

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.13
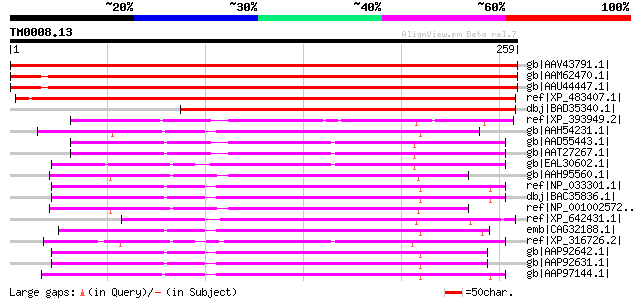
(259 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAV43791.1| At5g05670 [Arabidopsis thaliana] gi|53850473|gb|A... 385 e-106
gb|AAM62470.1| putative signal recognition particle receptor bet... 361 1e-98
gb|AAU44447.1| hypothetical protein AT2G18770 [Arabidopsis thali... 359 5e-98
ref|XP_483407.1| putative signal recognition particle receptor b... 347 1e-94
dbj|BAD35340.1| signal recognition particle receptor beta subuni... 237 3e-61
ref|XP_393949.2| PREDICTED: similar to ENSANGP00000020947 [Apis ... 117 3e-25
gb|AAH54231.1| Apmcf1-prov protein [Xenopus laevis] 114 3e-24
gb|AAD55443.1| GM04779p [Drosophila melanogaster] gi|23093878|gb... 111 2e-23
gb|AAT27267.1| RE55992p [Drosophila melanogaster] 111 2e-23
gb|EAL30602.1| GA17335-PA [Drosophila pseudoobscura] 111 2e-23
gb|AAH95560.1| Zgc:92746 protein [Danio rerio] 109 7e-23
ref|NP_033301.1| signal recognition particle receptor, B subunit... 109 9e-23
dbj|BAC35836.1| unnamed protein product [Mus musculus] gi|128425... 109 9e-23
ref|NP_001002572.1| zgc:92746 [Danio rerio] gi|49900692|gb|AAH76... 108 1e-22
ref|XP_642431.1| hypothetical protein DDB0205523 [Dictyostelium ... 108 1e-22
emb|CAG32188.1| hypothetical protein [Gallus gallus] 108 2e-22
ref|XP_316726.2| ENSANGP00000020947 [Anopheles gambiae str. PEST... 107 3e-22
gb|AAP92642.1| Cc1-8 [Rattus norvegicus] gi|33086606|gb|AAP92615... 107 3e-22
gb|AAP92631.1| Ba1-667 [Rattus norvegicus] 107 3e-22
gb|AAP97144.1| signal recognition particle receptor beta subunit... 107 3e-22
>gb|AAV43791.1| At5g05670 [Arabidopsis thaliana] gi|53850473|gb|AAU95413.1|
At5g05670 [Arabidopsis thaliana]
gi|9759092|dbj|BAB09661.1| signal recognition particle
receptor beta subunit-like protein [Arabidopsis
thaliana] gi|15239201|ref|NP_196186.1| expressed protein
[Arabidopsis thaliana] gi|42573277|ref|NP_974735.1|
expressed protein [Arabidopsis thaliana]
Length = 260
Score = 385 bits (990), Expect = e-106
Identities = 186/260 (71%), Positives = 225/260 (86%), Gaps = 1/260 (0%)
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLL-LSLRLFKRAKTNTVV 59
M+ LE K + +YL+++PP QLY A + +FTT+LL LS+RL +R K+NTV+
Sbjct: 1 MENLEDLKILAEQWSHQGIEYLQKIPPNQLYAAIGVLLFTTILLFLSIRLVRRTKSNTVL 60
Query: 60 LTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHS 119
L+GLTGSGKTVLFYQLRDGS+HQGTVTSMEPNEGTF+LHSE TKKGK+KPVH+VDVPGHS
Sbjct: 61 LSGLTGSGKTVLFYQLRDGSSHQGTVTSMEPNEGTFVLHSENTKKGKIKPVHLVDVPGHS 120
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
RLRPKL+E+LPQAA +VFVVDA++FLPNCRAASEYLY+ILT +VVKKKIP+L+ CNKTD
Sbjct: 121 RLRPKLEEFLPQAAAIVFVVDALEFLPNCRAASEYLYEILTNANVVKKKIPVLLCCNKTD 180
Query: 180 KVTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADA 239
K+TAHTKEFIR+Q+EKEI+KLRASRSAVS AD+ N+FT+G+ GE FSFT C NKVT A+A
Sbjct: 181 KLTAHTKEFIRKQMEKEIEKLRASRSAVSTADIANDFTIGIEGEVFSFTHCSNKVTVAEA 240
Query: 240 SGLTGEISQLEEFIREYVKP 259
SGLTGE Q+E+FIREY+KP
Sbjct: 241 SGLTGETIQIEDFIREYIKP 260
