

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.15
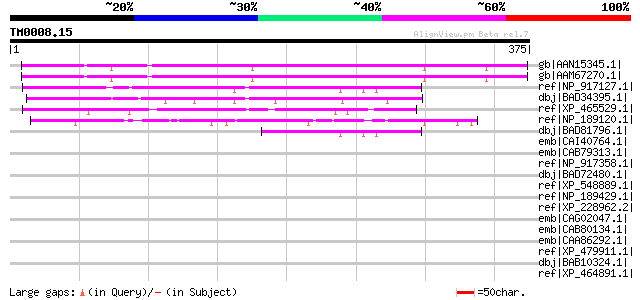
(375 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN15345.1| putative knotted-like homeobox protein [Arabidops... 280 7e-74
gb|AAM67270.1| unknown [Arabidopsis thaliana] 272 1e-71
ref|NP_917127.1| P0421H07.29 [Oryza sativa (japonica cultivar-gr... 135 2e-30
dbj|BAD34395.1| kelch repeat-containing F-box-like protein [Oryz... 125 2e-27
ref|XP_465529.1| kelch repeat-containing F-box-like protein [Ory... 106 1e-21
ref|NP_189120.1| F-box family protein [Arabidopsis thaliana] 84 5e-15
dbj|BAD81796.1| kelch repeat-containing F-box-like protein [Oryz... 51 5e-05
emb|CAI40764.1| novel protein [Homo sapiens] gi|21750834|dbj|BAC... 45 0.004
emb|CAB79313.1| putative protein [Arabidopsis thaliana] gi|44540... 45 0.005
ref|NP_917358.1| P0684E06.20 [Oryza sativa (japonica cultivar-gr... 44 0.008
dbj|BAD72480.1| fimbriata-like [Oryza sativa (japonica cultivar-... 44 0.008
ref|XP_548889.1| PREDICTED: similar to hypothetical protein FLJ3... 44 0.010
ref|NP_189429.1| kelch repeat-containing protein [Arabidopsis th... 42 0.023
ref|XP_228962.2| PREDICTED: similar to hypothetical protein FLJ3... 41 0.052
emb|CAG02047.1| unnamed protein product [Tetraodon nigroviridis] 41 0.052
emb|CAB80134.1| putative protein [Arabidopsis thaliana] gi|29110... 40 0.089
emb|CAA86292.1| scruin [Limulus polyphemus] gi|2497944|sp|Q25390... 40 0.12
ref|XP_479911.1| kelch repeat-containing protein -like [Oryza sa... 40 0.15
dbj|BAB10324.1| unnamed protein product [Arabidopsis thaliana] g... 40 0.15
ref|XP_464891.1| F-box-like protein [Oryza sativa (japonica cult... 39 0.26
>gb|AAN15345.1| putative knotted-like homeobox protein [Arabidopsis thaliana]
gi|21539423|gb|AAM53264.1| putative knotted-like
homeobox protein [Arabidopsis thaliana]
gi|8778589|gb|AAF79597.1| F28C11.3 [Arabidopsis
thaliana] gi|18395236|ref|NP_564193.1| kelch
repeat-containing F-box family protein [Arabidopsis
thaliana] gi|25402982|pir||G86367 protein F28C11.3
[imported] - Arabidopsis thaliana
gi|9295700|gb|AAF87006.1| F26F24.26 [Arabidopsis
thaliana]
Length = 394
Score = 280 bits (715), Expect = 7e-74
Identities = 160/380 (42%), Positives = 221/380 (58%), Gaps = 18/380 (4%)
Query: 9 EEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRAS 68
EE+ I GD+LE+ILS++PL+ L AC VS +W AVF SL +T+ PWL + Q
Sbjct: 16 EESSIDGDILESILSYLPLLDLDSACQVSKSWNRAVFYSLRRLKTM-PWLFVYNQRNSPP 74
Query: 69 HVIT--AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALH 126
+ + A AYDP+S W+E+ H S RSSHSTLLY LSPA F+FS DA H
Sbjct: 75 YTMATMAMAYDPKSEAWIELNTASSPVEHVSVA---RSSHSTLLYALSPARFSFSTDAFH 131
