

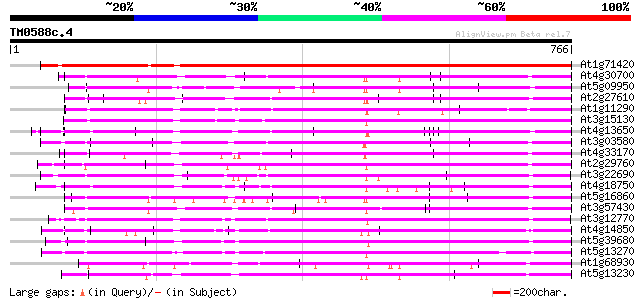
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588c.4
(766 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g71420 hypothetical protein 770 0.0
At4g30700 unknown protein 418 e-117
At5g09950 selenium-binding protein-like 417 e-116
At2g27610 putative selenium-binding protein 416 e-116
At1g11290 hypothetical protein 409 e-114
At3g15130 hypothetical protein 407 e-114
At4g13650 unknown protein 407 e-113
At3g03580 unknown protein (At3g03580) 406 e-113
At4g33170 putative protein 402 e-112
At2g29760 hypothetical protein 400 e-111
At3g22690 hypothetical protein 399 e-111
At4g18750 putative protein 398 e-111
At5g16860 putative protein 397 e-110
At3g57430 putative protein 396 e-110
At3g12770 unknown protein 394 e-110
At4g14850 unknown protein 394 e-109
At5g39680 unknown protein 385 e-107
At5g13270 putative protein 383 e-106
At1g68930 hypothetical protein 380 e-105
At5g13230 putative protein 379 e-105
>At1g71420 hypothetical protein
Length = 745
Score = 770 bits (1989), Expect = 0.0
Identities = 387/728 (53%), Positives = 513/728 (70%), Gaps = 18/728 (2%)
Query: 42 IRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILH 101
+R L G++ A+SL Y+ + S Q YA+LF ACA+ + L G+ LH+++L
Sbjct: 33 LRTLVRSGDIRRAVSLFYSAPVELQ-----SQQAYAALFQACAEQRNLLDGINLHHHMLS 87
Query: 102 KDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECF 161
++ L N L+NMY+KCG++ YAR VFD MP RN+VSWTALI+GY Q G +E F
Sbjct: 88 HPYCYSQNVILANFLINMYAKCGNILYARQVFDTMPERNVVSWTALITGYVQAGNEQEGF 147
Query: 162 YLFSGLLAHYRPNEFAFASLLSACEEHDIKCGMQVHAVALKISLDASVYVANALITMYSK 221
LFS +L+H PNEF +S+L++C G QVH +ALK+ L S+YVANA+I+MY +
Sbjct: 148 CLFSSMLSHCFPNEFTLSSVLTSCRYEP---GKQVHGLALKLGLHCSIYVANAVISMYGR 204
Query: 222 CSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIG 281
C G +AWT+F++++F+NL++WNSMIA FQ LG KAI +F M+ G+G
Sbjct: 205 CHDG------AAAYEAWTVFEAIKFKNLVTWNSMIAAFQCCNLGKKAIGVFMRMHSDGVG 258
Query: 282 FDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGG 341
FDRATLL++ SSL + S + + C LH L K+GL+++ EV TAL+K Y+ +
Sbjct: 259 FDRATLLNICSSLYKSSDLVPNEVS-KCCLQLHSLTVKSGLVTQTEVATALIKVYSEMLE 317
Query: 342 QISDCYRLFLDTSGKQDIVSWTAIITVLADQDPEQAFLLFCQLHRENFVPDWHTFSIALK 401
+DCY+LF++ S +DIV+W IIT A DPE+A LF QL +E PDW+TFS LK
Sbjct: 318 DYTDCYKLFMEMSHCRDIVAWNGIITAFAVYDPERAIHLFGQLRQEKLSPDWYTFSSVLK 377
Query: 402 ACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVS 461
ACA VT + AL++H+QVIK GF DTVL+N+LIHAYA+ GSL L +VFD+M D+VS
Sbjct: 378 ACAGLVTARHALSIHAQVIKGGFLADTVLNNSLIHAYAKCGSLDLCMRVFDDMDSRDVVS 437
