 [Arabidopsis thaliana] gi|18411012|ref|... 42 0.014
ref|XP_557628.1| ENSANGP00000026170 [Anopheles gambiae str. PEST... 42 0.018
pir||A47328 natural killer cell tumor-recognition protein - human 42 0.023
gb|AAH86820.1| Zgc:103497 [Danio rerio] gi|56797744|ref|NP_00100... 41 0.040
gb|AAT45385.1| sperm nuclear basic protein PL-I isoform PLIb [Sp... 41 0.040
>gb|AAD21443.1| expressed protein [Arabidopsis thaliana] gi|18491123|gb|AAL69530.1|
At2g36220/F2H17.17 [Arabidopsis thaliana]
gi|15450733|gb|AAK96638.1| At2g36220/F2H17.17
[Arabidopsis thaliana] gi|25408452|pir||B84778
hypothetical protein At2g36220 [imported] - Arabidopsis
thaliana gi|18404079|ref|NP_565838.1| expressed protein
[Arabidopsis thaliana]
Length = 263
Score = 166 bits (421), Expect = 5e-40
Identities = 116/269 (43%), Positives = 150/269 (55%), Gaps = 36/269 (13%)
Query: 9 SACAGGSIDRKITCETLANKTDPPERHPDS-------PPESFWLSKDEEHDWFDRNALYE 61
S C GGS RKI CETLA+ + P + +S PPES++LS D + +W NA ++
Sbjct: 12 SVCIGGSDHRKIVCETLADDSTIPPYYNNSAVSPSDFPPESYFLSNDAQLEWLSDNAFFD 71
Query: 62 RKESTKG------STSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIK 115
RK+S KG S +S +SQRF L K+ IIGLPKPQK F +AK RR H N +
Sbjct: 72 RKDSQKGNSGILNSNPNSNPSSQRFLLKSKASIIGLPKPQKTCFNEAKQRR-HAGKNRVI 130
Query: 116 LFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPG 175
L KR S K+ S EPSSPKVSC+GRVRS+R+RSR R+ + S ++ P
Sbjct: 131 L--KRVGSRIKTDISLLEPSSPKVSCIGRVRSRRERSR----RMHRQKSSRVE-----PV 179
Query: 176 RIGRKHGFFERFRAIFR--SGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAE 233
+K GF FRAIFR G + + RET ++ ++ S + A SV D
Sbjct: 180 NRVKKPGFMASFRAIFRIKGGCKDVSARETHTSTRNTHDIRSRLPAEADEKSVFD----- 234
Query: 234 SISSEP--PGLGGMMRFASGRRSESWGVG 260
EP PGLGGM RFASGRR++ G G
Sbjct: 235 --GGEPVVPGLGGMTRFASGRRADLLGGG 261
>gb|AAM61505.1| unknown [Arabidopsis thaliana]
Length = 263
Score = 165 bits (418), Expect = 1e-39
Identities = 115/269 (42%), Positives = 150/269 (55%), Gaps = 36/269 (13%)
Query: 9 SACAGGSIDRKITCETLANKTDPPERHPDS-------PPESFWLSKDEEHDWFDRNALYE 61
S C GGS RKI CETLA+ + P + +S PPES++LS D + +W NA ++
Sbjct: 12 SVCIGGSDHRKIVCETLADDSTIPPYYNNSAVSPSDFPPESYFLSNDAQLEWLSDNAFFD 71
Query: 62 RKESTKG------STSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIK 115
RK+S KG S +S +SQRF L K+ IIGLPKPQK F +AK RR H N +
Sbjct: 72 RKDSQKGNSGILNSNPNSNPSSQRFLLKSKASIIGLPKPQKTCFNEAKQRR-HAGKNRVI 130
Query: 116 LFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPG 175
L KR S K+ S EPSSPKVSC+GRVRS+R+RSR R+ + S ++ P
Sbjct: 131 L--KRVGSRIKTDISLLEPSSPKVSCIGRVRSRRERSR----RMHRQKSSRVE-----PV 179
Query: 176 RIGRKHGFFERFRAIFR--SGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAE 233
+K GF FRAIFR G + + RET ++ ++ S + A S+ D
Sbjct: 180 NRVKKPGFMASFRAIFRIKGGCKDVSARETHTSTRNTHDIRSRLPAEAVEKSIFD----- 234
Query: 234 SISSEP--PGLGGMMRFASGRRSESWGVG 260