Query: 127 LDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMES--NTWVMCQSMP 184
L W H PRVWR DPIVA VG+ +++AGG CDFE+D AVE++D+ES W C+SMP
Sbjct: 132 LTWQHVAPPRVWRIDPIVAVVGRSLIIAGGVCDFEEDRFAVELFDIESGDGAWERCESMP 191
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGYCVT-G 243
L S+ STWLSVAV+ E++ VTE+ SG+T F+ V+ W DLCP + Y + G
Sbjct: 192 DFLYESASSTWLSVAVSSEKMYVTEKRSGVTCSFNPVTRSWTKLLDLCPGECSLYSRSIG 251
Query: 244 MLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMT-EEVGEMPEELLEKFKGDSDE--LG 300
RL++AG++GD + ++LW V S + E +G MPE LEK +G + + L
Sbjct: 252 FSVNRLIMAGIIGDEYNPTGIELWEVIDSDESHLKFESIGSMPETYLEKLRGINSDWPLT 311
Query: 301 SLEVTWVGNFVYLRNTLEI-EELVMCEVVNGSRCEWRSVRNAAV-----DGGTRMVVCGS 354
S+ + VG+ VY+ E E+V E+ G C+WR++ NA R++V S
Sbjct: 312 SIVLNAVGDMVYVHGAAENGGEIVAAEIEGGKLCKWRTLPNADATWKKSHAAERVIVACS 371
Query: 355 EVRLEDLQSAMVSGIQTCRT 374
V DL+ A + + T
Sbjct: 372 NVGFSDLKLAFRNNLSFLST 391
>gb|AAM67270.1| unknown [Arabidopsis thaliana]
Length = 394
Score = 272 bits (696), Expect = 1e-71
Identities = 158/380 (41%), Positives = 219/380 (57%), Gaps = 18/380 (4%)
Query: 9 EEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRAS 68
E+ I GD+LE+ILS++PL+ L A VS +W AVF SL +T+ PWL + Q
Sbjct: 16 EQRSIDGDILESILSYLPLLDLDSASQVSKSWNRAVFYSLRRLKTM-PWLFVYNQRNSPP 74
Query: 69 HVIT--AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALH 126
+ + A AYDP+S W+E+K H S RSSHSTLLY LSPA F+FS DA H
Sbjct: 75 YTMATMAMAYDPKSEAWIELKTASSPVEHVSVA---RSSHSTLLYALSPARFSFSTDAFH 131
Query: 127 LDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMES--NTWVMCQSMP 184
L W H PRVWR DPIVA VG+ +++AGG CDFE+D AVE++D+ES W C+SMP
Sbjct: 132 LTWQHVSPPRVWRIDPIVAVVGRSLIIAGGVCDFEEDRFAVELFDIESGDGAWERCESMP 191
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGYCVT-G 243
L S+ STWLSVAV+ E++ VTE+ SG+T F+ V+ W DLCP + Y + G
Sbjct: 192 DFLYESASSTWLSVAVSSEKMYVTEKRSGVTCSFNPVTRXWTKLLDLCPGECSLYSRSIG 251
Query: 244 MLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMT-EEVGEMPEELLEKFKGDSDE--LG 300
RL++A ++GD + ++LW V S + E +G MPE LEK +G + + L
Sbjct: 252 FSVNRLIMAWIIGDEYNPTGIELWEVIDSDESHLKFESIGSMPETYLEKLRGINSDWPLT 311
Query: 301 SLEVTWVGNFVYLRNTLEI-EELVMCEVVNGSRCEWRSVRNAAV-----DGGTRMVVCGS 354
S+ + VG+ VY+ E E+V E+ G C+WR++ NA R++V S
Sbjct: 312 SIVLNAVGDMVYVHGAAENGGEIVAAEIEGGKLCKWRTLPNADATWKKSHAAERVIVACS 371
Query: 355 EVRLEDLQSAMVSGIQTCRT 374
V DL+ A + + T
Sbjct: 372 NVGFSDLKLAFRNNLSFLST 391
>ref|NP_917127.1| P0421H07.29 [Oryza sativa (japonica cultivar-group)]
Length = 426
Score = 135 bits (341), Expect = 2e-30
Identities = 104/303 (34%), Positives = 140/303 (45%), Gaps = 23/303 (7%)
Query: 10 EAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRASH 69
E ++GDVLE+++ VP L + VS W AV ++L PWL++ R