>gb|AAM62470.1| putative signal recognition particle receptor beta subunit
[Arabidopsis thaliana] gi|4185143|gb|AAD08946.1|
putative signal recognition particle receptor beta
subunit [Arabidopsis thaliana]
gi|60547709|gb|AAX23818.1| hypothetical protein
At2g18770 [Arabidopsis thaliana] gi|25411901|pir||D84568
hypothetical protein At2g18770 [imported] - Arabidopsis
thaliana gi|15224229|ref|NP_179467.1| expressed protein
[Arabidopsis thaliana]
Length = 260
Score = 361 bits (926), Expect = 1e-98
Identities = 176/260 (67%), Positives = 220/260 (83%), Gaps = 4/260 (1%)
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
+++L+ E+ S+ L ++++++PP QLY A + + T+ LLS+RLF+R K+NTV+L
Sbjct: 4 LEDLKIVAEQWSKQGL---EFVQKIPPPQLYTAIGVLLLATIWLLSIRLFRRTKSNTVLL 60
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSET-TKKGKVKPVHIVDVPGHS 119
+GL+GSGKTVLFYQLRDGS+HQG VTSMEPNEGTF+LH+E TKKGKVKPVH++DVPGHS
Sbjct: 61 SGLSGSGKTVLFYQLRDGSSHQGAVTSMEPNEGTFVLHNENNTKKGKVKPVHLIDVPGHS 120
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
RL KL+EYLP+AA VVFVVDA++FLPN RAASEYLYDILT SV+K K P+L+ CNKTD
Sbjct: 121 RLISKLEEYLPRAAAVVFVVDALEFLPNIRAASEYLYDILTNASVIKNKTPVLLCCNKTD 180
Query: 180 KVTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADA 239
KVTAHTKEFIR+Q+EKEIDKLR SRSA+S AD+ N+FTLG+ GE FSF+ C NKVT A+A
Sbjct: 181 KVTAHTKEFIRKQMEKEIDKLRVSRSAISTADIANDFTLGIEGEVFSFSHCHNKVTVAEA 240
Query: 240 SGLTGEISQLEEFIREYVKP 259
SGLTGE Q++EFIRE+VKP
Sbjct: 241 SGLTGETDQVQEFIREHVKP 260
>gb|AAU44447.1| hypothetical protein AT2G18770 [Arabidopsis thaliana]
Length = 260
Score = 359 bits (921), Expect = 5e-98
Identities = 175/260 (67%), Positives = 220/260 (84%), Gaps = 4/260 (1%)
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
+++L+ E+ S+ L ++++++PP QLY A + + T+ LLS+RLF+R K++TV+L
Sbjct: 4 LEDLKIVAEQWSKQGL---EFVQKIPPPQLYTAIGVLLLATIWLLSIRLFRRTKSDTVLL 60
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSET-TKKGKVKPVHIVDVPGHS 119
+GL+GSGKTVLFYQLRDGS+HQG VTSMEPNEGTF+LH+E TKKGKVKPVH++DVPGHS
Sbjct: 61 SGLSGSGKTVLFYQLRDGSSHQGAVTSMEPNEGTFVLHNENNTKKGKVKPVHLIDVPGHS 120
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
RL KL+EYLP+AA VVFVVDA++FLPN RAASEYLYDILT SV+K K P+L+ CNKTD
Sbjct: 121 RLISKLEEYLPRAAAVVFVVDALEFLPNIRAASEYLYDILTNASVIKNKTPVLLCCNKTD 180
Query: 180 KVTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADA 239
KVTAHTKEFIR+Q+EKEIDKLR SRSA+S AD+ N+FTLG+ GE FSF+ C NKVT A+A
Sbjct: 181 KVTAHTKEFIRKQMEKEIDKLRVSRSAISTADIANDFTLGIEGEVFSFSHCHNKVTVAEA 240
Query: 240 SGLTGEISQLEEFIREYVKP 259
SGLTGE Q++EFIRE+VKP
Sbjct: 241 SGLTGETDQVQEFIREHVKP 260
>ref|XP_483407.1| putative signal recognition particle receptor beta subunit
(SR-beta) [Oryza sativa (japonica cultivar-group)]
gi|42407742|dbj|BAD08889.1| putative signal recognition
particle receptor beta subunit (SR-beta) [Oryza sativa
(japonica cultivar-group)]
Length = 256
Score = 347 bits (891), Expect = 1e-94
Identities = 165/256 (64%), Positives = 212/256 (82%), Gaps = 2/256 (0%)
Query: 4 LEHWKEELSRLWL-TASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTG 62