EP PGLGGM RFASGRR++ G G
Sbjct: 235 --GGEPVVPGLGGMTRFASGRRADLLGGG 261
>emb|CAB89232.1| putative protein [Arabidopsis thaliana] gi|20465291|gb|AAM20049.1|
unknown protein [Arabidopsis thaliana]
gi|18377716|gb|AAL67008.1| unknown protein [Arabidopsis
thaliana] gi|15231297|ref|NP_190839.1| expressed protein
[Arabidopsis thaliana] gi|11357760|pir||T49024
hypothetical protein F3C22.110 - Arabidopsis thaliana
Length = 289
Score = 164 bits (414), Expect = 3e-39
Identities = 117/289 (40%), Positives = 155/289 (53%), Gaps = 45/289 (15%)
Query: 5 ETLVSACAGGSIDRKITCETLANKTDPPERHP----------DSPPESFWLSKDEEHDWF 54
+ + SAC GGS DRKI+CETLA+ + + D PPES+ LSK+ + +W
Sbjct: 3 QVVASACTGGS-DRKISCETLADDNEDSPHNSKIRPVSISAVDFPPESYSLSKEAQLEWL 61
Query: 55 DRNALYERKESTKGSTSSSTTN--------SQRFSLNLKSRIIGLPKPQKPSFTDAKNRR 106
+ NA +ERKES KG++S+ +N S R SL K+ II LPKPQK F +AK RR
Sbjct: 62 NDNAFFERKESQKGNSSAPISNPNTNPNSSSHRISLKSKASIIRLPKPQKTCFNEAKKRR 121
Query: 107 NHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSS 166
N + + + PKR S KS + +EP SPKVSC+GRVRSKRDRSR R++ +
Sbjct: 122 NCRIARTL-MIPKRIGSRLKSDPTLSEPCSPKVSCIGRVRSKRDRSR----RMQRQKSGR 176
Query: 167 IDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAA--EDSTVTNSVVKARDST- 223
++ ++ P + +K GFF FRAIFR+G K + + A D V+ V R ST
Sbjct: 177 TNSFKDKPVPV-KKPGFFASFRAIFRTGGGCKDLSASGAHAPRRDVVVSPPRVSVRRSTD 235
Query: 224 --------------ASVNDASFAESI-SSEP--PGLGGMMRFASGRRSE 255
N SI EP PGLGGM RF SGRR +
Sbjct: 236 IRGRLPPGDVGKSSPQRNSTGSRRSIDGGEPVLPGLGGMTRFTSGRRPD 284
>gb|AAX95235.1| conserved hypothetical protein [Oryza sativa (japonica
cultivar-group)] gi|62732731|gb|AAX94850.1| expressed
protein [Oryza sativa (japonica cultivar-group)]
Length = 241
Score = 82.0 bits (201), Expect = 2e-14
Identities = 81/240 (33%), Positives = 112/240 (45%), Gaps = 47/240 (19%)
Query: 39 PPESFWLSKDEEHDWFDRNALYERKESTKGSTS-----------SSTTNSQRFSLNLK-- 85
PPES L ++ W D +Y+R +S K +T+ + SQRFS NLK
Sbjct: 12 PPESTRLRIGDDIAWSDVGGVYDRDDSLKENTNPKCILKNHLPGAHNGGSQRFSGNLKPT 71
Query: 86 -SRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVT-------KSVTSHAEPSSP 137
+ IIG+ R+H P +FPK+ V+VT K+ EP+SP
Sbjct: 72 AAPIIGI----SGKLGQGGKNRHHPPA----MFPKK-VAVTGGGGRNPKAAVPEHEPTSP 122
Query: 138 KVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRK 197
KVSC+G+V S R+R+R R R + AG PG +G G F R + R+K
Sbjct: 123 KVSCIGKVLSDRERAR----RGRRPAGRMVPAGGCCPG-LG---GLFRRSHS-----RKK 169
Query: 198 KAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRFASGRRSESW 257
A D + S R V +A+ A + ++ PGLGGMMRFASGRR+ W
Sbjct: 170 NAVECVDQSPPPLPPWAS---RRGEPKEVKEATPAAA-AAMAPGLGGMMRFASGRRAADW 225
>ref|NP_915594.1| P0489B03.23 [Oryza sativa (japonica cultivar-group)]
Length = 300
Score = 73.9 bits (180), Expect = 4e-12
Identities = 83/291 (28%), Positives = 114/291 (38%), Gaps = 83/291 (28%)
Query: 5 ETLVSACAG-GSIDRKITCETLANKTD------------PPERHPDSPPESFWLSKDEEH 51