Sbjct: 41 EMDLYGDVLESVVERVPAADLAASARVSREWLRAVRAALRRRPRRLPWLVVHLHGRRRRT 100
Query: 70 VITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDW 129
AYDP S WV + P PS VR + + LS + A S D L D
Sbjct: 101 A----AYDPHSGAWVTV--PAARHDTPSHVRLVRGAGGDRVCALSLSGLAVSGDPLGKDV 154
Query: 130 HHA-PAPRVWRTDPIVARVGQRVVVAGGACDFE----DDPLAVEMYDMESNTWVMCQSMP 184
A AP VWR DP+ A VG RVV GGAC +D VE++ ES +W C MP
Sbjct: 155 CVALKAPGVWRVDPVFAAVGDRVVALGGACQLALGEGEDASVVEVH--ESGSWTACGPMP 212
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSV---GYCV 241
A L+ S+ +TWLSVA + V +T+ S+G FD +W L PD +V G
Sbjct: 213 AELRESAAATWLSVAATDQRVYLTDRSTGWASWFDPAKQQWGPTCRLRPDATVSTWGLAP 272
Query: 242 TGMLGKRLMVAGV--VGDGEDVKA---VKLWAVKG-GLGSGMTEEVGEMPEELLEK-FKG 294
+RL++ G G E K+ ++ W V G GL MP E+ E+ F
Sbjct: 273 GRGGAERLVLFGAKRCGRAEQAKSRVVIQAWEVDGDGLALSRGAAHDTMPGEMSERLFPR 332
Query: 295 DSD 297
D D
Sbjct: 333 DED 335
>dbj|BAD34395.1| kelch repeat-containing F-box-like protein [Oryza sativa (japonica
cultivar-group)] gi|50725309|dbj|BAD34311.1| kelch
repeat-containing F-box-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 394
Score = 125 bits (314), Expect = 2e-27
Identities = 95/309 (30%), Positives = 136/309 (43%), Gaps = 27/309 (8%)
Query: 13 IHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRASHVIT 72
+HGDVLE+ + VP L A VS W AV ++L PWL++ R +
Sbjct: 4 LHGDVLESAVERVPAPDLAAAALVSREWLRAVRAALRRRMLRLPWLVVHVIHLRGQRRLA 63
Query: 73 AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLY--------TLSPAEFAFSIDA 124
A AYDPRS W+ + P + H + + SH L+ LS + A + DA
Sbjct: 64 A-AYDPRSGAWLAVPTAPPAR-HGATSPPQPHSHVRLMRGASGDRVCALSLSGLAVARDA 121
Query: 125 LHLDWHHA----PAPRVWRTDPIVARVGQRVVVAGGACDFE----DDPLAVEMYDMESNT 176
L +D AP VWR DP++A VG RVV GGAC +D AVE++ E
Sbjct: 122 LGMDDDALVVALKAPGVWRVDPVLAAVGDRVVAMGGACRLALGDGEDTSAVEVH--ERGG 179
Query: 177 WVMCQSMPAMLKGS--SGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPD 234
W C ++P L+ S + +TWLS A + V V + ++G FD +W L PD
Sbjct: 180 WTHCGAVPTALRESAAAAATWLSTAATDQRVYVADRATGTASWFDPAKQQWGPTSRLRPD 239
Query: 235 QSVGY--CVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKG---GLGSGMTEEVGEMPEELL 289
+V G G ++ V + ++ W V G L G MP E+
Sbjct: 240 ATVSTWGLAAGRAGAEKIILFGVKHADSRVVIRSWEVDGDSLSLSHGAAAAHDTMPSEMS 299
Query: 290 EKFKGDSDE 298
E+ D+
Sbjct: 300 ERLFPHGDD 308
>ref|XP_465529.1| kelch repeat-containing F-box-like protein [Oryza sativa (japonica
cultivar-group)] gi|47497505|dbj|BAD19558.1| kelch
repeat-containing F-box-like protein [Oryza sativa
(japonica cultivar-group)] gi|47496883|dbj|BAD19847.1|
kelch repeat-containing F-box-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 401
Score = 106 bits (264), Expect = 1e-21
