

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0002.14
(340 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
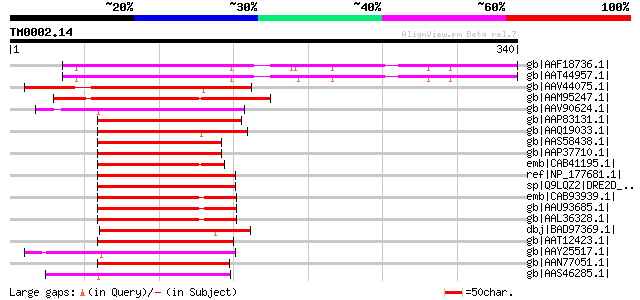
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF18736.1| AP2 domain transcription factor (ABI4:abscisic ac... 223 5e-57
gb|AAT44957.1| putative AP2/EREBP transcription factor [Arabidop... 221 2e-56
gb|AAV44075.1| hypothetical protein [Oryza sativa (japonica cult... 160 4e-38
gb|AAM95247.1| AP2 domain transcription factor [Zea mays] 151 3e-35
gb|AAV90624.1| DREB 2A [Pennisetum glaucum] 109 1e-22
gb|AAP83131.1| DREB [Glycine max] 108 3e-22
gb|AAQ19033.1| Ap22 [Oryza sativa (japonica cultivar-group)] gi|... 107 4e-22
gb|AAS58438.1| DREB2A [Thellungiella salsuginea] 107 4e-22
gb|AAP37710.1| At3g11020 [Arabidopsis thaliana] gi|6016692|gb|AA... 107 5e-22
emb|CAB41195.1| putative protein [Arabidopsis thaliana] gi|21592... 106 9e-22
ref|NP_177681.1| DRE-binding transcription factor, putative [Ara... 106 1e-21
sp|Q9LQZ2|DRE2D_ARATH Putative dehydration responsive element bi... 106 1e-21
emb|CAB93939.1| AP2-domain DNA-binding protein [Catharanthus ros... 105 1e-21
gb|AAU93685.1| DREB2A [Arabidopsis thaliana] 105 2e-21
gb|AAL36328.1| putative DREB2A protein [Arabidopsis thaliana] gi... 105 2e-21
dbj|BAD97369.1| EREBP/AP2 type transcription factor [Triticum ae... 104 3e-21
gb|AAT12423.1| DREBa transcription factor [Glycine max] 103 6e-21
gb|AAY25517.1| dehydration responsive element binding protein 1 ... 103 7e-21
gb|AAN77051.1| dehydration responsive element binding protein [L... 102 1e-20
gb|AAS46285.1| DREB-like protein 2 [Cynodon dactylon] 102 1e-20
>gb|AAF18736.1| AP2 domain transcription factor (ABI4:abscisic acid-insensitive 4 )
[Arabidopsis thaliana] gi|4587996|gb|AAD25937.1| ABI4
[Arabidopsis thaliana] gi|3282693|gb|AAC39489.1| AP2
domain family transcription factor homolog [Arabidopsis
thaliana] gi|25408675|pir||G84826 hypothetical protein
At2g40220 [imported] - Arabidopsis thaliana
gi|15225661|ref|NP_181551.1| abscisic acid-insensitive 4
(ABI4) [Arabidopsis thaliana]
Length = 328
Score = 223 bits (569), Expect = 5e-57
Identities = 148/329 (44%), Positives = 180/329 (53%), Gaps = 42/329 (12%)
Query: 36 NSNSSHNN--NITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKR 93
N +HNN + +T ++ST+ RK KGKGGPDN+KFRYRGVRQRSWGKWVAEIREPRKR
Sbjct: 17 NQTLTHNNPQSDSTTDSSTSSAQRKRKGKGGPDNSKFRYRGVRQRSWGKWVAEIREPRKR 76
Query: 94 TRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASS---SSS 150
TRKWLGTF+TAEDAARAYDRAA+ LYGSRAQLNL PS SS S SSSS S ASS SSS
Sbjct: 77 TRKWLGTFATAEDAARAYDRAAVYLYGSRAQLNLTPSSPSSVSSSSSSVSAASSPSTSSS 136
Query: 151 STQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGPFG--FYN---ANNGYMQLQPQQHHF 205
STQTLRPLLPRP+ V A GP+G F N N G L P F
Sbjct: 137 STQTLRPLLPRPAA----------ATVGGGANFGPYGIPFNNNIFLNGGTSMLCPSYGFF 186