E+LV AG G+ DRK++CET+ + PP PD PPES + +E
Sbjct: 7 ESLVCGVAGAGAGDRKVSCETVIAAGESGDASPPRMPPPPPPPDPDFPPESITIPIGDEV 66
Query: 52 DWFDRNALYERKESTKGSTS-SSTTNSQRFSLNLKSR---------------IIGLPKPQ 95
+ + N +Y+R +STKGST+ S + + KSR GLP
Sbjct: 67 AFSELNPIYDRDDSTKGSTNPKSAAGASSNPIPAKSRSNSTRIAGAPAAATTFFGLPASI 126
Query: 96 KPSFTDAKNRRNHKPCNNIKLFPKRSVS-------VTKSVTSHAEPSSPKVSCMGRVRSK 148
+P+FT +P L KRS S ++ EP SPKVSC+G+V S
Sbjct: 127 RPAFT------RRRPSQGRILPDKRSGSRGGGGGGSSRRGDGEEEPRSPKVSCIGKVLSD 180
Query: 149 RDR-SRSRSLR-LRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSA 206
R+R RSR R R + G + GR+H RKK + D
Sbjct: 181 RERYGRSRGRRWWRGLVAVLLCGGGCSCQGGGRRHA-------------RKKVALDEDHH 227
Query: 207 AEDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRFASGRRSESW 257
D + G+ M RF SGRR+ SW
Sbjct: 228 DGD--------------------------DDKQAGIAAMRRFKSGRRTASW 252
>gb|AAS67891.1| calcium/calmodulin protein kinase [Nicotiana tabacum]
gi|25053813|gb|AAN71903.1| calcium/calmodulin protein
kinase 1 [Nicotiana tabacum]
Length = 1415
Score = 73.9 bits (180), Expect = 4e-12
Identities = 46/129 (35%), Positives = 63/129 (48%), Gaps = 18/129 (13%)
Query: 32 PERHPDSPPESFWLSKDEEHDWFDRNALYERKEST-------KGSTSSSTTNSQRFSLNL 84
PE D PPESFW+ KD E DWFD NA +R S K + S + + + F+ L
Sbjct: 32 PENFLDLPPESFWIPKDSEQDWFDENATIQRMTSMMKLGFFGKANHHSKSFSHRSFTSTL 91
Query: 85 --------KSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSS 136
+ + LP+ +K S T+ ++ P + LF RS K + EP S
Sbjct: 92 FNHHQKPKSTSLFALPQSKKTSSTEGNLKQKKVPKS---LFRSRSEPGRKGIRHVREPGS 148
Query: 137 PKVSCMGRV 145
PKVSC+GRV
Sbjct: 149 PKVSCIGRV 157
>dbj|BAD29286.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|50251404|dbj|BAD28431.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 214
Score = 55.8 bits (133), Expect = 1e-06
Identities = 43/132 (32%), Positives = 63/132 (47%), Gaps = 23/132 (17%)
Query: 33 ERHPDSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSLNLKSR----- 87
E P SP ES WL E+ DW + A+ ER++STKG+++ + + + +R
Sbjct: 49 EYDPYSPAESLWLRIGEDIDWSEVGAVLEREDSTKGASNPKSAAACSCAGAPAARMPTCA 108
Query: 88 ----------IIGLP-KPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSS 136
I GLP +K S + RR + ++F +V V AEP S
Sbjct: 109 GGGGTAKAVVIAGLPAAARKASREHERRRRLGRARARARVFAGDAVEV-------AEPGS 161
Query: 137 PKVSCMGRVRSK 148
PKVSC+G VRS+
Sbjct: 162 PKVSCLGGVRSR 173
>dbj|BAD82641.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|56784813|dbj|BAD82034.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 249
Score = 53.1 bits (126), Expect = 8e-06
Identities = 40/125 (32%), Positives = 58/125 (46%), Gaps = 29/125 (23%)
Query: 5 ETLVSACAG-GSIDRKITCETLANKTD------------PPERHPDSPPESFWLSKDEEH 51
E+LV AG G+ DRK++CET+ + PP PD PPES + +E
Sbjct: 7 ESLVCGVAGAGAGDRKVSCETVIAAGESGDASPPRMPPPPPPPDPDFPPESITIPIGDEV 66