Query: 462 WNSMLKSYALHGKAKDALELFKKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNH 521
WNSMLK+Y+LHG+ L +F+K+D++PDS TF+ALLSACSHAG VEEG+ IF SM +
Sbjct: 438 WNSMLKAYSLHGQVDSILPVFQKMDINPDSATFIALLSACSHAGRVEEGLRIFRSMFEKP 497
Query: 522 GIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIA 581
+PQL+HYAC++D+ R + +EAE++I MPM PD+V+W +LLGSCRKHG TRL ++A
Sbjct: 498 ETLPQLNHYACVIDMLSRAERFAEAEEVIKQMPMDPDAVVWIALLGSCRKHGNTRLGKLA 557
Query: 582 AEKFKEL-DPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVH 640
A+K KEL +P NS+ Y+QMSNIY++EGSF EA L KEM RV+K+P LSW E+G +VH
Sbjct: 558 ADKLKELVEPTNSMSYIQMSNIYNAEGSFNEANLSIKEMETWRVRKEPDLSWTEIGNKVH 617
Query: 641 EFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTE-VEHKEDQLFHHSEKMA 699
EF SGG H P KEA+ L+ LI LKEMGYVPE+ A D E E +ED L HHSEK+A
Sbjct: 618 EFASGGRHRPDKEAVYRELKRLISWLKEMGYVPEMRSASQDIEDEEQEEDNLLHHSEKLA 677
Query: 700 LVFAIM-NEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNA 758
L FA+M + CG N+I+IMKN RIC DCHNFMKLAS L KEI++RDSNRFHHFK++
Sbjct: 678 LAFAVMEGRKSSDCGVNLIQIMKNTRICIDCHNFMKLASKLLGKEILMRDSNRFHHFKDS 737
Query: 759 TCSCNDYW 766
+CSCNDYW
Sbjct: 738 SCSCNDYW 745
>At4g30700 unknown protein
Length = 792
Score = 418 bits (1075), Expect = e-117
Identities = 243/699 (34%), Positives = 378/699 (53%), Gaps = 34/699 (4%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
TYA A + + + G +H + ++L L +++V MY K +E AR VFD
Sbjct: 121 TYAFAISAASGFRDDRAGRVIHGQAVVDG--CDSELLLGSNIVKMYFKFWRVEDARKVFD 178
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHY--RPNEFAFASLLSACEE-HDIK 191
+MP ++ + W +ISGY + + E +F L+ R + +L A E +++
Sbjct: 179 RMPEKDTILWNTMISGYRKNEMYVESIQVFRDLINESCTRLDTTTLLDILPAVAELQELR 238
Query: 192 CGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLIS 251
GMQ+H++A K + YV I++YSKC G +F+ ++++
Sbjct: 239 LGMQIHSLATKTGCYSHDYVLTGFISLYSKCGKIKMGS---------ALFREFRKPDIVA 289
Query: 252 WNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCF 311
+N+MI G+ G + ++ LF + SG +TL+S+ L +
Sbjct: 290 YNAMIHGYTSNGETELSLSLFKELMLSGARLRSSTLVSLVPVSGH----------LMLIY 339
Query: 312 VLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLAD 371
+H K+ +S V TAL Y+ L +I +LF D S ++ + SW A+I+
Sbjct: 340 AIHGYCLKSNFLSHASVSTALTTVYSKLN-EIESARKLF-DESPEKSLPSWNAMISGYTQ 397
Query: 372 QD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVL 430
E A LF ++ + F P+ T + L ACA VH V F+ +
Sbjct: 398 NGLTEDAISLFREMQKSEFSPNPVTITCILSACAQLGALSLGKWVHDLVRSTDFESSIYV 457
Query: 431 SNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DV 487
S ALI YA+ GS+A + ++FD M + V+WN+M+ Y LHG+ ++AL +F ++ +