Identities = 97/305 (31%), Positives = 127/305 (40%), Gaps = 34/305 (11%)
Query: 10 EAPIHGDVLETILSHVPLIHLVPA-CHVSNTWKHAVFSSLAHTRTIK---PWLIILTQST 65
++ +HGD+LE +L VP L + VS W+ A + R + P L+
Sbjct: 8 DSVLHGDLLECVLLRVPHGELTASPALVSREWRRAAREAHQRHRRRRRHLPCLVAHVHGA 67
Query: 66 RASHVITAHAYDPRSHVWVE--MKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSID 123
A + H YDPR+ W ++ L A A +Y LS A A S D
Sbjct: 68 AAGVGRSTHVYDPRAGAWASDGWRVAGALPVRRCACAG-----GDRVYALSLASMAVSED 122
Query: 124 ALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSM 183
A+ W P PRVWR DP+VA VG VVV GG C VE+ D E W C M
Sbjct: 123 AVGAAWRELPPPRVWRVDPVVAAVGPHVVVLGGGCGATAAAGVVEVLD-EGAGWATCPPM 181
Query: 184 PAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDL-CPD-------Q 235
PA L S W+S A + V V E +G FD + +W L P+ +
Sbjct: 182 PAPL----ASRWVSSAASERRVYVVERRTGWASWFDPAARQWGPARQLQLPEGNNTASVE 237
Query: 236 SVGYC-VT----GMLGKRLMVAGVVGDGEDVKAVKLWAVKGG-LGSGMTEEVGEMPEELL 289
S C VT G +RL+V G G+ V LW V G L MP E+
Sbjct: 238 SWAACGVTTSGGGGASERLLVLAGGGGGK----VSLWGVDGDTLLLDAEANNTSMPPEMS 293
Query: 290 EKFKG 294
E+ G
Sbjct: 294 ERLGG 298
>ref|NP_189120.1| F-box family protein [Arabidopsis thaliana]
Length = 383
Score = 84.3 bits (207), Expect = 5e-15
Identities = 93/347 (26%), Positives = 152/347 (43%), Gaps = 45/347 (12%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFS---SLAHTRTIKPWLIILTQSTRASHVIT 72
DV E+IL H+P+ LV VS W+ + S S + + + PWL + +S
Sbjct: 19 DVTESILYHLPIPSLVRFTLVSKQWRSLITSLPPSPSPSPSSPPWLFLFGIHNTSSFHNQ 78
Query: 73 AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHST-LLYTLSPAEFAFSIDALHLDWHH 131
+ A+DP S+ W ++LP PS++++ S L+T +P F+FS L +W
Sbjct: 79 SFAFDPLSNTW--LRLP------PSSSSSDHLVGSNRFLFTTAP-RFSFS-PILKPNWRF 128
Query: 132 APAPRVWRTDPIV------ARVGQRVVVAG-----GACDFEDDPLAVEMYDMESNTWVMC 180
R R +P++ + + ++V G G D E+ LAV++YD ++W +C
Sbjct: 129 TSPVRFPRINPLLTVFTTLSNSSKLILVGGSSRIGGLVDIEER-LAVQIYDPVLDSWELC 187
Query: 181 QSMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGM--TFCFDTVSMKWDGPYDLCPDQSVG 238
+PA + L+ A+ V + S +FC D S W L P +
Sbjct: 188 SPLPADFRSGQDHQTLTSALFKRRFYVFDNYSCFISSFCLD--SYTWSDVQTLKP-PGLS 244
Query: 239 YCVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMTEEVGEMPEELLEKFKGDSDE 298
+ L++ G+ G + LW+++ GS E+ MPE+LL DE
Sbjct: 245 FAYLNSCNGMLVLGGMCG-----FSFNLWSIEE--GSMEFSEIAVMPEDLLFGLVDSDDE 297
Query: 299 ---LGSLEVTWVGNFVYLRNTLEIEEL--VMCEVVNGSR--CEWRSV 338
SL+ GN VY+ N ++ +CE+ G C WR V
Sbjct: 298 DDKFRSLKCAGSGNLVYVFNDDCHKKFPACVCEIGGGENGICSWRRV 344
>dbj|BAD81796.1| kelch repeat-containing F-box-like protein [Oryza sativa (japonica
cultivar-group)] gi|56784189|dbj|BAD81574.1| kelch
repeat-containing F-box-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 216