Query: 206 HHQEVVQSQL----QPQLQQCRQPEPDVKGGGVVD-HVRSTSYQNQHSHHHQDQVVMQNY 260
Q+ Q+Q+ Q Q QQ + + + D + N S HH+
Sbjct: 187 PQQQQQQNQMVQMGQFQHQQYQNLHSNTNNNKISDIELTDVPVTNSTSFHHE-------- 238
Query: 261 PVLNHQQHNQNCMGEGVSD--NTLVGPSLASQNFVH-------IDAAPMDPDPGSGIGSP 311
L +Q C + N+L G +S + H + + +DP G GS
Sbjct: 239 VALGQEQGGSGCNNNSSMEDLNSLAGSVGSSLSITHPPPLVDPVCSMGLDPGYMVGDGSS 298
Query: 312 SIWPLTSTELEDYNPVCLWDYNDPFFLDF 340
+IWP E +N +WD+ DP +F
Sbjct: 299 TIWPFGGEEEYSHNWGSIWDFIDPILGEF 327
>gb|AAT44957.1| putative AP2/EREBP transcription factor [Arabidopsis thaliana]
Length = 327
Score = 221 bits (564), Expect = 2e-56
Identities = 146/328 (44%), Positives = 181/328 (54%), Gaps = 41/328 (12%)
Query: 36 NSNSSHNN--NITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKR 93
N +HNN + +T ++ST+ RK KGKGGPDN+KFRYRGVRQRSWGKWVAEIREPRKR
Sbjct: 17 NQTLTHNNPQSDSTTDSSTSSAQRKRKGKGGPDNSKFRYRGVRQRSWGKWVAEIREPRKR 76
Query: 94 TRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASS---SSS 150
TRKWLGTF+TAEDAARAYDRAA+ LYGSRAQLNL PS SS S SSSS S ASS SSS
Sbjct: 77 TRKWLGTFATAEDAARAYDRAAVYLYGSRAQLNLTPSSPSSVSSSSSSVSAASSPSTSSS 136
Query: 151 STQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGPFGF-YNAN---NGYMQLQPQQHHFH 206
STQTLRPLLPRP+ V A GP+G +N N NG + + F
Sbjct: 137 STQTLRPLLPRPAA----------ATVGGGANFGPYGIPFNNNIFLNGGTSMLCPSYGFF 186
Query: 207 HQEVVQSQL----QPQLQQCRQPEPDVKGGGVVD-HVRSTSYQNQHSHHHQDQVVMQNYP 261
Q+ Q+Q+ Q Q QQ + + + D + N S HH+
Sbjct: 187 PQQQQQNQMVQMGQFQHQQYQNLHSNTNNNKISDIELTDVPVANSTSFHHE--------V 238
Query: 262 VLNHQQHNQNCMGEGVSD--NTLVGPSLASQNFVH-------IDAAPMDPDPGSGIGSPS 312
L +Q C + N+L G +S + H + + +DP G GS +
Sbjct: 239 ALGQEQGGSGCNNNSSMEDLNSLAGSVGSSLSITHPPPLVDPVCSMGLDPGYMVGDGSST 298
Query: 313 IWPLTSTELEDYNPVCLWDYNDPFFLDF 340
IWP E +N +WD+ DP +F
Sbjct: 299 IWPFGGEEEYSHNWGSIWDFIDPILGEF 326
>gb|AAV44075.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 292
Score = 160 bits (406), Expect = 4e-38
Identities = 84/157 (53%), Positives = 102/157 (64%), Gaps = 15/157 (9%)
Query: 11 PPQEITKPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKF 70
PP E + T A + S+S +K GKGGP+N KF
Sbjct: 22 PPMEPSDDACTVAAPAAETAASSSGAGGGGGGGR----------TKKKAAGKGGPENGKF 71
Query: 71 RYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQ-- 128
RYRGVRQRSWGKWVAEIREPRKR+RKWLGTF+TAEDAARAYDRAA++LYG RA LNL
Sbjct: 72 RYRGVRQRSWGKWVAEIREPRKRSRKWLGTFATAEDAARAYDRAALLLYGPRAHLNLTAP 131
Query: 129 ---PSPTSSSSQSSSSSSRASSSSSSTQTLRPLLPRP 162
P P SS+ ++++SS +++SS+S LRPLLPRP
Sbjct: 132 PPLPPPPPSSAAAAAASSSSAASSTSAPPLRPLLPRP 168
>gb|AAM95247.1| AP2 domain transcription factor [Zea mays]
Length = 248
Score = 151 bits (381), Expect = 3e-35
Identities = 83/147 (56%), Positives = 102/147 (68%), Gaps = 5/147 (3%)
Query: 30 ETSNSENSNSSHNNNITTPNTSTTINNRKCKG-KGGPDNNKFRYRGVRQRSWGKWVAEIR 88
E SN+E++ ++ + P RK K KGGP+N KFRYRGVRQRSWGKWVAEIR