Sbjct: 458 STALIGMYAKCGSIAEARRLFDLMTKKNEVTWNTMISGYGLHGQGQEALNIFYEMLNSGI 517
Query: 488 HPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAE 547
P TF+ +L ACSHAGLV+EG EIFNSM +G P + HYACMVD+ GR G + A
Sbjct: 518 TPTPVTFLCVLYACSHAGLVKEGDEIFNSMIHRYGFEPSVKHYACMVDILGRAGHLQRAL 577
Query: 548 DLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEG 607
I M ++P S +W +LLG+CR H +T LAR +EK ELDP N +V +SNI+S++
Sbjct: 578 QFIEAMSIEPGSSVWETLLGACRIHKDTNLARTVSEKLFELDPDNVGYHVLLSNIHSADR 637
Query: 608 SFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLK 667
++ +A +R+ + ++ K PG + +E+G+ H FTSG HP + I +LE L G+++
Sbjct: 638 NYPQAATVRQTAKKRKLAKAPGYTLIEIGETPHVFTSGDQSHPQVKEIYEKLEKLEGKMR 697
Query: 668 EMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICA 727
E GY PE LAL+D E E +E + HSE++A+ F ++ G I+I+KN+R+C
Sbjct: 698 EAGYQPETELALHDVEEEERELMVKVHSERLAIAFGLIATE----PGTEIRIIKNLRVCL 753
Query: 728 DCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
DCH KL S + ++ IVVRD+NRFHHFK+ CSC DYW
Sbjct: 754 DCHTVTKLISKITERVIVVRDANRFHHFKDGVCSCGDYW 792
Score = 138 bits (347), Expect = 1e-32
Identities = 133/526 (25%), Positives = 231/526 (43%), Gaps = 56/526 (10%)
Query: 67 THSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHL 126
T + +S TY F L Q A ILH +ND+ L L S G +
Sbjct: 14 TAALISKNTYLDFFKRSTSISHLAQTHA--QIILHG---FRNDISLLTKLTQRLSDLGAI 68
Query: 127 EYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHYR------PNEFAFAS 180
YAR +F + R ++ + L+ G++ + E + + AH R PN +A
Sbjct: 69 YYARDIFLSVQRPDVFLFNVLMRGFS----VNESPHSSLSVFAHLRKSTDLKPNSSTYAF 124
Query: 181 LLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWT 239
+SA D + G +H A+ D+ + + + ++ MY K +DA
Sbjct: 125 AISAASGFRDDRAGRVIHGQAVVDGCDSELLLGSNIVKMYFKFW---------RVEDARK 175
Query: 240 MFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYC-SGIGFDRATLLSVFSSLNECS 298
+F M ++ I WN+MI+G++ + ++I++F + S D TLL + ++ E
Sbjct: 176 VFDRMPEKDTILWNTMISGYRKNEMYVESIQVFRDLINESCTRLDTTTLLDILPAVAE-- 233
Query: 299 AFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQD 358
EL + ++ +H LATKTG S V+T + Y+ G+I LF + K D
Sbjct: 234 -LQELRLGMQ----IHSLATKTGCYSHDYVLTGFISLYSKC-GKIKMGSALFREFR-KPD 286
Query: 359 IVSWTAII-TVLADQDPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHS 417
IV++ A+I ++ + E + LF +L T + + + A+H
Sbjct: 287 IVAYNAMIHGYTSNGETELSLSLFKELMLSGARLRSSTLVSLVPVSGHLML---IYAIHG 343
Query: 418 QVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKD 477
+K F +S AL Y++ + + ++FDE L SWN+M+ Y +G +D
Sbjct: 344 YCLKSNFLSHASVSTALTTVYSKLNEIESARKLFDESPEKSLPSWNAMISGYTQNGLTED 403