++ W + + +W+ A ++RQ PPEQ+Y+A A+ T LLL+++ K +K NT+VL+G
Sbjct: 1 MDEWVRQ-AEVWVGQAERWIRQQPPEQVYVAVAVVAVTVLLLVAVSCLKSSKANTIVLSG 59
Query: 63 LTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLR 122
L+GSGKT+LFYQLRDGSTHQGTVTSME N TF+LHSE ++GKVKPVH+VDVPGH+RL+
Sbjct: 60 LSGSGKTILFYQLRDGSTHQGTVTSMEQNNDTFVLHSELERRGKVKPVHVVDVPGHARLK 119
Query: 123 PKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVT 182
PKLDE LPQAAG+V+VVDA DFL AA+EYLYDILTK +VVKK++P+LI CNKTDKVT
Sbjct: 120 PKLDEVLPQAAGIVYVVDAQDFLSTMHAAAEYLYDILTKATVVKKRVPVLIFCNKTDKVT 179
Query: 183 AHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGL 242
AH+KEFI++QLEKE++KLR SR+A+S AD+T+E LG PGEAF+F+QC NKVT + +GL
Sbjct: 180 AHSKEFIKKQLEKELNKLRESRNAISSADITDEVKLGNPGEAFNFSQCQNKVTVTEGAGL 239
Query: 243 TGEISQLEEFIREYVK 258
TG +S +EEFIREYVK
Sbjct: 240 TGNVSAVEEFIREYVK 255
>dbj|BAD35340.1| signal recognition particle receptor beta subunit-like [Oryza
sativa (japonica cultivar-group)]
Length = 172
Score = 237 bits (604), Expect = 3e-61
Identities = 113/171 (66%), Positives = 138/171 (80%)
Query: 88 MEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPN 147
ME N TF+LHSE +K K+KPVH+VDVPGH+ L+PKLDE LPQAAG+VF VDA DFL
Sbjct: 1 MEQNNDTFVLHSELERKSKIKPVHVVDVPGHAGLKPKLDEVLPQAAGIVFAVDAQDFLST 60
Query: 148 CRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRSAV 207
+ +EYLYDILTK +VVKK+I +LI CNKTDKVTAH+KEFI++QLEKEI+KLR SR +
Sbjct: 61 MQVVAEYLYDILTKATVVKKRIHVLIFCNKTDKVTAHSKEFIKKQLEKEINKLRESRKDI 120
Query: 208 SDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFIREYVK 258
S AD T+E LG PGE F F+QC N+VT A+ +GLTG +S +E+FIREYVK
Sbjct: 121 SSADTTDEVKLGNPGETFYFSQCQNRVTVAEGAGLTGNVSAVEQFIREYVK 171
>ref|XP_393949.2| PREDICTED: similar to ENSANGP00000020947 [Apis mellifera]
Length = 244
Score = 117 bits (293), Expect = 3e-25
Identities = 80/236 (33%), Positives = 138/236 (57%), Gaps = 22/236 (9%)
Query: 32 IAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPN 91
IAA I + T++L ++ +R+ N+++LTGL+ +GKT+++ L S T TS++ N
Sbjct: 21 IAAVIVIIITIVLFAIWHKRRSIGNSILLTGLSDAGKTLIYAHLLC-SKFVKTHTSVKEN 79
Query: 92 EGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAGVVFVVDAVDFLPNCRA 150
G I+++ + K IVD+PGH RLR K D++ A G+V+V+D+V F + R
Sbjct: 80 IGDIIINNRSLK--------IVDIPGHERLRYKFFDQFKLSAKGLVYVIDSVTFQKDIRD 131
Query: 151 ASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRSA---V 207
+EYLY++L+ S+++KK P+LILCNK D+ A I+ LEKE++ LR ++++
Sbjct: 132 VAEYLYNLLS-DSIIQKK-PVLILCNKQDQTMAKGSVVIKTLLEKEMNLLRMTKTSQLEA 189
Query: 208 SDADVTNEFTLGVPGEAFSFTQCCNKVTTADASG------LTGEISQLEEFIREYV 257
+DA TN F LG + F F+ + A+ S + ++ QL ++++ +
Sbjct: 190 TDASATNIF-LGKQEKDFDFSHLDINIEFAECSAYNKDSETSADMEQLNNWLKKII 244
>gb|AAH54231.1| Apmcf1-prov protein [Xenopus laevis]
Length = 264
Score = 114 bits (285), Expect = 3e-24
Identities = 79/235 (33%), Positives = 125/235 (52%), Gaps = 15/235 (6%)
Query: 15 WLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFK-----RAKTNTVVLTGLTGSGKT 69
WL ++++Q Q ++ + ++LLS L+K R V+L GL SGKT
Sbjct: 12 WLPDLEFIKQELQRQDPTLLSVGIALIVVLLSFVLWKILRGSRISRRAVLLVGLCDSGKT 71