Query: 52 DWFDRNALYERKESTKGSTS-SSTTNSQRFSLNLKSR---------------IIGLPKPQ 95
+ + N +Y+R +STKGST+ S + + KSR GLP
Sbjct: 67 AFSELNPIYDRDDSTKGSTNPKSAAGASSNPIPAKSRSNSTRIAGAPAAATTFFGLPASI 126
Query: 96 KPSFT 100
+P+FT
Sbjct: 127 RPAFT 131
>ref|XP_638262.1| hypothetical protein DDB0218709 [Dictyostelium discoideum]
gi|60466725|gb|EAL64776.1| hypothetical protein
DDB0218709 [Dictyostelium discoideum]
Length = 1035
Score = 47.8 bits (112), Expect = 3e-04
Identities = 53/212 (25%), Positives = 87/212 (41%), Gaps = 9/212 (4%)
Query: 46 SKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNR 105
SK+ +H N ++T ++STT+ S +SR + +K S + N
Sbjct: 240 SKNSKHKKSSSNGNNNTNQTTSAIPTTSTTHKNSRS---RSRSRSKSRDRKKSHYN-NNN 295
Query: 106 RNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSK-RDRSRSRSLRLRNSRK 164
N NN K + +KS + S + + RS+ RDR RS R S
Sbjct: 296 NNSSSSNNNKSLTSKDKRRSKSRSRSKSKSKSRSRSRTKSRSRSRDRKRSDRYYRRRSSY 355
Query: 165 SSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTA 224
+S D G ++ + R ++ RS R + S + + T S RD+ +
Sbjct: 356 NSSDVGSDSRRSSKKFRNRSSRSKSKSRSRSRSSGRKHKSSRSRSRSTTKS---NRDTKS 412
Query: 225 SVNDASFAESISSEPPGLGGMMRFASGRRSES 256
S D S + S S+ G GG R S +R+++
Sbjct: 413 SRMDISRSRS-RSKSAGAGGRRRDRSPKRTKT 443
Score = 36.6 bits (83), Expect = 0.75
Identities = 32/105 (30%), Positives = 46/105 (43%), Gaps = 15/105 (14%)
Query: 62 RKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRS 121
RK + S S STT S R + KS + + + + S + R +R
Sbjct: 391 RKHKSSRSRSRSTTKSNR---DTKSSRMDISRSRSRSKSAGAGGR------------RRD 435
Query: 122 VSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSS 166
S ++ T+ S + + GR R KR RSR RS L + RKSS
Sbjct: 436 RSPKRTKTTDRSKSRSRSNSNGRSRKKRGRSRDRSRSLSSKRKSS 480
>ref|XP_323213.1| hypothetical protein [Neurospora crassa] gi|28918626|gb|EAA28297.1|
hypothetical protein [Neurospora crassa]
Length = 954
Score = 45.8 bits (107), Expect = 0.001
Identities = 40/137 (29%), Positives = 62/137 (45%), Gaps = 12/137 (8%)
Query: 54 FDRNALYERKESTKGSTSSSTTNSQRF--SLNLKSRIIGLPKPQKP-----SFTDAKNRR 106
FD NA+ R E+TK S + + T++ S N+ R + P P P S TD+K R
Sbjct: 187 FDGNAILSRIEATKLSAAPAPTSTTTIAHSNNISPRRVAPPPPPPPPALSRSNTDSKAAR 246
Query: 107 NHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSS 166
+ KP + PK ++S K++ + + S S ++ S+S S N R S
Sbjct: 247 SSKPSKS----PKSTIS-NKTMGASSFRHSASFSSAEQLPPSEKPSKSESSSSSNKRHSG 301
Query: 167 IDAGEENPGRIGRKHGF 183
PG + +K GF
Sbjct: 302 DGKESRVPGMLRKKSGF 318
>ref|XP_472403.1| OSJNBa0073L04.10 [Oryza sativa (japonica cultivar-group)]
gi|38346339|emb|CAE04668.2| OSJNBa0073L04.10 [Oryza
sativa (japonica cultivar-group)]
gi|38346320|emb|CAE02062.2| OJ000126_13.2 [Oryza sativa
(japonica cultivar-group)]
Length = 261
Score = 43.1 bits (100), Expect = 0.008
Identities = 44/162 (27%), Positives = 68/162 (41%), Gaps = 33/162 (20%)