Sbjct: 2 EASNNESAPTAEAAAGSGPAGG---EGRKGKAPKGGPENGKFRYRGVRQRSWGKWVAEIR 58
Query: 89 EPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASSS 148
EPRKR+RKWLGTF+TAEDAARAYDRAA++LYG RA LNL SP + + S +S++
Sbjct: 59 EPRKRSRKWLGTFATAEDAARAYDRAALLLYGPRAHLNL-TSPPPPTLAAPRSHPHSSAT 117
Query: 149 SSSTQTLRPLLPRPSGYGFSFSGSHLP 175
SS+ LRPLLPRP + S G+ P
Sbjct: 118 SSAPPALRPLLPRPPLHQLSSDGAPAP 144
>gb|AAV90624.1| DREB 2A [Pennisetum glaucum]
Length = 332
Score = 109 bits (273), Expect = 1e-22
Identities = 61/143 (42%), Positives = 84/143 (58%), Gaps = 7/143 (4%)
Query: 18 PTATAAITTTPSETSNSENSNSSHNNN-ITTPNTSTTINNRK--CKGKGGPDNNKFRYRG 74
PT+ AA+ +E N H+++ P + ++K KGKGGP+N YRG
Sbjct: 25 PTSVAAVIQRWAE----HNKQLEHDSDGAKRPRKAPAKGSKKGCMKGKGGPENTHCGYRG 80
Query: 75 VRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSS 134
VRQR+WGKWVAEIREP + R WLGTF TAEDAARAYD AA +YG A+ N +
Sbjct: 81 VRQRTWGKWVAEIREPNRVNRLWLGTFPTAEDAARAYDEAARAMYGELARTNFPSQKAVA 140
Query: 135 SSQSSSSSSRASSSSSSTQTLRP 157
SSQ++ + A + ++ + + P
Sbjct: 141 SSQAARVPTPAQVAPAAVEGVVP 163
>gb|AAP83131.1| DREB [Glycine max]
Length = 198
Score = 108 bits (269), Expect = 3e-22
Identities = 53/96 (55%), Positives = 66/96 (68%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N++ YRGVRQR+WGKWVAEIREP + R WLGTF TA AA AYD AA +Y
Sbjct: 53 KGKGGPENSRCNYRGVRQRTWGKWVAEIREPNRGNRLWLGTFPTAIGAALAYDEAAQAMY 112
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSSTQTL 155
GS A+LN SS S+ SS S +++ S+ +
Sbjct: 113 GSCARLNFPNVSVSSFSEESSKDSPSANHCGSSMAV 148
>gb|AAQ19033.1| Ap22 [Oryza sativa (japonica cultivar-group)]
gi|50948607|ref|XP_483831.1| AP2-domain DNA-binding
protein-like [Oryza sativa (japonica cultivar-group)]
gi|28071316|dbj|BAC56005.1| AP2-domain DNA-binding
protein-like [Oryza sativa (japonica cultivar-group)]
gi|42409075|dbj|BAD10326.1| AP2-domain DNA-binding
protein-like [Oryza sativa (japonica cultivar-group)]
Length = 230
Score = 107 bits (268), Expect = 4e-22
Identities = 57/112 (50%), Positives = 73/112 (64%), Gaps = 12/112 (10%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N + +RGVRQR+WGKWVAEIREP + R WLGTF+TA DAARAYD AA LY
Sbjct: 43 KGKGGPENQRCPFRGVRQRTWGKWVAEIREPNRGARLWLGTFNTALDAARAYDSAARALY 102
Query: 120 GSRAQLNL------------QPSPTSSSSQSSSSSSRASSSSSSTQTLRPLL 159
G A+LNL +P+ ++ S++ S SSS++ Q L +L
Sbjct: 103 GDCARLNLLLAAATAGAPPAAATPSVATPCSTNDDSNNSSSTTHQQQLTTML 154
>gb|AAS58438.1| DREB2A [Thellungiella salsuginea]
Length = 340
Score = 107 bits (268), Expect = 4e-22
Identities = 51/83 (61%), Positives = 60/83 (71%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGPDN + +RGVRQR WGKWVAEIREP + +R WLGTF TAE+AA AYD AA +Y
Sbjct: 67 KGKGGPDNGRCSFRGVRQRIWGKWVAEIREPNRGSRLWLGTFPTAEEAASAYDEAAKAMY 126
Query: 120 GSRAQLNLQPSPTSSSSQSSSSS 142
G A+LN S S + +SS S
Sbjct: 127 GPSARLNFPESTVSDVTSTSSQS 149
>gb|AAP37710.1| At3g11020 [Arabidopsis thaliana] gi|6016692|gb|AAF01519.1| DREB2B