Query: 70 VLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEY 128
+LF +L G T++ T TS+ N + + S+ K + +VD+PGH LR + L++Y
Sbjct: 72 LLFNRLLTG-TYKKTQTSITANSAAYKVKSD-----KGTSLTLVDLPGHESLRHQFLEQY 125
Query: 129 LPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEF 188
A ++FVVD+ F + +E LY +LT +V+K P+LI CNK D A + +
Sbjct: 126 KASARALLFVVDSSAFQREVKEVAELLYQLLTDVAVLKNAPPILIACNKQDISMAKSAKL 185
Query: 189 IRRQLEKEIDKLRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADAS 240
+++QLEKE++ LR +RSA + + LG G+ F FTQ KV + S
Sbjct: 186 VQQQLEKELNTLRVTRSAAPTTLEGSNSGVAQLGKKGKDFDFTQLPMKVKFLECS 240
>gb|AAD55443.1| GM04779p [Drosophila melanogaster] gi|23093878|gb|AAF50377.2|
CG33162-PA [Drosophila melanogaster]
gi|28574941|ref|NP_788485.1| CG33162-PA [Drosophila
melanogaster]
Length = 244
Score = 111 bits (278), Expect = 2e-23
Identities = 74/227 (32%), Positives = 120/227 (52%), Gaps = 16/227 (7%)
Query: 32 IAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPN 91
I A + F + + + + A +LTGL+ SGK+ +F QL G T TS++ N
Sbjct: 25 IVALLLGFIAVAIFVILRRRSAGRKDFLLTGLSESGKSAIFMQLIHGK-FPATFTSIKEN 83
Query: 92 EGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAGVVFVVDAVDFLPNCRA 150
G + S + + +VD+PGH R+R K L+ Y +A G+VFVVD+V + R
Sbjct: 84 VGDYRTGSASAR--------LVDIPGHYRVRDKCLELYKHRAKGIVFVVDSVTAHKDIRD 135
Query: 151 ASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRS----A 206
+++LY IL+ + + +L+LCNK D+ TA + + I+ LE E+ +R +RS +
Sbjct: 136 VADFLYTILSDSAT--QPCSVLVLCNKQDQTTAKSAQVIKSLLESELHTVRDTRSRKLQS 193
Query: 207 VSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFI 253
V D D + TLG PG F F+ + A+AS E+ L +++
Sbjct: 194 VGDEDGSKSITLGKPGRDFEFSHIAQNIQFAEASAKDTELDPLTDWL 240
>gb|AAT27267.1| RE55992p [Drosophila melanogaster]
Length = 268
Score = 111 bits (278), Expect = 2e-23
Identities = 74/227 (32%), Positives = 120/227 (52%), Gaps = 16/227 (7%)
Query: 32 IAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPN 91
I A + F + + + + A +LTGL+ SGK+ +F QL G T TS++ N
Sbjct: 49 IVALLLGFIAVAIFVILRRRSAGRKDFLLTGLSESGKSAIFMQLIHGK-FPATFTSIKEN 107
Query: 92 EGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAGVVFVVDAVDFLPNCRA 150
G + S + + +VD+PGH R+R K L+ Y +A G+VFVVD+V + R
Sbjct: 108 VGDYRTGSASAR--------LVDIPGHYRVRDKCLELYKHRAKGIVFVVDSVTAHKDIRD 159
Query: 151 ASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRS----A 206
+++LY IL+ + + +L+LCNK D+ TA + + I+ LE E+ +R +RS +
Sbjct: 160 VADFLYTILSDSAT--QPCSVLVLCNKQDQTTAKSAQVIKSLLESELHTVRDTRSRKLQS 217
Query: 207 VSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFI 253
V D D + TLG PG F F+ + A+AS E+ L +++
Sbjct: 218 VGDEDGSKSITLGKPGRDFEFSHIAQNIQFAEASAKDTELDPLTDWL 264
>gb|EAL30602.1| GA17335-PA [Drosophila pseudoobscura]
Length = 245
Score = 111 bits (277), Expect = 2e-23
Identities = 76/237 (32%), Positives = 121/237 (50%), Gaps = 16/237 (6%)
Query: 22 LRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTH 81
L ++ + +A + + + LR + A +LTGL SGK+ +F QL G
Sbjct: 16 LGEIDSGPILVALLLGFIAVAIFVILRR-RSAGRRDFLLTGLCESGKSAIFMQLLHGKLP 74