Query: 11 CAGGSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDR--NALYERKESTKG 68
C GG ++ CET+A D+P ES + + W + A+ ER STKG
Sbjct: 11 CGGGEAGSRVACETIALPGCS-----DAPAESRCVRIGDGAIWAELAGGAVLERDGSTKG 65
Query: 69 STSS----------------STTNSQRFSLNLKSRII--GLPKPQKPSFTDAKNRRNHKP 110
S++ S+ S+R + K+ ++ GLP + + ++ P
Sbjct: 66 SSNPKAAAASGKGKKGGPRPSSAESRRLPVTGKAAVVICGLPAGKMVA-----QKKRLSP 120
Query: 111 C--NNIKLFPKRS-VSVTKSVTSHAEPSSPKVSCMGRVRSKR 149
C + P + V S +P SPKVSC G VRS+R
Sbjct: 121 CLGRGWRRAPAAAGARVFASEAVETDPGSPKVSCFGAVRSER 162
>gb|AAM66948.1| unknown [Arabidopsis thaliana] gi|18394848|ref|NP_564111.1|
expressed protein [Arabidopsis thaliana]
Length = 240
Score = 43.1 bits (100), Expect = 0.008
Identities = 40/136 (29%), Positives = 61/136 (44%), Gaps = 28/136 (20%)
Query: 125 TKSVTSHAEPSSPKVSCMGRVRSKRDR------SRSRSLRLRNSRKSSIDAGEENPGRIG 178
TK AEP+SPKVSC+G+V+ R + ++L+ +S S + E+N
Sbjct: 66 TKYEAVFAEPTSPKVSCIGQVKLARPKCPEKKNKAPKNLKTASSLSSCVIKEEDN----- 120
Query: 179 RKHGFFERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSE 238
G F + + IF R R+++S A + AR+ + DA A
Sbjct: 121 ---GSFSKLKRIF--SMRSYPSRKSNSTA------FAAAAAREHPIAEVDAVTA------ 163
Query: 239 PPGLGGMMRFASGRRS 254
P LG M +FAS R +
Sbjct: 164 APSLGAMKKFASSREA 179
>gb|AAH75283.1| Pinin, desmosome associated protein [Xenopus tropicalis]
gi|54020926|ref|NP_001005707.1| pinin, desmosome
associated protein [Xenopus tropicalis]
Length = 691
Score = 43.1 bits (100), Expect = 0.008
Identities = 57/225 (25%), Positives = 80/225 (35%), Gaps = 34/225 (15%)
Query: 20 ITCETLANKTDPPERHPDS-------------PPESFWLSKDEEHDWFDRNALYERKEST 66
+ ET A K PER DS PP S+ EH + + KE
Sbjct: 453 LEAETEATKEPEPEREADSPERPQKELELMFPPPASYESRTQPEHHFNVDEEHFAEKEQI 512
Query: 67 KGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTK 126
T ++ PKP S AK R+ N K+ +S S +
Sbjct: 513 VAVTVPVAETQVEYA----------PKP--ASVHKAKRSRSRGRTKN-KMGKSKSSSSSS 559
Query: 127 SVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGRI-GRKHGFFE 185
S +S + SS S RS S S R+S SS + E+ R GR H +
Sbjct: 560 SSSSSSSSSSGSSSSSSSSRSSSSSSSSSGSSSRDSSSSSSSSSSESRSRSRGRGHARDD 619
Query: 186 RFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDAS 230
+ RR H+ D A + + +S RDS S + +S
Sbjct: 620 K-------RRRSVEHKRRDGAGVERSHKSSKGSGRDSKGSKDKSS 657
>pir||G86336 hypothetical protein F14O10.9 - Arabidopsis thaliana
gi|9558595|gb|AAF88158.1| Contains similarity to a
hypothetical protein T23E18.1 gi|6573705 from
Arabidopsis thaliana BAC T23E18 gb|AC009978
Length = 313
Score = 43.1 bits (100), Expect = 0.008
Identities = 40/136 (29%), Positives = 61/136 (44%), Gaps = 28/136 (20%)
Query: 125 TKSVTSHAEPSSPKVSCMGRVRSKRDR------SRSRSLRLRNSRKSSIDAGEENPGRIG 178
TK AEP+SPKVSC+G+V+ R + ++L+ +S S + E+N
Sbjct: 66 TKYEAVFAEPTSPKVSCIGQVKLARPKCPEKKNKAPKNLKTASSLSSCVIKEEDN----- 120
Query: 179 RKHGFFERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSE 238
