

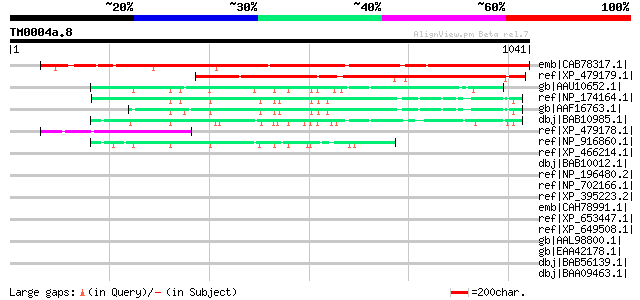
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004a.8
(1041 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB78317.1| putative protein [Arabidopsis thaliana] gi|45862... 930 0.0
ref|XP_479179.1| homeobox transcription factor Hox7-like protein... 617 e-175
gb|AAU10652.1| unknown protein [Oryza sativa (japonica cultivar-... 221 1e-55
ref|NP_174164.1| homeobox transcription factor, putative [Arabid... 209 4e-52
gb|AAF16763.1| F3M18.14 [Arabidopsis thaliana] gi|25403342|pir||... 200 2e-49
dbj|BAB10985.1| unnamed protein product [Arabidopsis thaliana] g... 186 5e-45
ref|XP_479178.1| unknown protein [Oryza sativa (japonica cultiva... 173 2e-41
ref|NP_916860.1| P0007F06.8 [Oryza sativa (japonica cultivar-gro... 170 2e-40
ref|XP_466214.1| putative DDT domain-containing protein [Oryza s... 48 0.002
dbj|BAB10012.1| unnamed protein product [Arabidopsis thaliana] 45 0.010
ref|NP_196480.2| DDT domain-containing protein [Arabidopsis thal... 45 0.010
ref|NP_702166.1| coatamer protein, beta subunit, putative [Plasm... 45 0.017
ref|XP_395223.2| PREDICTED: similar to bromodomain adjacent to z... 44 0.022
emb|CAH78991.1| conserved hypothetical protein [Plasmodium chaba... 43 0.063
ref|XP_653447.1| Viral A-type inclusion protein repeat, putative... 41 0.24
ref|XP_649508.1| conserved hypothetical protein [Entamoeba histo... 40 0.31
gb|AAL98800.1| ORF076L [infectious spleen and kidney necrosis vi... 40 0.41
gb|EAA42178.1| GLP_480_88069_85913 [Giardia lamblia ATCC 50803] 40 0.41
dbj|BAB56139.1| kinesin-like protein 2 [Giardia intestinalis] 40 0.41
dbj|BAA09463.1| troponin T [Halocynthia roretzi] 40 0.53
>emb|CAB78317.1| putative protein [Arabidopsis thaliana] gi|4586251|emb|CAB40992.1|
putative protein [Arabidopsis thaliana]
gi|15235464|ref|NP_193011.1| expressed protein
[Arabidopsis thaliana] gi|7452049|pir||T06633
hypothetical protein T20K18.100 - Arabidopsis thaliana
Length = 1108
Score = 930 bits (2404), Expect = 0.0
Identities = 523/1037 (50%), Positives = 675/1037 (64%), Gaps = 87/1037 (8%)
Query: 63 LGTVLRNDGPSIGREFDYLPSGHKSYN-----STCREDQQPAKRSKVSKGTGKGLMNASV 117
L V R DGPS+G EFD+LPSG + + S ++ Q+ A++ K+S+ +
Sbjct: 86 LAKVFRKDGPSLGSEFDHLPSGARKASWLGTSSVGQQKQKVARKRKISELMDHTSQDCIQ 145
Query: 118 KKHGIGKGLMNASVKKHGIGKGLMTIWRATNPDSGELPVGFGFADREVHLISNSKKSVRE 177
+ NA+V KHGIGKGLMT+WR NP+ ++ D L +S ++
Sbjct: 146 E---------NATVMKHGIGKGLMTVWRVMNPNRRDVSPCVDLLDERATLPQSSARN--P 194
Query: 178 NNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSLKEKCELSLDS 237
++ K + +LK KL KR+ + +R + + E N+ T+ + KE CEL+ D
Sbjct: 195 PHQKKKQRQLASILKQKLLQKRSTEKKRRSI---HREAELNKDETQRE-FKENCELAADG 250
Query: 238 EISEEGVDRISMLIDDEELELRELQERTNLLICSDHLAASGMLGCSLGK----------- 286
E+ +E IS L+DDEELE+RE ER N L CS H +SG GC L K
Sbjct: 251 EVFKETCQTISTLVDDEELEMRERHERGNPLTCSCHHPSSGSHGCFLCKGIAMRSSDSSL 310
Query: 287 ---DVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFKDSMLLGKIHVALLTLLLSDIEVE 343
D+L K+PP++V+M+ P L PW+SSPE VKKLFKDS+LLGKIH++LL LLL D+E E
Sbjct: 311 LFPDLLPKFPPNSVQMRMPFGLHPWNSSPESVKKLFKDSLLLGKIHLSLLKLLLLDVETE 370
Query: 344 LSNGFAPHLDKSCNFLALLHSVESQEYSPDFWRRSLNSLTWIEILRQVLVASGFGSKQGA 403
L G +L SC FLALL SVESQ D WR SLNSLTW E+LRQ+LVA+G+GS + A
Sbjct: 371 LERGSFSNLSISCKFLALLQSVESQILILDMWRNSLNSLTWTELLRQILVAAGYGSLKCA 430
Query: 404 LRREALSKEL--------------------------------NLLINYGISPGTLKGELF 431
++ E LSK+L L+ YG+ GTLKGELF
Sbjct: 431 VQSEELSKQLASTCFVLGDRSVICGELKALARLYFVIDDIHMKLMKKYGLRLGTLKGELF 490
Query: 432 KILSERGNNGCKVSDLAKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLR 491
++L+ +GNNG K+S+LA A ++A LNLA+ EE E+ ICSTL+SDITLFEKIS S YR+R
Sbjct: 491 RMLNGQGNNGLKISELADAPEVAVLNLATVPEERENSICSTLASDITLFEKISESTYRVR 550
Query: 492 MSTVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFESGSVDSNIRKLKRVKSHKTKSA 551
++ ++D D+ QSD +D GSVDDE DD + S GD+ E S + +RK+K K K KS
Sbjct: 551 VNCFSEDPDKSQSDSDDSGSVDDESDDC-SISSGDEIEHVSENPALRKVKCRKRRKHKSK 609
Query: 552 KLTVFNEIDESHPGEVWLLGLMESEYSDLNIEEKLNALAALTDLLSSGSSIRMKDPVKVT 611
V +EIDESHPGE WLLGLME EYSDL++EEKL+ AL DLLSSGS+IRM+D +
Sbjct: 610 MREVCSEIDESHPGEPWLLGLMEGEYSDLSVEEKLDVFVALIDLLSSGSTIRMEDLPRAV 669
Query: 612 ADYDSSIQLRGSGAKIKRSVVKK---PVPFWNQFGQMQRVKEGHLNSHPCPVDSSSLMSK 668
AD SI GSG KIKRS + P W G++ +K +S PVDSSS++
Sbjct: 670 ADCAPSIYSHGSGGKIKRSSSNQYSYPRGSWVHGGELYGIKALSKSSDSHPVDSSSIVGA 729
Query: 669 FRSHEPSFEKGKGSTDSHPIQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGH 728
F G + + HP+QSVYLGSDRR+NRYWLFLG CNA+DPGH+ V+FESSEDGH
Sbjct: 730 FAKLA-----GNRANNVHPMQSVYLGSDRRFNRYWLFLGTCNANDPGHRCVFFESSEDGH 784