Query: 82 QGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAGVVFVVD 140
+ T TS++ N G + + G V +VD+PGH R+R K D Y +A G+VFVVD
Sbjct: 75 E-TFTSIKENVGDY-------QAGGHSSVRLVDIPGHYRVRDKCFDLYKHKAKGIVFVVD 126
Query: 141 AVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKL 200
+V + R ++ LY IL + + +L+LCNK D+ TA + + I+ LEKE+ +
Sbjct: 127 SVTVQKDIRDVADTLYTILADSAT--QPCSVLVLCNKQDQTTAKSAQVIKSLLEKELHTV 184
Query: 201 RASRS----AVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFI 253
R +RS +V D DV TLG PG F F + ++S E++ L +++
Sbjct: 185 RDTRSRKLQSVGDDDVNKPVTLGKPGRDFEFAHISQNIQFVESSAKEKELNTLTDWM 241
>gb|AAH95560.1| Zgc:92746 protein [Danio rerio]
Length = 266
Score = 109 bits (273), Expect = 7e-23
Identities = 76/223 (34%), Positives = 115/223 (51%), Gaps = 15/223 (6%)
Query: 21 YLRQVPPEQLYIAAAIAVFTTLLLLSLRLF-----KRAKTNTVVLTGLTGSGKTVLFYQL 75
+L+++ E I V L++L++ F N V+L GL SGKT+LF +L
Sbjct: 21 FLKEIQNEDAVFLVGIFVALALVILTIVFFGNFWGSSKVRNAVLLLGLCDSGKTLLFTRL 80
Query: 76 RDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAG 134
G + T TS+ + T+ SE ++DVPGH LR ++ ++Y A
Sbjct: 81 LLGKFVR-TQTSITESSATYKSKSERGSSWT-----LIDVPGHESLRTQIVEKYKDVARA 134
Query: 135 VVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLE 194
+VFVVD+ F + R +E+LY ILT + K L++ CNK D A + + I++QLE
Sbjct: 135 IVFVVDSSTFQKDVRDVAEFLYSILTDSILAKNAPTLVVACNKQDITMAKSAKLIQQQLE 194
Query: 195 KEIDKLRASRSAV---SDADVTNEFTLGVPGEAFSFTQCCNKV 234
KE++ LR +RSA D V LG G+ F F+Q N+V
Sbjct: 195 KELNTLRLTRSAALSSQDGAVGGSVYLGKKGKDFEFSQLANRV 237
>ref|NP_033301.1| signal recognition particle receptor, B subunit [Mus musculus]
gi|1351115|sp|P47758|SRPB_MOUSE Signal recognition
particle receptor beta subunit (SR-beta)
gi|13277831|gb|AAH03798.1| Srprb protein [Mus musculus]
gi|600886|gb|AAA69976.1| signal recognition particle
receptor beta subunit gi|26326439|dbj|BAC26963.1|
unnamed protein product [Mus musculus]
gi|12853827|dbj|BAB29860.1| unnamed protein product [Mus
musculus] gi|12841684|dbj|BAB25311.1| unnamed protein
product [Mus musculus] gi|12836352|dbj|BAB23618.1|
unnamed protein product [Mus musculus]
Length = 269
Score = 109 bits (272), Expect = 9e-23
Identities = 79/244 (32%), Positives = 125/244 (50%), Gaps = 18/244 (7%)
Query: 22 LRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGST 80
L+Q P L +A A+ AV TL+ +++ V+ GL SGKT+LF +L G
Sbjct: 28 LQQRDPTLLSVAVALLAVLLTLVFWKFIWSRKSSQRAVLFVGLCDSGKTLLFVRLLTGQ- 86
Query: 81 HQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAGVVFVV 139
++ T TS+ + + +++ + + ++D+PGH LR +L D + A VVFVV
Sbjct: 87 YRDTQTSITDSSAIYKVNNN-----RGNSLTLIDLPGHESLRFQLLDRFKSSARAVVFVV 141
Query: 140 DAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDK 199
D+ F + +E+LY +L +K LLI CNK D A + + I++QLEKE++
Sbjct: 142 DSAAFQREVKDVAEFLYQVLIDSMALKNSPSLLIACNKQDIAMAKSAKLIQQQLEKELNT 201
Query: 200 LRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG-------EISQL 249
LR +RSA D+ T LG G+ F F+Q KV + S G +I L
Sbjct: 202 LRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGGRGDTGSADIQDL 261
Query: 250 EEFI 253
E+++
Sbjct: 262 EKWL 265