G F + + IF R R+++S A + AR+ + DA A
Sbjct: 121 ---GSFSKLKRIF--SMRSYPSRKSNSTA------FAAAAAREHPIAEVDAVTA------ 163
Query: 239 PPGLGGMMRFASGRRS 254
P LG M +FAS R +
Sbjct: 164 APSLGAMKKFASSREA 179
>gb|AAM65454.1| unknown [Arabidopsis thaliana]
Length = 252
Score = 42.4 bits (98), Expect = 0.014
Identities = 40/128 (31%), Positives = 61/128 (47%), Gaps = 28/128 (21%)
Query: 133 EPSSPKVSCMGRVR--------SKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFF 184
EP+SPKVSC+G+++ K++++ S + + +S E+ GR+ + F
Sbjct: 62 EPTSPKVSCIGQIKLGKSKCPTGKKNKAPSSLIPKISKTSTSSLTKEDEKGRLSKIKSIF 121
Query: 185 ERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGG 244
A R+ R K+H SAA++ VT VV STA+V P LG
Sbjct: 122 SFSPASGRNTSR-KSHPTAVSAADEHPVT--VV----STAAV-------------PSLGQ 161
Query: 245 MMRFASGR 252
M +FAS R
Sbjct: 162 MKKFASSR 169
>gb|AAO24552.1| At1g76070 [Arabidopsis thaliana] gi|18411012|ref|NP_565125.1|
expressed protein [Arabidopsis thaliana]
gi|6573705|gb|AAF17625.1| T23E18.1 [Arabidopsis
thaliana]
Length = 272
Score = 42.4 bits (98), Expect = 0.014
Identities = 40/128 (31%), Positives = 61/128 (47%), Gaps = 28/128 (21%)
Query: 133 EPSSPKVSCMGRVR--------SKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFF 184
EP+SPKVSC+G+++ K++++ S + + +S E+ GR+ + F
Sbjct: 82 EPTSPKVSCIGQIKLGKSKCPTGKKNKAPSSLIPKISKTSTSSLTKEDEKGRLSKIKSIF 141
Query: 185 ERFRAIFRSGRRKKAHRETDSAAEDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGG 244
A R+ R K+H SAA++ VT VV STA+V P LG
Sbjct: 142 SFSPASGRNTSR-KSHPTAVSAADEHPVT--VV----STAAV-------------PSLGQ 181
Query: 245 MMRFASGR 252
M +FAS R
Sbjct: 182 MKKFASSR 189
>ref|XP_557628.1| ENSANGP00000026170 [Anopheles gambiae str. PEST]
gi|55239687|gb|EAL40209.1| ENSANGP00000026170 [Anopheles
gambiae str. PEST]
Length = 346
Score = 42.0 bits (97), Expect = 0.018
Identities = 38/143 (26%), Positives = 61/143 (42%), Gaps = 11/143 (7%)
Query: 87 RIIGLPKPQKPSFTDAKNRRNHKPCNNIKLF-PKRSVSV--TKSVTSHAEPSSPKVSCMG 143
R L + KP K + H P N I+ + P S + SV+ HAEPS P
Sbjct: 138 RAESLDEHGKPIEVGLKTKNYHDPLNVIRRYVPSASFKLPSATSVSRHAEPSVPCPDTSP 197
Query: 144 RVRSKRD------RSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRK 197
V+SK ++ + R R SS D + R + ++ + + R+K
Sbjct: 198 AVQSKTQYTALIGQTGRKESRKRKLSSSSSDTSRHSKKRKKKHKKHSDKKKK--KEKRKK 255
Query: 198 KAHRETDSAAEDSTVTNSVVKAR 220
K H+++ S ++ST V KA+
Sbjct: 256 KKHKKSSSDEDESTEDEEVAKAK 278
>pir||A47328 natural killer cell tumor-recognition protein - human
Length = 1403
Score = 41.6 bits (96), Expect = 0.023
Identities = 59/212 (27%), Positives = 89/212 (41%), Gaps = 30/212 (14%)
Query: 37 DSPPESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSL-NLKSR--IIGLPK 93
DSPP S W K E W YER + K T+ +SL N+K K
Sbjct: 666 DSPPPSRW--KPGEKPW---KPSYERIQEMKAKTTHLLPIQSTYSLANIKETGSSSSYHK 720
Query: 94 PQKPSFTDAK-----NRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSK 148