Query: 729 WEVIDTEEALCTLLSVLDDRGKREALLIESLERRQTSLCRFMSRVKVNGIGTGCMSHSDQ 788
WEVI+ +EAL LLSVLDDRG+REA LIESLE+R++ LC+ M +V T +
Sbjct: 785 WEVINNKEALRALLSVLDDRGRREARLIESLEKRESFLCQAMLSRQVTQSETAHFT---- 840
Query: 789 SELDMVTEDSNSPVSDVDN-LNLSDSAKSLPSA--GAVVIEAGKGVEEQVQKWIRVQEYD 845
D+V EDS+SPVSD+DN L L++ A S+ A+V E G E+ + W +QE+D
Sbjct: 841 ---DIVREDSSSPVSDIDNNLCLNEIANDQFSSQHAAIVFEIGSKREKSLL-WSLIQEFD 896
Query: 846 SWIWNSFYLDLNVVKYGRRSYLDSLARCKSCHDLYWRDERHCKICHMTFELDFDLEERYA 905
WIW +F +LN VK+ RRSYLDSL RCKSCHDLYWRDE+HCKICH TFE+D DLEERYA
Sbjct: 897 DWIWANFNFNLNSVKHRRRSYLDSLTRCKSCHDLYWRDEKHCKICHATFEVDIDLEERYA 956
Query: 906 IHIATCREKENSKTHPNHKVLSSQIQSLKAAIYAIESVMPEDSLVGAWRKSAHKLWVKRL 965
IH ATC KE T P+HKVLSSQ+QSLKAA+YAIES MPED+L+GAWRKSAH+LW KRL
Sbjct: 957 IHAATCMRKEECDTFPDHKVLSSQLQSLKAAVYAIESAMPEDALIGAWRKSAHRLWAKRL 1016
Query: 966 RRTSTLVELVQVLTDFVGAINESWLFQCKFP-DGMVEEIIASFASMPHTSSALALWLVKL 1024
RR+S++ E+ QV+ DFVGAINE WL+ C ++ EII F SMP T+SA+ALWLVKL
Sbjct: 1017 RRSSSVSEITQVIGDFVGAINEEWLWHCSDQGQTLMGEIINCFPSMPQTTSAIALWLVKL 1076
Query: 1025 DTIIAPYLDRVYLQKKQ 1041
DT+IAPY+++ ++ Q
Sbjct: 1077 DTLIAPYVEKAPPERDQ 1093
>ref|XP_479179.1| homeobox transcription factor Hox7-like protein [Oryza sativa
(japonica cultivar-group)] gi|33146626|dbj|BAC79914.1|
homeobox transcription factor Hox7-like protein [Oryza
sativa (japonica cultivar-group)]
gi|33146880|dbj|BAC79878.1| homeobox transcription factor
Hox7-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 706
Score = 617 bits (1592), Expect = e-175
Identities = 341/689 (49%), Positives = 450/689 (64%), Gaps = 46/689 (6%)
Query: 373 DFWRRSLNSLTWIEILRQVLVASGFGSKQGALRREALSKELNLLINYGISPGTLKGELFK 432
+FW +SLNSLTW+EILRQVLVASGFGSK L R+ +KE N ++ YG+ P TLKGELF
Sbjct: 2 NFWIKSLNSLTWVEILRQVLVASGFGSKHHMLNRDFFNKEKNQMVKYGLRPRTLKGELFA 61
Query: 433 ILSERGNNGCKVSDLAKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLRM 492
+LS++G+ G KVS+LAK+ +I +L+++ST E+E LI STLSSDITLFEKI+ SAYRLR+
Sbjct: 62 LLSKKGSGGLKVSELAKSPEIVDLSISST--EIEQLIYSTLSSDITLFEKIAPSAYRLRV 119
Query: 493 STVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFESGSVDSNIRKLKRVKSHKTKSAK 552
K ++ SD ED GSVDD D S D S + ++ R K K
Sbjct: 120 DPRIKGKEDSGSDTEDSGSVDDHSDASSGADESDGSHEMSFSEHEHRILRRK-WKNGHEN 178
Query: 553 LTVFNEIDESHPGEVWLLGLMESEYSDLNIEEKLNALAALTDLLSSGSSI-RMKDPVKVT 611
+ +EIDES+ GE WLLGLME EYSDL+I+EKL+ L AL D++S S R+++P +V
Sbjct: 179 VNRCSEIDESYSGERWLLGLMEGEYSDLSIDEKLDCLVALMDVVSGADSAPRLEEPSRVV 238
Query: 612 ADYDSSIQLRGSGAKIKRS---VVKKPVPFWNQFGQMQRVKEG-HLNSHPCPVDSSSLMS 667
+ Q SG KIK+S + + +N G M + H S
Sbjct: 239 PSIPRA-QPHVSGGKIKKSTRNICQSSDECFNASGSMYGLDSSMHEQSR----------- 286
Query: 668 KFRSHEPSFEKGKGSTDS---HPIQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESS 724
RS + G+ T + H Q V LGSDRRYN YWLFLGPC ADDPGH+RVYFESS
Sbjct: 287 SLRSRDYVAYSGRNDTSTGVAHQPQVVLLGSDRRYNNYWLFLGPCRADDPGHRRVYFESS 346
Query: 725 EDGHWEVIDTEEALCTLLSVLDDRGKREALLIESLERRQTSLCRFM-----SRVKVNGIG 779
EDGHWEVID+ + L +LL+ LD RG REA L+ S+++RQT L M +R V
Sbjct: 347 EDGHWEVIDSPQELLSLLASLDSRGTREAYLLASMKKRQTCLFEAMKKHYENRDAVQPAM 406
Query: 780 TGCMSHSDQSELD-----MVTEDSNSPVSDVDNLNL--SDSAKSLPSAGAVVIEAGKGVE 832
SHS+ S D + + D SP SD+DN ++ + + + ++ A+ IE G+ +
Sbjct: 407 PSDTSHSETSSGDGASPKLSSGDGASPTSDIDNASVPTNPAENMINASSAIAIEVGRRGD 466
Query: 833 EQVQKWIRVQEYDSWIWNSFYLDLNVVKYGRRSYLDSLARCKSCHDLYWRDERHCKICHM 892
E++ KW R Q +D WIW SFY L VK G++S+ +SL RC+SCHDLYWRDE+HC+ICH
Sbjct: 467 EKILKWERSQTFDKWIWTSFYSCLTAVKCGKKSFKESLVRCESCHDLYWRDEKHCRICHS 526
Query: 893 TFELDFDLEERYAIHIATCREKENSKTHPNHKVLSSQIQSLKAAIYAIESVMPEDSLVGA 952
TFE+ FDLEERYAIH+ATCR+ E+ PNHKVL SQ+Q+LKAAI+AIE+ MPE + G
Sbjct: 527 TFEVSFDLEERYAIHVATCRDPEDVYDVPNHKVLPSQLQALKAAIHAIEAHMPEAAFAGL 586
Query: 953 WRKSAHKLWVKRLRRTSTLVELVQVLTDFVGAINESWLFQ-------CKFPDGMVEEIIA 1005
W KS+HKLWVKRLRRTS+L EL+QVL DFVGA++E WL++ C + +++I+
Sbjct: 587 WMKSSHKLWVKRLRRTSSLAELLQVLVDFVGAMDEDWLYKSSSSVSFCSY----LDDIVI 642
Query: 1006 SFASMPHTSSALALWLVKLDTIIAPYLDR 1034
F +MP T+SA+ALW+VKLD +I PYL+R
Sbjct: 643 YFQTMPQTTSAVALWVVKLDALITPYLER 671
>gb|AAU10652.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 1397
Score = 221 bits (563), Expect = 1e-55
Identities = 246/1049 (23%), Positives = 418/1049 (39%), Gaps = 233/1049 (22%)
Query: 163 REVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVT 222
RE + +++ RE + + + + + + + KR K + +
Sbjct: 6 REEQVRREMERNDRERRKEEERLLREKQKEEERFQREQRREHKRMEKYLLKQSLRAEKIR 65
Query: 223 RNQSLKEKCELSLDSEISEEGVDR-----ISMLIDDEELELRELQERTNLLICSDHLAAS 277
+ + L+++ E + +E R L++DE LEL EL R+ L L +
Sbjct: 66 QKEELRKEKEAARQKAANERATARRIAREYMELMEDERLELMELVSRSKGLPSMLSLDSD 125