Score = 51.2 bits (121), Expect = 5e-05
Identities = 41/125 (32%), Positives = 58/125 (45%), Gaps = 10/125 (8%)
Query: 183 MPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSV---GY 239
MPA L+ S+ +TWLSVA + V +T+ S+G FD +W L PD +V G
Sbjct: 1 MPAELRESAAATWLSVAATDQRVYLTDRSTGWASWFDPAKQQWGPTCRLRPDATVSTWGL 60
Query: 240 CVTGMLGKRLMVAGV--VGDGEDVKA---VKLWAVKG-GLGSGMTEEVGEMPEELLEK-F 292
+RL++ G G E K+ ++ W V G GL MP E+ E+ F
Sbjct: 61 APGRGGAERLVLFGAKRCGRAEQAKSRVVIQAWEVDGDGLALSRGAAHDTMPGEMSERLF 120
Query: 293 KGDSD 297
D D
Sbjct: 121 PRDED 125
>emb|CAI40764.1| novel protein [Homo sapiens] gi|21750834|dbj|BAC03848.1| unnamed
protein product [Homo sapiens]
gi|23397572|ref|NP_695002.1| hypothetical protein
LOC257240 [Homo sapiens]
Length = 644
Score = 45.1 bits (105), Expect = 0.004
Identities = 39/138 (28%), Positives = 56/138 (40%), Gaps = 25/138 (18%)
Query: 75 AYDPRSHVWVEM-KLPHPLKGHPSATAAVRSSHSTLLYTL---SPAEFAFS--------- 121
A+D +H W + +LP PL GH TA L+ L SP+ A S
Sbjct: 338 AFDVYNHRWRSLTQLPTPLLGHSVCTAG------NFLFVLGGESPSGSASSPLADDSRVV 391
Query: 122 ------IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESN 175
D W PA R R VG+R++ GG + +VEMYD+ +
Sbjct: 392 TAQVHRYDPRFHAWTEVPAMREARAHFWCGAVGERLLAVGGLGAGGEVLASVEMYDLRRD 451
Query: 176 TWVMCQSMPAMLKGSSGS 193
W ++P L G +G+
Sbjct: 452 RWTAAGALPRALHGHAGA 469
>emb|CAB79313.1| putative protein [Arabidopsis thaliana] gi|4454027|emb|CAA23024.1|
putative protein [Arabidopsis thaliana]
gi|67633756|gb|AAY78802.1| kelch repeat-containing F-box
family protein [Arabidopsis thaliana]
gi|15236530|ref|NP_194089.1| kelch repeat-containing
F-box family protein [Arabidopsis thaliana]
gi|7448249|pir||T05590 hypothetical protein F9D16.50 -
Arabidopsis thaliana
Length = 383
Score = 44.7 bits (104), Expect = 0.005
Identities = 46/156 (29%), Positives = 67/156 (42%), Gaps = 24/156 (15%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLK 188
WH AP+ RV R P + R+ V GG CD D +E++D ++ TW Q +
Sbjct: 159 WHEAPSMRVGRVFPSACTLDGRIYVTGG-CDNLDTMNWMEIFDTKTQTWEFLQIPSEEI- 216
Query: 189 GSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPD------QSVGYCVT 242
GS +LSV+ G V V + +T+ KW D+C + S YCV
Sbjct: 217 -CKGSEYLSVSYQG-TVYVKSDEKDVTYKMH--KGKW-READICMNNGWSLSSSSSYCVV 271
Query: 243 GMLGKRLMVAGVVGDGE----DVKAVKLWAVKGGLG 274
+ R +GE D+K + W + GLG
Sbjct: 272 ENVFYRYC------EGEIRWYDLKN-RAWTILKGLG 300
>ref|NP_917358.1| P0684E06.20 [Oryza sativa (japonica cultivar-group)]
Length = 406
Score = 43.9 bits (102), Expect = 0.008
Identities = 21/77 (27%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSS---LAHTRTIKPWLIILTQSTRASHVIT 72
D+LE I + +P++ ++ + V W ++SS H +PW + T + A+
Sbjct: 50 DILERIFTFLPIVSMIRSTAVCKRWHDIIYSSRFLWTHMLPQRPWYFMFTSNESAA---- 105