transcription factor [Arabidopsis thaliana]
gi|26449820|dbj|BAC42033.1| putative DREB2B
transcription factor [Arabidopsis thaliana]
gi|4126708|dbj|BAA36706.1| DREB2B [Arabidopsis thaliana]
gi|48427910|sp|O82133|DRE2B_ARATH Dehydration responsive
element binding protein 2B (DREB2B protein)
gi|15228427|ref|NP_187713.1| DRE-binding protein
(DREB2B) [Arabidopsis thaliana]
gi|3738232|dbj|BAA33795.1| DREB2B [Arabidopsis thaliana]
Length = 330
Score = 107 bits (267), Expect = 5e-22
Identities = 53/83 (63%), Positives = 60/83 (71%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGPDN+ +RGVRQR WGKWVAEIREP+ TR WLGTF TAE AA AYD AA +Y
Sbjct: 66 KGKGGPDNSHCSFRGVRQRIWGKWVAEIREPKIGTRLWLGTFPTAEKAASAYDEAATAMY 125
Query: 120 GSRAQLNLQPSPTSSSSQSSSSS 142
GS A+LN S S + +SS S
Sbjct: 126 GSLARLNFPQSVGSEFTSTSSQS 148
>emb|CAB41195.1| putative protein [Arabidopsis thaliana] gi|21592849|gb|AAM64799.1|
AP2 transcription factor-like protein [Arabidopsis
thaliana] gi|48428176|sp|Q9SVX5|DRE2F_ARATH Dehydration
responsive element binding protein 2F (DREB2F protein)
gi|15230344|ref|NP_191319.1| AP2 domain-containing
transcription factor, putative [Arabidopsis thaliana]
Length = 277
Score = 106 bits (265), Expect = 9e-22
Identities = 53/85 (62%), Positives = 64/85 (74%), Gaps = 1/85 (1%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
+GKGGP N +YRGVRQR+WGKWVAEIREP+KR R WLG+F+TAE+AA AYD AA+ LY
Sbjct: 16 RGKGGPQNALCQYRGVRQRTWGKWVAEIREPKKRARLWLGSFATAEEAAMAYDEAALKLY 75
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSR 144
G A LNL P ++ S S+S R
Sbjct: 76 GHDAYLNL-PHLQRNTRPSLSNSQR 99
>ref|NP_177681.1| DRE-binding transcription factor, putative [Arabidopsis thaliana]
Length = 197
Score = 106 bits (264), Expect = 1e-21
Identities = 52/92 (56%), Positives = 64/92 (69%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
+GKGGPDN Y+GVRQR+WGKWVAEIREP + R WLGTF T+ +AA AYD AA LY
Sbjct: 21 RGKGGPDNASCTYKGVRQRTWGKWVAEIREPNRGARLWLGTFDTSREAALAYDSAARKLY 80
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSS 151
G A LNL S S +SS +S+ + SS++
Sbjct: 81 GPEAHLNLPESLRSYPKTASSPASQTTPSSNT 112
>sp|Q9LQZ2|DRE2D_ARATH Putative dehydration responsive element binding protein 2D (DREB2D
protein) gi|51969946|dbj|BAD43665.1| transcription
factor DREB2A like protein [Arabidopsis thaliana]
gi|9369375|gb|AAF87124.1| F10A5.29 [Arabidopsis
thaliana]
Length = 206
Score = 106 bits (264), Expect = 1e-21
Identities = 52/92 (56%), Positives = 64/92 (69%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
+GKGGPDN Y+GVRQR+WGKWVAEIREP + R WLGTF T+ +AA AYD AA LY
Sbjct: 30 RGKGGPDNASCTYKGVRQRTWGKWVAEIREPNRGARLWLGTFDTSREAALAYDSAARKLY 89
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSS 151
G A LNL S S +SS +S+ + SS++
Sbjct: 90 GPEAHLNLPESLRSYPKTASSPASQTTPSSNT 121
>emb|CAB93939.1| AP2-domain DNA-binding protein [Catharanthus roseus]