Query: 278 GMLGCSLGKDVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFK----------------- 320
+ + +L ++P + V++K P ++PW SS + + L
Sbjct: 126 TLQQLDSFRGMLRQFPSEIVRLKVPFSIKPWTSSEDNIGNLLMVWKFFITFADVLGIPSF 185
Query: 321 ------------DSMLLGKIHVALLTLLLSDIE-----VELSNGFAPHLDKSCNFLALLH 363
DS LLG++HVALL ++ DIE +++G + ++
Sbjct: 186 TLDEFVQSLHDYDSRLLGELHVALLKSIIKDIEDVARTPSVASGMTAN--PGGGHPQIVE 243
Query: 364 SVESQEYSPDFWRRSLNSLTWIEILRQVLVASGFG------------------------- 398
++ W+R LN LTW EILRQ +++G G
Sbjct: 244 GAYDWGFNILAWQRHLNLLTWPEILRQFGLSAGLGPQLRKRNAENVNNHDDNEGRNGEDV 303
Query: 399 -----SKQGALRREALSKELNL----LINYGISPGTLKGELFKILSERGNNGCKVSDLAK 449
S A+ A KE + ++PGT+K F +LS G+ G + ++A+
Sbjct: 304 ISILRSGSAAVNAAAKMKERGYGNRRRSRHRLTPGTVKFAAFHVLSLEGSQGLTILEVAE 363
Query: 450 ATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAY--------------------- 488
Q + L +T++ E+ I + LS D LFE+ + S Y
Sbjct: 364 KIQKSGLRDLTTSKTPEASISAALSRDSKLFERTAPSTYCVKTPYRKDPADSEAVLAAAR 423
Query: 489 ---RLRMSTVTKDDDECQSDMEDYGSVDD-ELDDSDTCSCGDDFE--------------- 529
R+ +T++ + +E + D++D +D E DD+D GD+
Sbjct: 424 EKIRVFQNTIS-ECEEVEKDVDDAERDEDSECDDADDDPDGDEVNIEEKDVKTSLVKAQD 482
Query: 530 ----------------------------SGSVDSNIRKLKRVKSHKTKS---AKLTVFN- 557
S S++R L + +T S A+++ N
Sbjct: 483 GGMPTAVGDIKKETNSIVNSLTTPLIHTKSSESSSLRTLDKSVQVRTTSDLPAEISSDNH 542
Query: 558 ----------EIDESHPGEVWLLGLMESEYSDLNIEEKLNALAALTDLLSSGSSIR---- 603
EIDES+ GE W+ GL E +Y DL++EE+LNAL AL + + G+SIR
Sbjct: 543 EGASDSAQDAEIDESNQGESWVQGLAEGDYCDLSVEERLNALVALIGVATEGNSIRAVLE 602
Query: 604 --------MKDPVKVTADYD--------SSIQLRGSGAKIKRSVVKKPVPFWNQFGQMQR 647
+K + A D SS SG +K V ++ + +
Sbjct: 603 ERLEAASALKKQMWAEAQLDKRRSREEFSSKMQYDSGMGLKTDVDQQNTLAESNLTPVHN 662
Query: 648 VKE-----GHLNSHPCPVDSSS-------------------------------LMSKFRS 671
+ + G L ++ PVD S K RS
Sbjct: 663 LVKDSNGNGSLVNNELPVDQQSQPNACSVVHERNGVRQEFSANPENLSGQQYVTSEKTRS 722
Query: 672 HEPSFEKGKGSTDSHPIQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEV 731
S+ G + H +S+ LG DRR NRYW F + DDPG R++FE S DG+W +
Sbjct: 723 QLKSYI-GHKAEQLHVYRSLPLGQDRRRNRYWQFSTSASPDDPGSGRIFFE-SRDGYWRL 780
Query: 732 IDTEEALCTLLSVLDDRGKREALLIESLERRQTSLCRFMSRVKVNGI--GTGCMSHSDQS 789
ID+ E L+S LD RG RE+ L L+ + + + R + I G + + S
Sbjct: 781 IDSIETFDALVSSLDTRGIRESHLHSMLQSIEPTFKEAIGRKRCASIEPSAGRVLKNGTS 840
Query: 790 ELDMVTEDSNSPVSDVDNLNLSDSAKSLPSAGAVVIEAGKGVEEQVQKWIRVQEYDSWIW 849
E+ + SN S L+ + ++ + + IE G+ E+ R + W+W
Sbjct: 841 EI-ISPNHSNEFGSPCSTLSGVATDSAMAYSDSFRIELGRNDVEKTAISERADLFIKWMW 899
Query: 850 N--SFYLDLNVVKYGRRSYLDSLARCKSCHDLYWRDERHCKICHMTFELDFDLEERYAIH 907
+ + +K+G++ + + C C+ +Y +E HC CH TF+ ++ E H
Sbjct: 900 KECNNHQPTCAMKHGKKRCSELIQCCDFCYQIYLAEETHCASCHKTFKSIHNISE----H 955
Query: 908 IATCREKENSKTHPNHKVLSSQ------IQSLKAAIYAIESVMPEDSLVGAWRKSAHKLW 961
+ C EK +T PN K+ S ++ LK + +E+ +P ++L W K W
Sbjct: 956 SSQCEEKR--RTDPNWKMQISDYSVPVGLRLLKLLLATVEASVPAEALEPFWTDVYRKSW 1013
Query: 962 VKRLRRTSTLVELVQVLTDFVGAINESWL 990
+L TS+ E+ ++LT GAI +L
Sbjct: 1014 GVKLYSTSSTKEVFEMLTILEGAIRRDFL 1042
>ref|NP_174164.1| homeobox transcription factor, putative [Arabidopsis thaliana]
Length = 1703
Score = 209 bits (532), Expect = 4e-52
Identities = 248/1063 (23%), Positives = 418/1063 (38%), Gaps = 227/1063 (21%)
Query: 164 EVHLISNSKKSVRENNRSSKTATMNRMLKN-KLQNKRNNLQDKRKLLMQRKAGESNQHVT 222
E + +++ RE + + R+ + +LQ ++ ++R+ +QR+ + +
Sbjct: 394 EERMRKEMERNERERRKEEERLMRERIKEEERLQREQRREVERREKFLQRENERAEKKKQ 453
Query: 223 RNQSLKEKCELSLDSEISEEGVDRISM----LIDDEELELRELQERTNLLICSDHLAASG 278
+++ +EK + I + RI+ LI+DE+LEL EL + L L
Sbjct: 454 KDEIRREKDAIRRKLAIEKATARRIAKESMDLIEDEQLELMELAAISKGLPSVLQLDHDT 513
Query: 279 MLGCSLGKDVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFK------------------ 320
+ + +D L +PP ++++K P + PW S E V L
Sbjct: 514 LQNLEVYRDSLSTFPPKSLQLKMPFAISPWKDSDETVGNLLMVWRFLISFSDVLDLWPFT 573
Query: 321 -----------DSMLLGKIHVALLTLLLSDIE-------VELSNGFAPHLDKSCNFLALL 362
DS LLG+IHV LL ++ D+E + N + ++
Sbjct: 574 LDEFIQAFHDYDSRLLGEIHVTLLRSIIRDVEDVARTPFSGIGNNQYTTANPEGGHPQIV 633
Query: 363 HSVESQEYSPDFWRRSLNSLTWIEILRQVLVASGFGSK---------------------- 400
+ + W++ LN LTW EILRQ+ +++GFG K
Sbjct: 634 EGAYAWGFDIRSWKKHLNPLTWPEILRQLALSAGFGPKLKKKHSRLTNTGDKDEAKGCED 693
Query: 401 ------QGALRREALS--KELNLLI----NYGISPGTLKGELFKILSERGNNGCKVSDLA 448
G A + +E LL + ++PGT+K F +LS G+ G V +LA
Sbjct: 694 VISTIRNGTAAESAFASMREKGLLAPRKSRHRLTPGTVKFAAFHVLSLEGSKGLTVLELA 753
Query: 449 KATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLRMSTVTKDDD-------- 500
Q + L +T++ E+ I L+ D+ LFE+I+ S Y +R V D
Sbjct: 754 DKIQKSGLRDLTTSKTPEASISVALTRDVKLFERIAPSTYCVRAPYVKDPKDGEAILADA 813
Query: 501 --------------------ECQSDMEDYGSVDDELDDSDTCSCGDDFE----------- 529