+K S +D + R+ + + RS S ++S T S S R RS
Sbjct: 721 REKNSESDQSTYSKYSDRSSESSPRSRSRSSRSRSYSRSYTR----SRSLASSHSRSRSP 776
Query: 149 RDRSRSR---SLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDS 205
RS SR S + SR SS + + GR ++ R R+ SG++ + S
Sbjct: 777 SSRSHSRNKYSDHSQCSRSSSYTSISSDDGRRAKR-----RLRS---SGKKNSVSHKKHS 828
Query: 206 AAEDSTVTNSVVKARDSTASVNDASFAESISS 237
++ + T+ + VK RD ++ V ++ES SS
Sbjct: 829 SSSEKTLHSKYVKGRDRSSCVR--KYSESRSS 858
>gb|AAH86820.1| Zgc:103497 [Danio rerio] gi|56797744|ref|NP_001008732.1|
hypothetical protein LOC494133 [Danio rerio]
Length = 355
Score = 40.8 bits (94), Expect = 0.040
Identities = 47/176 (26%), Positives = 83/176 (46%), Gaps = 13/176 (7%)
Query: 59 LYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFP 118
L E K + S S+S + S R +SR + Q S + +++R H + + +
Sbjct: 182 LVEDKPRRRRSYSASRSRS-RSRRRSRSRSRRSSRSQSRSRSRSRSRSKHSRSRSGRKYR 240
Query: 119 KRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRS-LRLRNSRKSSIDAGEENPGRI 177
RS S KS H++ S K R R+++ RSRSRS + ++ R S + E++ R
Sbjct: 241 SRSRSSRKS---HSKSHSKKSRSRSRSRTEKSRSRSRSRSKAKSERDSRSRSREKSTSRK 297
Query: 178 G-------RKHGFFERFRAIFRSGRRKKAHRETDSAAEDS-TVTNSVVKARDSTAS 225
R++G ER ++ RS K+ + +S + S + + S ++R +AS
Sbjct: 298 SRSRSASPRENGDGERVKSTSRSPSPKENRHQLESPRKRSASRSRSKSRSRSRSAS 353
>gb|AAT45385.1| sperm nuclear basic protein PL-I isoform PLIb [Spisula solidissima]
Length = 455
Score = 40.8 bits (94), Expect = 0.040
Identities = 42/170 (24%), Positives = 71/170 (41%), Gaps = 22/170 (12%)
Query: 62 RKESTKGSTSSSTTNSQRFSLN-----LKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKL 116
+ +STK STS+ + S++ S + K R + + + +K RR +
Sbjct: 43 KSKSTKRSTSTKRSKSRKRSASKKRSASKKRSASKKRSRTSKRSASKKRRRSR------- 95
Query: 117 FPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGR 176
KRS S +S + + S S R KR S+ RS + SR S A ++ R
Sbjct: 96 --KRSASKKRSASKRRKSSKRSASKKRRRSRKRSASKKRSASKKRSRSSKRSASKKR--R 151
Query: 177 IGRKHGFFERFRAIFRSGRR------KKAHRETDSAAEDSTVTNSVVKAR 220
RK ++ ++ RS +R +K+ + + S +V S K R
Sbjct: 152 RSRKRSASKKRKSTKRSAKRSASKKPRKSRKRSASKKRSKSVKRSASKKR 201
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.126 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 445,614,557
Number of Sequences: 2540612
Number of extensions: 17308631
Number of successful extensions: 47464
Number of sequences better than 10.0: 322
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 321
Number of HSP's that attempted gapping in prelim test: 46724
Number of HSP's gapped (non-prelim): 661
length of query: 268
length of database: 863,360,394
effective HSP length: 126
effective length of query: 142
effective length of database: 543,243,282
effective search space: 77140546044
effective search space used: 77140546044
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0010.15