>dbj|BAC35836.1| unnamed protein product [Mus musculus] gi|12842535|dbj|BAB25638.1|
unnamed protein product [Mus musculus]
Length = 269
Score = 109 bits (272), Expect = 9e-23
Identities = 79/244 (32%), Positives = 125/244 (50%), Gaps = 18/244 (7%)
Query: 22 LRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGST 80
L+Q P L +A A+ AV TL+ +++ V+ GL SGKT+LF +L G
Sbjct: 28 LQQRDPTLLSVAVALLAVLLTLVFWKFIWSRKSSQRAVLFVGLCDSGKTLLFVRLLTGQ- 86
Query: 81 HQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAGVVFVV 139
++ T TS+ + + +++ + + ++D+PGH LR +L D + A VVFVV
Sbjct: 87 YRDTQTSITDSSAIYKVNNN-----RGNSLTLIDLPGHESLRFQLLDRFKSSARAVVFVV 141
Query: 140 DAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDK 199
D+ F + +E+LY +L +K LLI CNK D A + + I++QLEKE++
Sbjct: 142 DSAAFQREVKDVAEFLYQVLIDSMALKNSPSLLIACNKQDIAMAKSAKLIQQQLEKELNT 201
Query: 200 LRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG-------EISQL 249
LR +RSA D+ T LG G+ F F+Q KV + S G +I L
Sbjct: 202 LRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGGRGDTGSADIQDL 261
Query: 250 EEFI 253
E+++
Sbjct: 262 EKWL 265
>ref|NP_001002572.1| zgc:92746 [Danio rerio] gi|49900692|gb|AAH76224.1| Zgc:92746 [Danio
rerio]
Length = 266
Score = 108 bits (271), Expect = 1e-22
Identities = 76/223 (34%), Positives = 115/223 (51%), Gaps = 15/223 (6%)
Query: 21 YLRQVPPEQLYIAAAIAVFTTLLLLSLRLF-----KRAKTNTVVLTGLTGSGKTVLFYQL 75
+L+++ E I V L++L++ F N V+L GL SGKT+LF +L
Sbjct: 21 FLKEIQNEDAVFLVGIFVALALVILTIVFFGNFWGSSKVRNAVLLLGLCDSGKTLLFTRL 80
Query: 76 RDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAG 134
G + T TS+ + T+ SE ++DVPGH LR ++ ++Y A
Sbjct: 81 LLGKFVR-TQTSITESSATYKSKSERGNSWT-----LIDVPGHESLRTQIVEKYKDVARA 134
Query: 135 VVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLE 194
+VFVVD+ F + R +E+LY ILT + K L++ CNK D A + + I++QLE
Sbjct: 135 IVFVVDSSIFQKDVRDVAEFLYSILTDSILAKNAPTLVVACNKQDITMAKSAKLIQQQLE 194
Query: 195 KEIDKLRASRSAV---SDADVTNEFTLGVPGEAFSFTQCCNKV 234
KE++ LR +RSA D V LG G+ F F+Q N+V
Sbjct: 195 KELNTLRLTRSAALSSQDGAVGGSVYLGKKGKDFEFSQLANRV 237
>ref|XP_642431.1| hypothetical protein DDB0205523 [Dictyostelium discoideum]
gi|60470464|gb|EAL68444.1| hypothetical protein
DDB0205523 [Dictyostelium discoideum]
Length = 290
Score = 108 bits (270), Expect = 1e-22
Identities = 64/207 (30%), Positives = 110/207 (52%), Gaps = 14/207 (6%)
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPG 117
+ + GL+ +GKT L L + T TS+ N G +I ++ K + I+DVPG
Sbjct: 88 IAILGLSNAGKTALLLNLTNVDKKISTHTSITTNNGVYITENK-------KKLPIIDVPG 140
Query: 118 HSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNK 177
+ + + L + L +A +++V+D F+ N ++YLYDILT SV +KKIP+L+ NK
Sbjct: 141 NGKAKASLPKILSNSACIIYVIDGTTFIDNSTQEAQYLYDILTNESVYQKKIPVLVFNNK 200
Query: 178 TDKVTAHTKEFIRRQLEKEIDKLRASRSA----VSDADVTNEFTLGVPGEAFSFTQCCNK 233
D + E ++ LE+E+D LR +R A + + + LG+ G F F N
Sbjct: 201 MDLDSTIDTEQVKNILERELDDLRRTRGATPIVLGQEEDKKDIYLGIEGTPFQFDHLPND 260
Query: 234 V--TTADASGLTGEISQLEEFIREYVK 258
V + AS GE+ ++++ I+ +++
Sbjct: 261 VQFSNGSASPSNGELKEIDD-IKNFIQ 286
>emb|CAG32188.1| hypothetical protein [Gallus gallus]