E D E D E+DD T +
Sbjct: 814 RKKIRAFENGFTGPEDVNDLERDEDFEIDIDEDPEVDDLATLASASKSAVLGEANVLSGK 873
Query: 530 ---------SGSVDSNIRK---------LKRVKSHKTKSAKLTVFNE----IDESHPGEV 567
V S + K +K + ++ K TV IDES+ G+
Sbjct: 874 GVDTMFCDVKADVKSELEKEFSSPPPSTMKSIVPQHSERHKNTVVGGVDAVIDESNQGQS 933
Query: 568 WLLGLMESEYSDLNIEEKLNALAALTDLLSSGSSIR------------MKDPVKVTADYD 615
W+ GL E +Y L++EE+LNAL AL + + G+SIR +K + A D
Sbjct: 934 WIQGLTEGDYCHLSVEERLNALVALVGIANEGNSIRTGLEDRMEAANALKKQMWAEAQLD 993
Query: 616 SS-------IQLRGSGAKIKRSVVKKPV-----------------------PFWNQFGQM 645
+S + L+ + S + P+ P + +
Sbjct: 994 NSCMRDVLKLDLQNLASSKTESTIGLPIIQSSTRERDSFDRDPSQLLDETKPLEDLSNDL 1053
Query: 646 QRVK-EGHLNSHPCPVDSSSLMSKFRSHEPSFEKGKGSTDSHPIQSVYLGSDRRYNRYWL 704
+ E L + + + SK + G + + +P +S+ LG DRR+NRYW
Sbjct: 1054 HKSSAERALINQDANISQENYASKRSRSQLKSYIGHKAEEVYPYRSLPLGQDRRHNRYWH 1113
Query: 705 FLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKREALLIESLERRQT 764
F + DP R+ F DG W +ID+EEA L++ LD RG RE+ L L++ +
Sbjct: 1114 FAVSVSKSDPC-SRLLFVELHDGKWLLIDSEEAFDILVASLDMRGIRESHLRIMLQKIEG 1172
Query: 765 SL----CRFMSRVKVNGIGTGCMSHSDQSELDMVTEDSNSPVSDVDNLNLSDSAKSLPSA 820
S C+ + + + +++S ++ DS SP S + + SDS ++ S
Sbjct: 1173 SFKENACKDIKLARNPFL-------TEKSVVNHSPTDSVSPSSSAISGSNSDSMETSTS- 1224
Query: 821 GAVVIEAGKGVEEQVQKWIRVQEYDSWIWNSFYLDLNVV--KYGRRSYLDSLARCKSCHD 878
+ ++ G+ E R ++ W+W Y L KYG++ + LA C +C
Sbjct: 1225 --IRVDLGRNDTENKNLSKRFHDFQRWMWTETYSSLPSCARKYGKKRS-ELLATCDACVA 1281
Query: 879 LYWRDERHCKICHMTFELDFDLEERYAIHIATCREKENSKTHPNHKVLSSQIQSLKAAIY 938
Y + C CH ++ D E +A L ++ LK +
Sbjct: 1282 SYLSEYTFCSSCHQRLDV-VDSSEILDSGLAV-------------SPLPFGVRLLKPLLV 1327
Query: 939 AIESVMPEDSLVGAWRKSAHKLWVKRLRRTSTLVELVQVLTDFVGAINESWLFQCKFPDG 998
+E+ +P+++L W + K W RL +S+ EL+QVLT AI + L F
Sbjct: 1328 FLEASVPDEALESFWTEDQRKKWGFRLNTSSSPGELLQVLTSLESAIKKESL-SSNFMS- 1385
Query: 999 MVEEIIASFAS-------------MPHTSSALALWLVKLDTII 1028
+E++ + + +P T SA+AL L +LD I
Sbjct: 1386 -AKELLGAANAEADDQGSVDVLPWIPKTVSAVALRLSELDASI 1427
>gb|AAF16763.1| F3M18.14 [Arabidopsis thaliana] gi|25403342|pir||E86410 protein
F3M18.14 [imported] - Arabidopsis thaliana
Length = 1819
Score = 200 bits (508), Expect = 2e-49
Identities = 240/992 (24%), Positives = 387/992 (38%), Gaps = 238/992 (23%)
Query: 239 ISEEGVDRISMLIDDEELELRELQERTNLLICSDHLAASGMLGCSLGKDVLVKYPPDTVK 298
I++E +D LI+DE+LEL EL + L L + + +D L +PP +++
Sbjct: 588 IAKESMD----LIEDEQLELMELAAISKGLPSVLQLDHDTLQNLEVYRDSLSTFPPKSLQ 643
Query: 299 MKKPIHLQPWDSSPELVKKLFK-----------------------------DSMLLGKIH 329
+K P + PW S E V L DS LLG+IH
Sbjct: 644 LKMPFAISPWKDSDETVGNLLMVWRFLISFSDVLDLWPFTLDEFIQAFHDYDSRLLGEIH 703
Query: 330 VALLTLLLSDIEVELSNGFA----------------PHLDKSCNFLALLHSVESQEYSPD 373
V LL ++ D+E F+ P + + F S + +
Sbjct: 704 VTLLRSIIRDVEDVARTPFSGIGNNQYTTANPEGGHPQIVEGVAFFV---SAYAWGFDIR 760
Query: 374 FWRRSLNSLTWIEILRQVLVASGFGSK----------------------------QGALR 405
W++ LN LTW EILRQ+ +++GFG K G
Sbjct: 761 SWKKHLNPLTWPEILRQLALSAGFGPKLKKKHSRLTNTGDKDEAKGCEDVISTIRNGTAA 820
Query: 406 REALS--KELNLLI----NYGISPGTLKGELFKILSERGNNGCKVSDLAKATQIAELNLA 459
A + +E LL + ++PGT+K F +LS G+ G V +LA Q + L
Sbjct: 821 ESAFASMREKGLLAPRKSRHRLTPGTVKFAAFHVLSLEGSKGLTVLELADKIQKSGLRDL 880
Query: 460 STTEELESLICSTLSSDITLFEKISSSAYRLRMSTVTKDDD------------------- 500
+T++ E+ I L+ D+ LFE+I+ S Y +R V D
Sbjct: 881 TTSKTPEASISVALTRDVKLFERIAPSTYCVRAPYVKDPKDGEAILADARKKIRAFENGF 940
Query: 501 ---------ECQSDMEDYGSVDDELDDSDTCSCGDDFE--------------------SG 531
E D E D E+DD T +
Sbjct: 941 TGPEDVNDLERDEDFEIDIDEDPEVDDLATLASASKSAVLGEANVLSGKGVDTMFCDVKA 1000
Query: 532 SVDSNIRK---------LKRVKSHKTKSAKLTVFNE----IDESHPGEVWLLGLMESEYS 578
V S + K +K + ++ K TV IDES+ G+ W+ GL E +Y
Sbjct: 1001 DVKSELEKEFSSPPPSTMKSIVPQHSERHKNTVVGGVDAVIDESNQGQSWIQGLTEGDYC 1060
Query: 579 DLNIEEKLNALAALTDLLSSGSSIR------------MKDPVKVTADYDSS-------IQ 619
L++EE+LNAL AL + + G+SIR +K + A D+S +
Sbjct: 1061 HLSVEERLNALVALVGIANEGNSIRTGLEDRMEAANALKKQMWAEAQLDNSCMRDVLKLD 1120
Query: 620 LRGSGAKIKRSVVKKPV-----------------------PFWNQFGQMQRVK-EGHLNS 655
L+ + S + P+ P + + + E L +
Sbjct: 1121 LQNLASSKTESTIGLPIIQSSTRERDSFDRDPSQLLDETKPLEDLSNDLHKSSAERALIN 1180
Query: 656 HPCPVDSSSLMSKFRSHEPSFEKGKGSTDSHPIQSVYLGSDRRYNRYWLFLGPCNADDPG 715
+ + SK + G + + +P +S+ LG DRR+NRYW F + DP
Sbjct: 1181 QDANISQENYASKRSRSQLKSYIGHKAEEVYPYRSLPLGQDRRHNRYWHFAVSVSKSDPC 1240
Query: 716 HKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKREALLIESLERRQTSL----CRFMS 771
R+ F DG W +ID+EEA L++ LD RG RE+ L L++ + S C+ +
Sbjct: 1241 -SRLLFVELHDGKWLLIDSEEAFDILVASLDMRGIRESHLRIMLQKIEGSFKENACKDIK 1299