Query: 73 AHAYDPRSHVWVEMKLP 89
+AYDP W +++LP
Sbjct: 106 GYAYDPILRKWYDLELP 122
>dbj|BAD72480.1| fimbriata-like [Oryza sativa (japonica cultivar-group)]
gi|55773855|dbj|BAD72393.1| fimbriata-like [Oryza sativa
(japonica cultivar-group)]
Length = 411
Score = 43.9 bits (102), Expect = 0.008
Identities = 21/77 (27%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSS---LAHTRTIKPWLIILTQSTRASHVIT 72
D+LE I + +P++ ++ + V W ++SS H +PW + T + A+
Sbjct: 55 DILERIFTFLPIVSMIRSTAVCKRWHDIIYSSRFLWTHMLPQRPWYFMFTSNESAA---- 110
Query: 73 AHAYDPRSHVWVEMKLP 89
+AYDP W +++LP
Sbjct: 111 GYAYDPILRKWYDLELP 127
>ref|XP_548889.1| PREDICTED: similar to hypothetical protein FLJ34960 [Canis
familiaris]
Length = 1028
Score = 43.5 bits (101), Expect = 0.010
Identities = 39/138 (28%), Positives = 54/138 (38%), Gaps = 25/138 (18%)
Query: 75 AYDPRSHVWVEM-KLPHPLKGHPSATAAVRSSHSTLLYTL---SPAEFAFS--------- 121
A+D +H W + +LP PL GH TA L+ L SP E A S
Sbjct: 719 AFDVYNHRWRSLTRLPAPLLGHSVCTAG------NFLFVLGGESPPEAASSPPADGPRPV 772
Query: 122 ------IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESN 175
D W PA R R VG+ ++ GG +VEMYD+ +
Sbjct: 773 TAEVHRYDPRFHAWTALPAMREARAHFWCGAVGEGLLAVGGLGADGQALASVEMYDLRRD 832
Query: 176 TWVMCQSMPAMLKGSSGS 193
W ++P L G +G+
Sbjct: 833 RWTAAAALPRALHGHAGA 850
>ref|NP_189429.1| kelch repeat-containing protein [Arabidopsis thaliana]
Length = 294
Score = 42.4 bits (98), Expect = 0.023
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 5/122 (4%)
Query: 120 FSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVM 179
F ID LH + P+ RV R V ++ V GG + VE++D+E TW
Sbjct: 65 FVIDCLHGTFQFLPSMRVPRGCAAFGIVDGKIYVIGGYNKADSLDNWVEVFDLEKQTW-- 122
Query: 180 CQSMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGY 239
+S + L V +++ + + +G+ FD W+ + L D VG
Sbjct: 123 -ESFSGLCNEELSKITLKSVVMNKKIYIMDRGNGIV--FDPKKGVWERDFLLDRDWVVGS 179
Query: 240 CV 241
CV
Sbjct: 180 CV 181
>ref|XP_228962.2| PREDICTED: similar to hypothetical protein FLJ34960 [Rattus
norvegicus]
Length = 645
Score = 41.2 bits (95), Expect = 0.052
Identities = 34/132 (25%), Positives = 54/132 (40%), Gaps = 13/132 (9%)
Query: 75 AYDPRSHVWVEM-KLPHPLKGHPSATA----------AVRSSHSTLLY--TLSPAEFAFS 121
++D +H W + +LP PL GH T ++ S S+ L + S
Sbjct: 339 SFDVYNHSWHTLTQLPTPLLGHSVCTIGNFLFVLGGESLSGSPSSSLADGSRSVTALVHR 398
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
D W PA R R VG+ ++ GG + +VEMYD+ + WV
Sbjct: 399 YDPRFHTWTEVPAMREARAYFWCGVVGESLLAVGGLGTCGEALASVEMYDLRRDRWVAAG 458
Query: 182 SMPAMLKGSSGS 193
+P + G +G+
Sbjct: 459 ELPRAVHGHAGA 470
>emb|CAG02047.1| unnamed protein product [Tetraodon nigroviridis]
Length = 579
Score = 41.2 bits (95), Expect = 0.052