Query: 478 ALELFK---KLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYA--- 531
A+ LF+ K + P+ T +LSAC+ G + G + H +V D +
Sbjct: 404 AISLFREMQKSEFSPNPVTITCILSACAQLGALSLGKWV-------HDLVRSTDFESSIY 456
Query: 532 ---CMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGE 574
++ +Y + G I+EA L M K + V W++++ HG+
Sbjct: 457 VSTALIGMYAKCGSIAEARRLFDLM-TKKNEVTWNTMISGYGLHGQ 501
Score = 75.1 bits (183), Expect = 1e-13
Identities = 60/274 (21%), Positives = 128/274 (45%), Gaps = 13/274 (4%)
Query: 321 GLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLA-DQDPEQAFL 379
G +++ ++T L + ++LG I +FL + D+ + ++ + ++ P +
Sbjct: 47 GFRNDISLLTKLTQRLSDLGA-IYYARDIFLSVQ-RPDVFLFNVLMRGFSVNESPHSSLS 104
Query: 380 LFCQLHRE-NFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAY 438
+F L + + P+ T++ A+ A + F ++ +H Q + G + +L + ++ Y
Sbjct: 105 VFAHLRKSTDLKPNSSTYAFAISAASGFRDDRAGRVIHGQAVVDGCDSELLLGSNIVKMY 164
Query: 439 ARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL----DVHPDSTTF 494
+ + + +VFD M D + WN+M+ Y + +++++F+ L D+TT
Sbjct: 165 FKFWRVEDARKVFDRMPEKDTILWNTMISGYRKNEMYVESIQVFRDLINESCTRLDTTTL 224
Query: 495 VALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMP 554
+ +L A + + G++I +S++ G + LY + GKI L
Sbjct: 225 LDILPAVAELQELRLGMQI-HSLATKTGCYSHDYVLTGFISLYSKCGKIKMGSALFREF- 282
Query: 555 MKPDSVIWSSLLGSCRKHGETRLARIAAEKFKEL 588
KPD V +++++ +GET L + FKEL
Sbjct: 283 RKPDIVAYNAMIHGYTSNGETEL---SLSLFKEL 313
>At5g09950 selenium-binding protein-like
Length = 995
Score = 417 bits (1071), Expect = e-116
Identities = 249/704 (35%), Positives = 404/704 (57%), Gaps = 47/704 (6%)
Query: 81 HACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRN 140
++ A+ L++G +H +++ + + + N LVNMY+KCG + AR VF M ++
Sbjct: 321 YSLAEEVGLKKGREVHGHVITTG-LVDFMVGIGNGLVNMYAKCGSIADARRVFYFMTDKD 379
Query: 141 IVSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEEHD-IKCGMQVHA 198
VSW ++I+G Q G E + + H P F S LS+C K G Q+H
Sbjct: 380 SVSWNSMITGLDQNGCFIEAVERYKSMRRHDILPGSFTLISSLSSCASLKWAKLGQQIHG 439
Query: 199 VALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAG 258
+LK+ +D +V V+NAL+T+Y++ GY ++ +F SM + +SWNS+I
Sbjct: 440 ESLKLGIDLNVSVSNALMTLYAET------GYL---NECRKIFSSMPEHDQVSWNSIIGA 490
Query: 259 F--QFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCL 316
R L + A+ F + +G +R T SV S+++ S F EL + H L
Sbjct: 491 LARSERSLPE-AVVCFLNAQRAGQKLNRITFSSVLSAVSSLS-FGELGKQI------HGL 542
Query: 317 ATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIIT------VLA 370
A K + E AL+ Y G ++ C ++F + ++D V+W ++I+ +LA