Query: 772 RVKVNGIGTGCMSHSDQSELDMVTEDSNSPVSDVDNLNLSDSAKSLPSAGAVVIEAGKGV 831
+ + +++S ++ DS SP S + + SDS ++ S + ++ G+
Sbjct: 1300 LARNPFL-------TEKSVVNHSPTDSVSPSSSAISGSNSDSMETSTS---IRVDLGRND 1349
Query: 832 EEQVQKWIRVQEYDSWIWNSFYLDLNVV--KYGRRSYLDSLARCKSCHDLYWRDERHCKI 889
E R ++ W+W Y L KYG++ + LA C +C Y + C
Sbjct: 1350 TENKNLSKRFHDFQRWMWTETYSSLPSCARKYGKKRS-ELLATCDACVASYLSEYTFCSS 1408
Query: 890 CHMTFELDFDLEERYAIHIATCREKENSKTHPNHKVLSSQIQSLKAAIYAIESVMPEDSL 949
CH ++ D E +A L ++ LK + +E+ +P+++L
Sbjct: 1409 CHQRLDV-VDSSEILDSGLAV-------------SPLPFGVRLLKPLLVFLEASVPDEAL 1454
Query: 950 VGAWRKSAHKLWVKRLRRTSTLVELVQVLTDFVGAINESWLFQCKFPDGMVEEIIASFAS 1009
W + K W RL +S+ EL+QVLT AI + L F +E++ + +
Sbjct: 1455 ESFWTEDQRKKWGFRLNTSSSPGELLQVLTSLESAIKKESL-SSNFMS--AKELLGAANA 1511
Query: 1010 -------------MPHTSSALALWLVKLDTII 1028
+P T SA+AL L +LD I
Sbjct: 1512 EADDQGSVDVLPWIPKTVSAVALRLSELDASI 1543
>dbj|BAB10985.1| unnamed protein product [Arabidopsis thaliana]
gi|15241428|ref|NP_199231.1| homeobox transcription
factor, putative [Arabidopsis thaliana]
Length = 1694
Score = 186 bits (471), Expect = 5e-45
Identities = 250/1085 (23%), Positives = 420/1085 (38%), Gaps = 259/1085 (23%)
Query: 163 REVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVT 222
RE + ++ RE + + R+L+ K + + L+++ + L QR+ +
Sbjct: 360 REEQIRKEMERQDRERRKEEE-----RLLREKQREEERYLKEQMREL-QRREKFLKKETI 413
Query: 223 RNQSLKEKCELSLDSEISE----------EGVDRISM-LIDDEELELRELQERTNLLICS 271
R + +++K E+ + E++ + + SM LI+DE LEL E+ T L
Sbjct: 414 RAEKMRQKEEMRKEKEVARLKAANERAIARKIAKESMELIEDERLELMEVAALTKGLPSM 473
Query: 272 DHLAASGMLGCSLGKDVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFK----------- 320
L + +D +PP +VK+KKP ++PW+ S E V L
Sbjct: 474 LALDFETLQNLDEYRDKQAIFPPTSVKLKKPFAVKPWNGSDENVANLLMVWRFLITFADV 533
Query: 321 ------------------DSMLLGKIHVALLTLLLSDIEV---ELSNGFAPHLDKSCNFL 359
D L+G+IH+ LL ++ DIE LS G + + + N
Sbjct: 534 LGLWPFTLDEFAQAFHDYDPRLMGEIHIVLLKTIIKDIEGVVRTLSTGVGANQNVAANPG 593
Query: 360 ALLHSVESQEYSPDF----WRRSLNSLTWIEILRQVLVASGFGSKQGALRREALS----- 410
V Y+ F WR++LN TW EILRQ+ +++G G + + +S
Sbjct: 594 GGHPHVVEGAYAWGFDIRSWRKNLNVFTWPEILRQLALSAGLGPQLKKMNIRTVSVHDDN 653
Query: 411 ---KELNLLIN--------------------------YGISPGTLKGELFKILSERGNNG 441
N++ N + ++PGT+K F +LS G G
Sbjct: 654 EANNSENVIFNLRKGVAAENAFAKMQERGLSNPRRSRHRLTPGTVKFAAFHVLSLEGEKG 713
Query: 442 CKVSDLAKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLRMSTVTKDDDE 501
+ ++A+ Q + L +T+ E+ + + LS D LFE+++ S Y +R S KD +
Sbjct: 714 LNILEVAEKIQKSGLRDLTTSRTPEASVAAALSRDTKLFERVAPSTYCVRAS-YRKDAGD 772
Query: 502 CQ--------------SDMEDYGSVDD-ELDDSDTCSCGDD------------------- 527
+ S + D VDD E D+ G+D
Sbjct: 773 AETIFAEARERIRAFKSGITDVEDVDDAERDEDSESDVGEDPEVDVNLKKEDPNPLKVEN 832
Query: 528 -------FESGSVDS-----------------NIRKLKRVKSHKTKSAKLTVFNE----- 558
E+G +D+ ++ KR + +S + V N
Sbjct: 833 LIGVEPLLENGKLDTVPMKTELGLPLTPSLPEEMKDEKRDDTLADQSLEDAVANGEDSAC 892
Query: 559 IDESHPGEVWLLGLMESEYSDLNIEEKLNALA--------------ALTDLLSSGSSI-- 602
DES GE W+ GL+E +YS+L+ EE+LNAL AL + L S++
Sbjct: 893 FDESKLGEQWVQGLVEGDYSNLSSEERLNALVALIGIATEGNTIRIALEERLEVASALKK 952
Query: 603 ----------RMKDPVKVTADYDS--------SIQLRGSGAKIKRSVVKKPVPFWNQFG- 643
R K+ + A+Y S +I SG + S P+ +
Sbjct: 953 QMWGEVQLDKRWKEESLIRANYLSYPTAKPGLNIATPASGNQESSSADVTPISSQDPVSL 1012
Query: 644 --------------QMQRVKEGHLN-----SHPCPVDSSSLMSKFRSHEPSFEKGKGSTD 684
Q+Q G N D L ++ +++ G + +
Sbjct: 1013 PQIDVNNVIAGPSLQLQENVPGVENLQYQQQQGYTADRERLRAQLKAYV-----GYKAEE 1067
Query: 685 SHPIQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSV 744
+ +S+ LG DRR NRYW F + +DPG R++ E +DG W +ID+EEA L+
Sbjct: 1068 LYVYRSLPLGQDRRRNRYWRFSASASRNDPGCGRIFVE-LQDGRWRLIDSEEAFDYLVKS 1126
Query: 745 LDDRGKREALLIESLERRQTSLCRFMSRVKVNGIGTGCMSHSDQSELDMVTEDSNSPVSD 804
LD RG RE+ L L + + S + R G +S S S+ + +S
Sbjct: 1127 LDVRGVRESHLHFMLLKIEASFKEALRRNVAANPGVCSISSSLDSD--------TAEIST 1178
Query: 805 VDNLNLSDSAKSLPSAGAVVIEAGKGVEEQVQKWIRVQEYDSWIWNSFY--LDLNVVKYG 862
+ L DS E+ R ++ W+W++ L+ KYG
Sbjct: 1179 TFKIELGDS----------------NAVERCSVLQRFHSFEKWMWDNMLHPSALSAFKYG 1222
Query: 863 RRSYLDSLARCKSCHDLYWRDERHCKICHMTFELDFDLEERYAIHIATCREKENSKTHPN 922
+ C+ C +L++ + C C E +A +A + +N +
Sbjct: 1223 AKQSSPLFRICRICAELHFVGDICCPSCGQMHAGPDVGELCFAEQVA--QLGDNLRRGDT 1280
Query: 923 HKVLSSQIQS------LKAAIYAIESVMPEDSLVGAWRKSAHKLWVKRLRRTSTLVELVQ 976
+L S I S LK + +E+ +P + L W ++ K W +L +S+ +L Q
Sbjct: 1281 GFILRSSILSPLRIRLLKVQLALVEASLPPEGLEAFWTENLRKSWGMKLLSSSSHEDLYQ 1340
Query: 977 VLTDFVGAINESWLFQCKFPD-----GMVEEIIASFAS--------MPHTSSALALWLVK 1023
VLT A+ +L F G+ E +AS + +P T+ +AL L
Sbjct: 1341 VLTTLEAALKRDFL-SSNFETTSELLGLQEGALASDLTCGVNVLPWIPKTAGGVALRLFD 1399