Identities = 29/113 (25%), Positives = 43/113 (37%), Gaps = 6/113 (5%)
Query: 72 TAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDWHH 131
+ YD R + W + P P+ +T R +Y L E+ F D W H
Sbjct: 388 SVECYDARDNCWTAVS-PMPVAMEFHSTLEYRDR----IYVLQ-GEYFFCFDPRKDYWSH 441
Query: 132 APAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMP 184
V R+ + A + G C VE+YD+E NTW + +P
Sbjct: 442 LAPMSVPRSQGLAALYKNCIYYIAGICRNHQRTFTVEVYDIEKNTWSRRRDLP 494
>emb|CAB80134.1| putative protein [Arabidopsis thaliana] gi|2911045|emb|CAA17555.1|
putative protein [Arabidopsis thaliana]
gi|15235260|ref|NP_195143.1| kelch repeat-containing
F-box family protein [Arabidopsis thaliana]
gi|7486288|pir||T05419 hypothetical protein F28A23.70 -
Arabidopsis thaliana
Length = 293
Score = 40.4 bits (93), Expect = 0.089
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 4/95 (4%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
+D+ W AP+ RV R P V + ++ V GG CD D +E++D ++ TW Q
Sbjct: 113 MDSRSHTWREAPSMRVPRMFPSVCTLDGKIYVMGG-CDNLDSTNWMEVFDTKTQTWEFLQ 171
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTF 216
+ G GS + SV G V V E +T+
Sbjct: 172 IPSEEIFG--GSAYESVRYEG-TVYVWSEKKDVTY 203
>emb|CAA86292.1| scruin [Limulus polyphemus] gi|2497944|sp|Q25390|SCRA_LIMPO
Alpha-scruin gi|1093326|prf||2103269A scrulin
Length = 918
Score = 40.0 bits (92), Expect = 0.12
Identities = 29/112 (25%), Positives = 44/112 (38%), Gaps = 6/112 (5%)
Query: 120 FSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVM 179
F + W R R V + +R+ V GG + AVEMY+ E + W
Sbjct: 158 FQLTVQTKQWRKRADMRYARAHHNVTVMDERIFVFGGRDSNGEILAAVEMYEPEMDQWTT 217
Query: 180 CQSMPAMLKGSSGSTWLSVAVAGEEVLVTEES------SGMTFCFDTVSMKW 225
S+P + GS+ + + V +E S FCFD ++ KW
Sbjct: 218 LASIPEPMMGSAIANNEGIIYVIGGVTTNKEKKPEGNLSNKIFCFDPLNNKW 269
Score = 37.4 bits (85), Expect = 0.75
Identities = 27/78 (34%), Positives = 35/78 (44%), Gaps = 10/78 (12%)
Query: 155 GGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLKGSS-----GSTWLSVAVAGEEVLVTE 209
GG D +VE YD ESN W M +SMP+ G + G WL + G + TE
Sbjct: 701 GGRDDIGRLVSSVESYDHESNEWTMEKSMPSPRMGMAAVAHGGYIWL---LGGLTSMTTE 757
Query: 210 ESSGM--TFCFDTVSMKW 225
E + C+D V W
Sbjct: 758 EPPVLDDVLCYDPVFKHW 775
>ref|XP_479911.1| kelch repeat-containing protein -like [Oryza sativa (japonica
cultivar-group)] gi|51964660|ref|XP_507114.1| PREDICTED
OJ1163_G08.35 gene product [Oryza sativa (japonica
cultivar-group)] gi|42407718|dbj|BAD08866.1| kelch
repeat-containing protein -like [Oryza sativa (japonica
cultivar-group)]
Length = 410
Score = 39.7 bits (91), Expect = 0.15
Identities = 43/162 (26%), Positives = 62/162 (37%), Gaps = 18/162 (11%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
I A DW PA V R D ++G + V G + + V++Y+ SNTW