Length = 376
Score = 105 bits (263), Expect = 1e-21
Identities = 53/93 (56%), Positives = 66/93 (69%), Gaps = 3/93 (3%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N+ +YRGVRQR+WGKWVAEIREP + +R WLGTF A +AA AYD AA +Y
Sbjct: 69 KGKGGPENSHCKYRGVRQRTWGKWVAEIREPNRGSRLWLGTFRNAIEAALAYDEAARAMY 128
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSST 152
G A+LNL P +S+ SSS S S ++T
Sbjct: 129 GPCARLNL---PNYRASEESSSLPTTSGSDTTT 158
>gb|AAU93685.1| DREB2A [Arabidopsis thaliana]
Length = 173
Score = 105 bits (262), Expect = 2e-21
Identities = 51/93 (54%), Positives = 68/93 (72%), Gaps = 3/93 (3%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N++ +RGVRQR WGKWVAEIREP + +R WLGTF TA++AA AYD AA +Y
Sbjct: 67 KGKGGPENSRCSFRGVRQRIWGKWVAEIREPNRGSRLWLGTFPTAQEAASAYDEAAKAMY 126
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSST 152
G A+LN P S +S+ +S+SS++ + T
Sbjct: 127 GPLARLNF---PRSDASEVTSTSSQSEVCTVET 156
>gb|AAL36328.1| putative DREB2A protein [Arabidopsis thaliana]
gi|10176753|dbj|BAB09984.1| DREB2A [Arabidopsis
thaliana] gi|4126706|dbj|BAA36705.1| DREB2A [Arabidopsis
thaliana] gi|50981013|gb|AAT91350.1| DREB-like protein
[Oryza sativa (indica cultivar-group)]
gi|15239107|ref|NP_196160.1| DRE-binding protein
(DREB2A) [Arabidopsis thaliana]
gi|48427909|sp|O82132|DRE2A_ARATH Dehydration responsive
element binding protein 2A (DREB2A protein)
gi|42742501|gb|AAS45279.1| dehydration responsive
element binding protein [Fraxinus pennsylvanica]
gi|3738230|dbj|BAA33794.1| DREB2A [Arabidopsis thaliana]
Length = 335
Score = 105 bits (262), Expect = 2e-21
Identities = 51/93 (54%), Positives = 68/93 (72%), Gaps = 3/93 (3%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N++ +RGVRQR WGKWVAEIREP + +R WLGTF TA++AA AYD AA +Y
Sbjct: 67 KGKGGPENSRCSFRGVRQRIWGKWVAEIREPNRGSRLWLGTFPTAQEAASAYDEAAKAMY 126
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSST 152
G A+LN P S +S+ +S+SS++ + T
Sbjct: 127 GPLARLNF---PRSDASEVTSTSSQSEVCTVET 156
>dbj|BAD97369.1| EREBP/AP2 type transcription factor [Triticum aestivum]
Length = 344
Score = 104 bits (260), Expect = 3e-21
Identities = 52/112 (46%), Positives = 73/112 (64%), Gaps = 11/112 (9%)
Query: 61 GKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYG 120
GKGGP+N + +RGVRQR+WGKWVAEIREP + +R WLGTF TAEDAARAYD AA +YG
Sbjct: 78 GKGGPENTQCGFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYG 137
Query: 121 SRAQLNLQPSPTSSSS-----------QSSSSSSRASSSSSSTQTLRPLLPR 161
+ A+ N P + + + +S+S ++++S++ + LPR
Sbjct: 138 ALARTNFPVHPAQAPAVAVPAAIEGVVRGASASCESTTTSTNHSDVASSLPR 189
>gb|AAT12423.1| DREBa transcription factor [Glycine max]
Length = 211
Score = 103 bits (258), Expect = 6e-21
Identities = 51/91 (56%), Positives = 64/91 (70%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N + YRGVRQR+WGKWVAEIREP + +R WLGTF TA AA AYD AA+ +Y