Query: 1024 LDTII 1028
D+ I
Sbjct: 1400 FDSSI 1404
>ref|XP_479178.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|33146625|dbj|BAC79913.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|33146879|dbj|BAC79877.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 330
Score = 173 bits (439), Expect = 2e-41
Identities = 110/305 (36%), Positives = 163/305 (53%), Gaps = 12/305 (3%)
Query: 63 LGTVLRNDGPSIGREFDYLPSGHKSYNSTCREDQQPAKRSKVSKGTGKGLMNASVKKHGI 122
L V R DGP +G EFD LP + +D + + K + ++ + ++ +
Sbjct: 26 LRKVFRKDGPPLGSEFDPLPHSAPGHLRDTTDDHFYQNQRVIKK---RKIVEPTTQRSSL 82
Query: 123 GKGLMNASVKKHGIGKGLMTIWRATNPDSGELPVGFGFADREVHLISNSKKSVRENNRSS 182
G N V+KHG GKGLMT+W A S ++ G F D + +S+R +
Sbjct: 83 PCG-DNGPVRKHGAGKGLMTVWHAMYSHSSKIQDGSNFIDE-----TGCLRSLRPLDDCG 136
Query: 183 KTATMN--RMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSLKEKCELSLDSEIS 240
+ + ++++ K+ ++ ++ R +RK S + E C LS+D S
Sbjct: 137 RIEDCDDGKLIQKKVLARKKVVKRTRPPSNKRKVPSSRVTDPKKHPPME-CHLSVDESQS 195
Query: 241 EEGVDRISMLIDDEELELRELQERTNLLICSDHLAASGMLGCSLGKDVLVKYPPDTVKMK 300
L+DDEELELRELQ N L CS HL++SG GC L KD+L ++PP +VKMK
Sbjct: 196 PVLQANQVTLVDDEELELRELQAGPNPLRCSAHLSSSGRHGCPLCKDLLSRFPPSSVKMK 255
Query: 301 KPIHLQPWDSSPELVKKLFKDSMLLGKIHVALLTLLLSDIEVELSNGFAPHLDKSCNFLA 360
+P +PW SSPE+VKKLF+DSMLLG++HV LL LLL + E ++ F P K C FL+
Sbjct: 256 QPFSTRPWGSSPEMVKKLFQDSMLLGEVHVNLLKLLLLNTERGSNDVFVPRSSKDCRFLS 315
Query: 361 LLHSV 365
++ V
Sbjct: 316 FVNFV 320
>ref|NP_916860.1| P0007F06.8 [Oryza sativa (japonica cultivar-group)]
Length = 1790
Score = 170 bits (431), Expect = 2e-40
Identities = 196/834 (23%), Positives = 330/834 (39%), Gaps = 240/834 (28%)
Query: 163 REVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRK------LLMQRKAGE 216
RE + ++ RE + + R+L+ + + + L+++R+ MQ+++
Sbjct: 516 REEQMRKEMERHDRERRKEEE-----RLLRERQREQERFLREQRREHERMEKFMQKQSRR 570
Query: 217 SNQHVTRNQSLKEKCELSLDSEISEEGVDR-----ISMLIDDEELELRELQERTNLLICS 271
+ + + + KEK E + +E R L++DE LEL EL ++ L
Sbjct: 571 AEKQRQKEELRKEK-EAARQKAANERATARRIAREYMELVEDECLELMELAAQSKGLPSM 629
Query: 272 DHLAASGMLGCSLGKDVLVKYPPDTVKMKKPIHLQPWDSSPELVKKLFK----------- 320
L + + + +L +PP+ V++K+P ++PW S + V L
Sbjct: 630 LSLDSDTLQQLDSFRGMLTPFPPEPVRLKEPFSIKPWTVSEDNVGNLLMVWKFSITFADV 689
Query: 321 ------------------DSMLLGKIHVALLTLLLSDIE-VELSNGFAPHLDKSCNFLAL 361
DS LLG++H+ALL ++ DIE V + A ++ + +
Sbjct: 690 LGLSSVTFDEFVQSLHDYDSRLLGELHIALLKSIIKDIEDVSRTPSVALAVNPAGGHPQI 749
Query: 362 LHSVESQEYSPDFWRRSLNSLTWIEILRQVLVASGFGSK--------------------- 400
+ + ++ W+R LN LTW EILRQ +++GFG +
Sbjct: 750 VEGAYAWGFNIRSWQRHLNVLTWPEILRQFALSAGFGPQLKKRNAEDVYYRDDNEGHDGQ 809
Query: 401 ---------QGALRREALSKELNLL----INYGISPGTLKGELFKILSERGNNGCKVSDL 447
A+ AL KE + ++PGT+K F +LS G+ G + ++
Sbjct: 810 DVISTLRNGSAAVHAAALMKERGYTHRRRSRHRLTPGTVKFAAFHVLSLEGSKGLTILEV 869
Query: 448 AKATQIAELNLASTTEELESLICSTLSSDITLFEKISSSAYRLRMSTVTKD--------- 498
A+ Q + L +T++ E+ I + LS D LFE+ + S Y ++ S KD
Sbjct: 870 AERIQKSGLRDLTTSKTPEASIAAALSRDTKLFERTAPSTYCVK-SPYRKDPADSEVVLS 928
Query: 499 --------------DDECQSDMEDYGSVDD-ELDDSDTCSCGDDFE-------------- 529
D E + + D +D E DD+D GDD
Sbjct: 929 SAREKIRAFQNVISDSEAEKEANDAERDEDSECDDADDDPDGDDVNIDVGDGKDPLIGVK 988
Query: 530 -------SGSVDSNIRKLKRVKSHKTKSAKLTVFN------------------------- 557
+ VDS R+ ++V + T+S+ LT
Sbjct: 989 EQDGVPITTIVDSTKREKEKVDA-LTQSSDLTTSGKEAPKPSLGKPSSANTSSDSPVRAS 1047
Query: 558 --------------EIDESHPGEVWLLGLMESEYSDLNIEEKLNALAALTDL-------- 595
EIDES+ GE W+ GL E +Y DL++EE+LNAL AL +
Sbjct: 1048 SEYHEVPPTDAEDKEIDESNQGESWVHGLAEGDYCDLSVEERLNALVALVSVANEGNFIR 1107
Query: 596 ------LSSGSSI-------------RMKDPVKVTADYDSSIQLRGSGAKIKRSVVKKPV 636
L S +++ R K+ Y+S++ L+ + + P
Sbjct: 1108 AVLEERLESANALKKQMLAEAQLDKRRSKEEFAGRVQYNSNMNLKAD-VNQENATESTPT 1166
Query: 637 PFWNQFGQMQRVKEGHLNSHPCPVDSSS---LMSKFRSHEPSFEKG------KGSTDSHP 687
PF N + H + + VD+++ + + S+E+ + D+
Sbjct: 1167 PFHNV--------DKHNDGNTGVVDNNNNEIIDHNSNAANASYERNGLGQDIAATPDTLS 1218
Query: 688 IQ---------------------------SVYLGSDRRYNRYWLFLGPCNADDPGHKRVY 720
+Q S+ LG DRR NRYW F + +DPG R++
Sbjct: 1219 VQQYAYADKTRSQLRAYIGHRAEQLFVYRSLPLGQDRRRNRYWQFSTSASPNDPGSGRIF 1278
Query: 721 FESSEDGHWEVIDTEEALCTLLSVLDDRGKREALLIESLERRQTSLCRFMSRVK 774
FE DG+W V+DTEEA +L++ LD RG REA L L+R + + + R K
Sbjct: 1279 FEC-RDGYWRVLDTEEAFDSLVASLDTRGSREAQLHSMLQRIEPTFKEAIKRKK 1331
>ref|XP_466214.1| putative DDT domain-containing protein [Oryza sativa (japonica
cultivar-group)] gi|46389867|dbj|BAD15468.1| putative
DDT domain-containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 617
Score = 47.8 bits (112), Expect = 0.002
Identities = 27/85 (31%), Positives = 42/85 (48%), Gaps = 8/85 (9%)