Length = 258
Score = 108 bits (269), Expect = 2e-22
Identities = 79/227 (34%), Positives = 118/227 (51%), Gaps = 13/227 (5%)
Query: 26 PPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTV 85
P + A + V T LL L R+ V+L GL +GKT+LF +L G ++ T
Sbjct: 21 PASLSVLLALLVVALTFLLWRLAQGARSTRRAVLLLGLCDAGKTLLFARLLTGR-YRDTQ 79
Query: 86 TSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAGVVFVVDAVDF 144
TS+ + + + ++ K V ++D+PGH LR + L+ + A +VFVVD+V F
Sbjct: 80 TSITDSSAVYRVSND-----KGTNVTLIDLPGHESLRLQFLERFKAAARAIVFVVDSVAF 134
Query: 145 LPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASR 204
+ +E+LY +L +V+K LLI CNK D A + + I++QLEKE++ LR +R
Sbjct: 135 QREVKDVAEFLYQVLVDSTVLKNAPALLIACNKQDVTMAKSAKLIQQQLEKELNTLRVTR 194
Query: 205 SAVS---DADVT-NEFTLGVPGEAFSFTQCCNKVTTADAS--GLTGE 245
SA DA T LG G+ F F+Q KV + S G GE
Sbjct: 195 SAAPTSLDASATGGPAQLGKKGKDFDFSQLPMKVEFVECSARGSKGE 241
>ref|XP_316726.2| ENSANGP00000020947 [Anopheles gambiae str. PEST]
gi|55239444|gb|EAA11816.2| ENSANGP00000020947 [Anopheles
gambiae str. PEST]
Length = 244
Score = 107 bits (268), Expect = 3e-22
Identities = 81/243 (33%), Positives = 132/243 (53%), Gaps = 20/243 (8%)
Query: 18 ASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKT--NTVVLTGLTGSGKTVLFYQL 75
AS L ++ + IA A+ + T ++L L+KR KT + V+ TGL SGKT LF L
Sbjct: 12 ASIKLGELNYTPVLIALAVVLLTIVVLF---LWKRKKTVRSAVLFTGLCDSGKTYLFAHL 68
Query: 76 RDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAG 134
G + T TS++ N G+F T++G+V + +VDVPG+ RLR K DEY A
Sbjct: 69 CLGGARE-TFTSIKENVGSF-----KTERGRV--LKMVDVPGNERLRGKFFDEYKNIAKA 120
Query: 135 VVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLE 194
+V+++D+V + R +++LY IL + K+P+++LCNK D+ A T+ I+ LE
Sbjct: 121 IVYMIDSVTVQKDIRDVADFLYTILVDKAT--SKVPVVVLCNKQDETLAKTETAIKSMLE 178
Query: 195 KEIDKLRASR----SAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLE 250
KEI+ +R +R +V ++ ++ F F F Q +V S + + + L
Sbjct: 179 KEINIVRQTRRSQLQSVDNSSSSDTFLGKSASVDFEFEQLGQRVRFVPCSAMEEQFAGLT 238
Query: 251 EFI 253
+F+
Sbjct: 239 KFL 241
>gb|AAP92642.1| Cc1-8 [Rattus norvegicus] gi|33086606|gb|AAP92615.1| Ab2-417
[Rattus norvegicus]
Length = 979
Score = 107 bits (268), Expect = 3e-22
Identities = 74/228 (32%), Positives = 117/228 (50%), Gaps = 11/228 (4%)
Query: 22 LRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGST 80
L+Q P L +A A+ AV TL+ +++ V+ GL SGKT+LF +L G
Sbjct: 738 LQQRDPTLLSVAVAVLAVLLTLVFWKFIWSRKSSQRAVLFVGLCDSGKTLLFVRLLTGQ- 796
Query: 81 HQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAGVVFVV 139
++ T TS+ + + +++ + + ++D+PGH LR + LD + A VVFVV
Sbjct: 797 YRDTQTSITDSSAIYKVNNN-----RGNSLTLIDLPGHESLRLQFLDRFKSSARAVVFVV 851
Query: 140 DAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDK 199
D+ F + +E+LY +L +K L+ CNK D A + + I++QLEKE++
Sbjct: 852 DSATFQREVKDVAEFLYQVLIDSMALKNTPAFLVACNKQDIAMAKSAKLIQQQLEKELNT 911
Query: 200 LRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG 244
LR +RSA D+ T LG G+ F F+Q KV + S G
Sbjct: 912 LRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGG 959