Sbjct: 55 KGKGGPENLRCNYRGVRQRTWGKWVAEIREPNRGSRLWLGTFPTAISAALAYDEAAMAMY 114
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSS 150
G A+LN S+ S+ S +S A++ S
Sbjct: 115 GFCARLNFPNVQVSTFSEEPSRNSPAAAYQS 145
>gb|AAY25517.1| dehydration responsive element binding protein 1 [Hordeum vulgare
subsp. vulgare]
Length = 278
Score = 103 bits (257), Expect = 7e-21
Identities = 60/147 (40%), Positives = 83/147 (55%), Gaps = 8/147 (5%)
Query: 11 PPQEITKPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCK-----GKGGP 65
P QE TK TT P + + N + N S + K GKGGP
Sbjct: 13 PGQERTKKVRRR--TTGPDSVAETIKKWKEQNQKLQQENGSRKAPAKGSKKGCMAGKGGP 70
Query: 66 DNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQL 125
+N+ YRGVRQR+WGKWVAEIREP + R WLG+F TA +AARAYD AA +YG+ A++
Sbjct: 71 ENSNCAYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGATARV 130
Query: 126 NL-QPSPTSSSSQSSSSSSRASSSSSS 151
N + SP ++S + + S S+ +++
Sbjct: 131 NFPEHSPDANSGCTMAPSLLTSNGATA 157
>gb|AAN77051.1| dehydration responsive element binding protein [Lycopersicon
esculentum]
Length = 300
Score = 102 bits (255), Expect = 1e-20
Identities = 50/88 (56%), Positives = 66/88 (74%)
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
+GKGGP+N + +YRGVRQR WGKWVAEIREP++ +R WLGTF TA +AA AYD AA +Y
Sbjct: 70 RGKGGPENWRCKYRGVRQRIWGKWVAEIREPKRGSRLWLGTFGTAIEAALAYDDAARAMY 129
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASS 147
G A+LNL S S +++S+S ++S
Sbjct: 130 GPCARLNLPNYACDSVSWATTSASASAS 157
>gb|AAS46285.1| DREB-like protein 2 [Cynodon dactylon]
Length = 250
Score = 102 bits (255), Expect = 1e-20
Identities = 53/126 (42%), Positives = 73/126 (57%), Gaps = 2/126 (1%)
Query: 25 TTTPSETSNSENSNSSHNNNITTPNTSTTINNRK--CKGKGGPDNNKFRYRGVRQRSWGK 82
+T P + + HN I + ++K GKGGP+N YRGVRQR+WGK
Sbjct: 25 STGPDSIAETIKRWKEHNQKIHEDRKAPAKGSKKGCMAGKGGPENGNCAYRGVRQRTWGK 84
Query: 83 WVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSS 142
WVAEIREP + R WLG+F TA +AA AYD AA +YG A++N S ++S +S+
Sbjct: 85 WVAEIREPNRGNRLWLGSFPTALEAAHAYDEAARAMYGPTARVNFSESSADANSGCTSAL 144
Query: 143 SRASSS 148
S +S+
Sbjct: 145 SLLASN 150
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.127 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 657,098,089
Number of Sequences: 2540612
Number of extensions: 30248660
Number of successful extensions: 508607
Number of sequences better than 10.0: 3446
Number of HSP's better than 10.0 without gapping: 1919
Number of HSP's successfully gapped in prelim test: 1631
Number of HSP's that attempted gapping in prelim test: 396436
Number of HSP's gapped (non-prelim): 43856
length of query: 340
length of database: 863,360,394
effective HSP length: 128
effective length of query: 212
effective length of database: 538,162,058
effective search space: 114090356296
effective search space used: 114090356296
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0002.14