Sbjct: 78 IPAPRWDWEEMPAAPVPRLDGYSVQIGDLLYVFAGYENLDHVHSHVDVYNFTSNTWTGRF 137
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSG--------MTFCFDTVSMKWDGPYDLCP 233
MP + S L +A G + G F DTV+ +W ++L P
Sbjct: 138 DMPKEMANSH----LGIATDGRYIYALTGQFGPQCRSPINRNFVVDTVTKEW---HELPP 190
Query: 234 DQSVGYCVTGMLGK-RLMVAGVVGDGEDVKAVKLW--AVKGG 272
Y L + RL V G + ++ W AVK G
Sbjct: 191 LPVPRYAPATQLWRGRLHVMGGGKEDRHEPGLEHWSLAVKDG 232
>dbj|BAB10324.1| unnamed protein product [Arabidopsis thaliana]
gi|15239760|ref|NP_199711.1| kelch repeat-containing
F-box family protein [Arabidopsis thaliana]
Length = 372
Score = 39.7 bits (91), Expect = 0.15
Identities = 25/93 (26%), Positives = 47/93 (49%), Gaps = 8/93 (8%)
Query: 91 PLKGHPSATAAVRSSHSTLLYTL------SPAEFAFSIDALHLDWHHAPAPRVWRTDPIV 144
P+ H S ++++ + S + Y + +P+ +D WH AP+ R+ R P
Sbjct: 114 PIPNHHSHSSSIVAIGSNI-YAIGGSIENAPSSKVSILDCRSHTWHEAPSMRMKRNYPAA 172
Query: 145 ARVGQRVVVAGGACDFEDDPLAVEMYDMESNTW 177
V ++ VAGG +F D +E++D+++ TW
Sbjct: 173 NVVDGKIYVAGGLEEF-DSSKWMEVFDIKTQTW 204
>ref|XP_464891.1| F-box-like protein [Oryza sativa (japonica cultivar-group)]
gi|47497917|dbj|BAD20123.1| F-box-like protein [Oryza
sativa (japonica cultivar-group)]
gi|47497893|dbj|BAD20077.1| F-box-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 405
Score = 38.9 bits (89), Expect = 0.26
Identities = 44/185 (23%), Positives = 73/185 (38%), Gaps = 24/185 (12%)
Query: 10 EAPIHGDVLETILSHVPLIHLVPACHVSNTWK---HAVFSSLAHTRTIKPWLIILTQSTR 66
+A + D+LE +LS +P+ ++ + V W HA + + KPW + T S
Sbjct: 41 DAVLPDDLLEKVLSFLPVASVIRSGSVCKRWHEIVHARRQTWSKMVPQKPWYFMFTCSEE 100
Query: 67 ASHVITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSI--DA 124
A ++ YDP W P K T SS S L+ + + + I +
Sbjct: 101 A---VSGFTYDPSLRKWYGFDFPCIEK-----TTWSISSSSGLVCLMDSEDRSRIIVCNP 152
Query: 125 LHLDWHH---APAPRVWRTDPI---VARVGQRVVVAGGAC-----DFEDDPLAVEMYDME 173
+ DW AP + + V R + +VA C ++ + +Y+ E
Sbjct: 153 ITKDWKRLVDAPGGKSADYSALAISVTRTSHQYMVAVARCNQVPSEYYQWEFTIHLYESE 212
Query: 174 SNTWV 178
NTWV
Sbjct: 213 INTWV 217
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.133 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 680,893,669
Number of Sequences: 2540612
Number of extensions: 29191587
Number of successful extensions: 63685
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 89
Number of HSP's that attempted gapping in prelim test: 63557
Number of HSP's gapped (non-prelim): 182
length of query: 375
length of database: 863,360,394
effective HSP length: 129
effective length of query: 246
effective length of database: 535,621,446
effective search space: 131762875716
effective search space used: 131762875716
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0008.15