>gb|AAP92631.1| Ba1-667 [Rattus norvegicus]
Length = 980
Score = 107 bits (268), Expect = 3e-22
Identities = 74/228 (32%), Positives = 117/228 (50%), Gaps = 11/228 (4%)
Query: 22 LRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGST 80
L+Q P L +A A+ AV TL+ +++ V+ GL SGKT+LF +L G
Sbjct: 739 LQQRDPTLLSVAVAVLAVLLTLVFWKFIWSRKSSQRAVLFVGLCDSGKTLLFVRLLTGQ- 797
Query: 81 HQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAGVVFVV 139
++ T TS+ + + +++ + + ++D+PGH LR + LD + A VVFVV
Sbjct: 798 YRDTQTSITDSSAIYKVNNN-----RGNSLTLIDLPGHESLRLQFLDRFKSSARAVVFVV 852
Query: 140 DAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDK 199
D+ F + +E+LY +L +K L+ CNK D A + + I++QLEKE++
Sbjct: 853 DSATFQREVKDVAEFLYQVLIDSMALKNTPAFLVACNKQDIAMAKSAKLIQQQLEKELNT 912
Query: 200 LRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG 244
LR +RSA D+ T LG G+ F F+Q KV + S G
Sbjct: 913 LRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGG 960
>gb|AAP97144.1| signal recognition particle receptor beta subunit [Homo sapiens]
gi|40850903|gb|AAH65299.1| Signal recognition particle
receptor, beta subunit [Homo sapiens]
gi|31340540|sp|Q9Y5M8|SRPRB_HUMAN Signal recognition
particle receptor beta subunit (SR-beta) (Protein
APMCF1) gi|22761735|dbj|BAC11675.1| unnamed protein
product [Homo sapiens] gi|24528583|gb|AAD34888.3| APMCF1
[Homo sapiens]
Length = 271
Score = 107 bits (268), Expect = 3e-22
Identities = 79/249 (31%), Positives = 126/249 (49%), Gaps = 18/249 (7%)
Query: 17 TASDYLRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQL 75
T L+Q P L + A+ AV TL+ L +R+ V+L GL SGKT+LF +L
Sbjct: 25 TLRQELQQTDPTLLSVVVAVLAVLLTLVFWKLIRSRRSSQRAVLLVGLCDSGKTLLFVRL 84
Query: 76 RDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAG 134
G ++ T TS+ + + +++ + + ++D+PGH LR + L+ + A
Sbjct: 85 LTG-LYRDTQTSITDSCAVYRVNNN-----RGNSLTLIDLPGHESLRLQFLERFKSSARA 138
Query: 135 VVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLE 194
+VFVVD+ F + +E+LY +L +K LI CNK D A + + I++QLE
Sbjct: 139 IVFVVDSAAFQREVKDVAEFLYQVLIDSMGLKNTPSFLIACNKQDIAMAKSAKLIQQQLE 198
Query: 195 KEIDKLRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG------- 244
KE++ LR +RSA D+ T LG G+ F F+Q KV + S G
Sbjct: 199 KELNTLRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGGRGDVGSA 258
Query: 245 EISQLEEFI 253
+I LE+++
Sbjct: 259 DIQDLEKWL 267
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 410,895,224
Number of Sequences: 2540612
Number of extensions: 15963028
Number of successful extensions: 56745
Number of sequences better than 10.0: 1354
Number of HSP's better than 10.0 without gapping: 195
Number of HSP's successfully gapped in prelim test: 1159
Number of HSP's that attempted gapping in prelim test: 55809
Number of HSP's gapped (non-prelim): 1373
length of query: 259
length of database: 863,360,394
effective HSP length: 125
effective length of query: 134
effective length of database: 545,783,894
effective search space: 73135041796
effective search space used: 73135041796
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0008.13