Query: 688 IQSVYLGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDD 747
I+S LG DR YNRYW F R++ ES++ W T+E L L+S L+
Sbjct: 497 IRSSPLGKDRHYNRYWFF--------RREGRLFVESADSKEWGYYSTKEELDVLMSSLNV 548
Query: 748 RGKREALLIESLERRQTSLCRFMSR 772
+G RE L L++ + + + +
Sbjct: 549 KGLRERALKRQLDKLYSKISNALEK 573
>dbj|BAB10012.1| unnamed protein product [Arabidopsis thaliana]
Length = 723
Score = 45.4 bits (106), Expect = 0.010
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Query: 693 LGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKRE 752
LG DR YNRYW F + R++ E+S+ W +E L L+ L+ +G+RE
Sbjct: 608 LGKDRDYNRYWWFRS--------NGRIFVENSDSEEWGYYTAKEELDALMGSLNRKGERE 659
Query: 753 ALLIESLERRQTSLCRFMSR 772
L LE +C + +
Sbjct: 660 LSLYTQLEIFYDRICSTLQK 679
>ref|NP_196480.2| DDT domain-containing protein [Arabidopsis thaliana]
Length = 723
Score = 45.4 bits (106), Expect = 0.010
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Query: 693 LGSDRRYNRYWLFLGPCNADDPGHKRVYFESSEDGHWEVIDTEEALCTLLSVLDDRGKRE 752
LG DR YNRYW F + R++ E+S+ W +E L L+ L+ +G+RE
Sbjct: 608 LGKDRDYNRYWWFRS--------NGRIFVENSDSEEWGYYTAKEELDALMGSLNRKGERE 659
Query: 753 ALLIESLERRQTSLCRFMSR 772
L LE +C + +
Sbjct: 660 LSLYTQLEIFYDRICSTLQK 679
>ref|NP_702166.1| coatamer protein, beta subunit, putative [Plasmodium falciparum
3D7] gi|23497345|gb|AAN36890.1| coatamer protein, beta
subunit, putative [Plasmodium falciparum 3D7]
Length = 1394
Score = 44.7 bits (104), Expect = 0.017
Identities = 49/193 (25%), Positives = 90/193 (46%), Gaps = 21/193 (10%)
Query: 365 VESQEYSPDFWRRSLNSLTWIEILRQVL--VASGFGSKQGALRREALSKELNLLINYGIS 422
+ EY R L+ + +++IL ++ + G + +R+ A++ ++ ++G+
Sbjct: 111 ISPNEYVRGSTLRLLSKIKYLKILDPLMETITKNLGHRHSYVRKNAITCIHTIIKHHGVD 170
Query: 423 --PGTLKGELFKILSERGNNGCKVSDLAKATQIAELN----LASTTEELESLICSTLSSD 476
P +K E+ KIL G+ K S LA T I L + S ++L L
Sbjct: 171 IIPNAVK-EVEKILFLEGDISTKRSALAMLTDIDPLTTLKYILSLNDQLYDTADVILLEV 229
Query: 477 ITLFEK-----ISSSAYRLRMSTVTKDDDECQSDMEDYGSVDDELDDSDTCSCGDDFESG 531
I LF+K + +Y L ++ KDDD+ +D E+ DE DD GDD
Sbjct: 230 IHLFKKLYIPHVFDDSYLL-INDEDKDDDDENND-ENNDKESDEEDDH-----GDDHNDY 282
Query: 532 SVDSNIRKLKRVK 544
+++SN+++ ++ K
Sbjct: 283 NINSNLKEEEKYK 295
>ref|XP_395223.2| PREDICTED: similar to bromodomain adjacent to zinc finger domain,
1A isoform a [Apis mellifera]
Length = 1320
Score = 44.3 bits (103), Expect = 0.022
Identities = 31/90 (34%), Positives = 42/90 (46%), Gaps = 15/90 (16%)
Query: 691 VYLGSDRRYNRYWLFL--------------GPCNADDPGHKRVYFESSEDGHWEVIDTEE 736
VYLGSDR Y RYW FL G C +D V+++ SE W +E
Sbjct: 613 VYLGSDRAYRRYWRFLSIPGIFVENDEWWPGNCISDGDKECPVHWKRSE-LKWSFFGKQE 671
Query: 737 ALCTLLSVLDDRGKREALLIESLERRQTSL 766
+ L++ L RG RE L ++ + TSL
Sbjct: 672 DIEALVNSLSKRGIREGELRNNIIQEMTSL 701
>emb|CAH78991.1| conserved hypothetical protein [Plasmodium chabaudi]
Length = 420
Score = 42.7 bits (99), Expect = 0.063
Identities = 32/125 (25%), Positives = 53/125 (41%), Gaps = 3/125 (2%)
Query: 150 DSGELPVGFGFADREVHLISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLL 209
+S E VG ++ L N KS ++NN+ T N+ KNK + NLQ K
Sbjct: 170 ESSEYEVGRLKKKKKKKLKKNDGKS-KKNNQDQPNLTENKSHKNKKETSNTNLQQKTTQN 228
Query: 210 MQRKAGESNQHV--TRNQSLKEKCELSLDSEISEEGVDRISMLIDDEELELRELQERTNL 267
Q + + T +K++ L+ +E + E + + + +DE L + E NL
Sbjct: 229 CQSNENDKEPIIGETSKSGIKKQDNLASSAEENVENAENVENVENDEVTNLESMLENNNL 288
Query: 268 LICSD 272
D
Sbjct: 289 FDSCD 293
>ref|XP_653447.1| Viral A-type inclusion protein repeat, putative [Entamoeba
histolytica HM-1:IMSS] gi|56470397|gb|EAL48061.1| Viral
A-type inclusion protein repeat, putative [Entamoeba
histolytica HM-1:IMSS]
Length = 1813
Score = 40.8 bits (94), Expect = 0.24
Identities = 26/114 (22%), Positives = 58/114 (50%), Gaps = 15/114 (13%)
Query: 168 ISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSL 227
+SN V + N T + K ++ N+ N++++++K + + K NQ + N+ +
Sbjct: 1233 LSNGSDGVSKLNEE---LTQTKQEKEEINNELNSIKEEKKRIEEEK----NQIINENKEI 1285
Query: 228 KE--------KCELSLDSEISEEGVDRISMLIDDEELELRELQERTNLLICSDH 273
KE K EL + E +EG +++ I+ + ++E++E+ +IC ++
Sbjct: 1286 KEEKEKIEEEKKELLKEIEKEKEGNNQLQNEINTIQTRMKEIEEKNQEIICDNN 1339
Score = 40.4 bits (93), Expect = 0.31
Identities = 25/114 (21%), Positives = 58/114 (49%), Gaps = 15/114 (13%)
Query: 168 ISNSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSL 227
+SN + + N T + K ++ N+ N++++++K + + K NQ + N+ +
Sbjct: 351 LSNGSDGISKLNEE---LTQTKQEKEEINNELNSIKEEKKRIEEEK----NQIINENKEI 403
Query: 228 KE--------KCELSLDSEISEEGVDRISMLIDDEELELRELQERTNLLICSDH 273
KE K EL + E +EG +++ I+ + ++E++E+ +IC ++
Sbjct: 404 KEEKEKIEEEKKELLKEIEKEKEGNNQLQNEINTIQTRMKEIEEKNQEIICDNN 457
Score = 35.8 bits (81), Expect = 7.7
Identities = 23/99 (23%), Positives = 51/99 (51%), Gaps = 5/99 (5%)
Query: 170 NSKKSVRENNRSSKTATMNRMLKNKLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQSLKE 229
N K ++ + K ++ +K + +N+ N L+++++ + K N + NQ ++E
Sbjct: 475 NQIKEEKQKTENEKNELVD--VKTQKENELNKLKEEKEQIFNEKTTIEN---SLNQIVEE 529
Query: 230 KCELSLDSEISEEGVDRISMLIDDEELELRELQERTNLL 268
K +L+ + E ++ +D I +ELE+ ++ E N L
Sbjct: 530 KNKLTEEKESIKQELDSIKADNSTKELEINKINEEKNQL 568
>ref|XP_649508.1| conserved hypothetical protein [Entamoeba histolytica HM-1:IMSS]
gi|56465967|gb|EAL44122.1| conserved hypothetical
protein [Entamoeba histolytica HM-1:IMSS]
Length = 762
Score = 40.4 bits (93), Expect = 0.31
Identities = 28/117 (23%), Positives = 58/117 (48%), Gaps = 9/117 (7%)
Query: 152 GELPVGFGFADREVHLISNSKKSVRENNRSSKTATMNRML---KNKLQNKRNNLQDKRKL 208
GEL D E+H KK + E + + A+ +RM K +++ ++N +++K
Sbjct: 413 GELEEEKQRIDEEMHKFEEEKKRIEEEINNKRGASASRMYFAKKQEIELQQNKIENK--- 469
Query: 209 LMQRKAGESNQHVTRNQS-LKEKCELSLDSEISEEGVDRISMLIDDEELELRELQER 264
Q++A + + +NQS +K K E + E + +D+ + ++E+ E+Q +
Sbjct: 470 --QKEATTNKMKIKKNQSEIKRKQEEIIKKEKEIKEIDKRKNIQEEEQKRQEEIQRK 524
>gb|AAL98800.1| ORF076L [infectious spleen and kidney necrosis virus]
gi|19881481|ref|NP_612298.1| ORF076L [infectious spleen
and kidney necrosis virus]
Length = 990
Score = 40.0 bits (92), Expect = 0.41
Identities = 43/161 (26%), Positives = 71/161 (43%), Gaps = 16/161 (9%)
Query: 881 WRDERHCKICHMTFELDFDLEERYAIHIATCREKENSKTHPNHKVLSSQIQSLKAAIYAI 940
W RH E + Y +A ++ N THP+ VL + + +++ +
Sbjct: 8 WNGARH--------EAQLHAMQTYDDFVAQLAKEHN--THPSFMVLEN-VHNMQTLLERR 56
Query: 941 ESVMPEDSLVGAWRKS---AHKLWVKRLRRTSTLVELVQVLTDFVGAINESWLFQCKFPD 997
ESV D+L A+R++ A T L+ + V+T + A F+ PD
Sbjct: 57 ESVAVYDALPYAYRRTEFAAPNFTKVASDETRVLLSHIFVVTHALRASTPDMYFRMMIPD 116
Query: 998 GMVEEIIASFASMPHTSSALALWL-VKLDTIIAPYLDRVYL 1037
G+V+ I A A P+ ++ L L V++D AP +D V L
Sbjct: 117 GIVQNIDAVLADTPNITARLRRMLGVQVDR-HAPVVDDVTL 156
>gb|EAA42178.1| GLP_480_88069_85913 [Giardia lamblia ATCC 50803]
Length = 718
Score = 40.0 bits (92), Expect = 0.41
Identities = 43/157 (27%), Positives = 70/157 (44%), Gaps = 31/157 (19%)
Query: 194 KLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQ---SLKEKCE-LSLDSEISEEGVDRISM 249
K Q K Q+K L Q + +H T+ S EK E L D +E ++ ++
Sbjct: 500 KAQQKLTTRQEKHNALQQALQNKQTEHQTKTAEILSAVEKLERLKADISKTEAEINEVNQ 559
Query: 250 LIDD-----------EELELRELQERTNLLICSDHLAASGMLGCSLGK-DVLVKYPPDTV 297
IDD E EL+E+ +R+ LL D + + + C + K + L +Y +T+
Sbjct: 560 EIDDITEQHALSIEEERRELKEVDKRSALL---DAIIQTFIPQCEVAKAEALAEYDEETM 616
Query: 298 KMKKPIHLQPWDSSPELVKKLFKDSMLLGKI-HVALL 333
K W SPE V + + MLL ++ H+ +L
Sbjct: 617 K---------WAISPEKVDR--EHQMLLKRVRHMQIL 642
>dbj|BAB56139.1| kinesin-like protein 2 [Giardia intestinalis]
Length = 619
Score = 40.0 bits (92), Expect = 0.41
Identities = 43/157 (27%), Positives = 70/157 (44%), Gaps = 31/157 (19%)
Query: 194 KLQNKRNNLQDKRKLLMQRKAGESNQHVTRNQ---SLKEKCE-LSLDSEISEEGVDRISM 249
K Q K Q+K L Q + +H T+ S EK E L D +E ++ ++
Sbjct: 401 KAQQKLTTRQEKHNALQQALQNKQTEHQTKTAEILSAVEKLERLKADISKTEAEINEVNQ 460
Query: 250 LIDD-----------EELELRELQERTNLLICSDHLAASGMLGCSLGK-DVLVKYPPDTV 297
IDD E EL+E+ +R+ LL D + + + C + K + L +Y +T+
Sbjct: 461 EIDDITEQHALSIEEERRELKEVDKRSALL---DAIIQTFIPQCEVAKAEALAEYDEETM 517
Query: 298 KMKKPIHLQPWDSSPELVKKLFKDSMLLGKI-HVALL 333
K W SPE V + + MLL ++ H+ +L
Sbjct: 518 K---------WAISPEKVDR--EHQMLLKRVRHMQIL 543
>dbj|BAA09463.1| troponin T [Halocynthia roretzi]
Length = 248
Score = 39.7 bits (91), Expect = 0.53
Identities = 31/113 (27%), Positives = 53/113 (46%), Gaps = 14/113 (12%)
Query: 166 HLISNSKKSVRENNRSSKTATMNRML----------KNKLQNKRNNLQDKRKLLMQRKAG 215
H +K +E K A N L KNK ++ + K+KLL +RK
Sbjct: 109 HKEEEDEKKRKEEEEKKKAAIANMSLHYGGYLARAEKNKPNKRQTEREKKKKLLAERKKN 168
Query: 216 ESNQHVTRNQSLKEKCELSLD--SEISEEGVDRISMLIDDEELELRELQERTN 266
S H++ ++ L+EK + D + EE +D + ID ++ +L +L++R N
Sbjct: 169 ISVDHLSPDK-LREKAQELWDLLYSLEEEKID-YEVRIDRQKYDLNQLRQRVN 219
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,754,799,553
Number of Sequences: 2540612
Number of extensions: 75178132
Number of successful extensions: 243483
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 243090
Number of HSP's gapped (non-prelim): 378
length of query: 1041
length of database: 863,360,394
effective HSP length: 138
effective length of query: 903
effective length of database: 512,755,938
effective search space: 463018612014
effective search space used: 463018612014
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 81 (35.8 bits)
Lotus: description of TM0004a.8