Sbjct: 543 ALKNNIADEATTENALIACYGKCG-EMDGCEKIFSRMAERRDNVTWNSMISGYIHNELLA 601
Query: 371 DQDPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVL 430
F+L ++F+ ++ L A A T ++ + VH+ ++ + D V+
Sbjct: 602 KALDLVWFMLQTGQRLDSFM-----YATVLSAFASVATLERGMEVHACSVRACLESDVVV 656
Query: 431 SNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFK--KLD-- 486
+AL+ Y++ G L + + F+ M + SWNSM+ YA HG+ ++AL+LF+ KLD
Sbjct: 657 GSALVDMYSKCGRLDYALRFFNTMPVRNSYSWNSMISGYARHGQGEEALKLFETMKLDGQ 716
Query: 487 VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEA 546
PD TFV +LSACSHAGL+EEG + F SMSD++G+ P+++H++CM D+ GR G++ +
Sbjct: 717 TPPDHVTFVGVLSACSHAGLLEEGFKHFESMSDSYGLAPRIEHFSCMADVLGRAGELDKL 776
Query: 547 EDLIHTMPMKPDSVIWSSLLGS-CRKHG-ETRLARIAAEKFKELDPKNSLGYVQMSNIYS 604
ED I MPMKP+ +IW ++LG+ CR +G + L + AAE +L+P+N++ YV + N+Y+
Sbjct: 777 EDFIEKMPMKPNVLIWRTVLGACCRANGRKAELGKKAAEMLFQLEPENAVNYVLLGNMYA 836
Query: 605 SEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIG 664
+ G + + RK+M+D+ VKK+ G SWV + VH F +G HP + I +L+ L
Sbjct: 837 AGGRWEDLVKARKKMKDADVKKEAGYSWVTMKDGVHMFVAGDKSHPDADVIYKKLKELNR 896
Query: 665 QLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVF--AIMNEGNLPCGGNVIKIMKN 722
++++ GYVP+ ALYD E E+KE+ L +HSEK+A+ F A LP I+IMKN
Sbjct: 897 KMRDAGYVPQTGFALYDLEQENKEEILSYHSEKLAVAFVLAAQRSSTLP-----IRIMKN 951
Query: 723 IRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
+R+C DCH+ K S + ++I++RDSNRFHHF++ CSC+D+W
Sbjct: 952 LRVCGDCHSAFKYISKIEGRQIILRDSNRFHHFQDGACSCSDFW 995
Score = 180 bits (456), Expect = 3e-45
Identities = 151/547 (27%), Positives = 253/547 (45%), Gaps = 50/547 (9%)
Query: 106 IQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFS 165
+ D++L N+L+N Y + G AR VFD+MP RN VSW ++SGY++ G +E
Sbjct: 32 LDKDVYLCNNLINAYLETGDSVSARKVFDEMPLRNCVSWACIVSGYSRNGEHKEALVFLR 91
Query: 166 GLLAH-YRPNEFAFASLLSACEE---HDIKCGMQVHAVALKISLDASVYVANALITMYSK 221
++ N++AF S+L AC+E I G Q+H + K+S V+N LI+MY K
Sbjct: 92 DMVKEGIFSNQYAFVSVLRACQEIGSVGILFGRQIHGLMFKLSYAVDAVVSNVLISMYWK 151
Query: 222 CSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIG 281
C G GY A F +E +N +SWNS+I+ + G A R+F+ M G
Sbjct: 152 CIGSV--GY------ALCAFGDIEVKNSVSWNSIISVYSQAGDQRSAFRIFSSMQYDG-- 201
Query: 282 FDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGG 341
R T + S + + E ++ L + C K+GL++++ V + LV ++A G
Sbjct: 202 -SRPTEYTFGSLVTTACSLTEPDVRLLE--QIMCTIQKSGLLTDLFVGSGLVSAFAK-SG 257
Query: 342 QISDCYRLFLDTSGKQDIVSWTAIITVLADQDPEQAFLLFCQLHRENFVPDWHTFSIALK 401
+S ++F + + ++ ++ + E+A LF ++ V +
Sbjct: 258 SLSYARKVFNQMETRNAVTLNGLMVGLVRQKWGEEATKLFMDMNSMIDVSPESYVILLSS 317
Query: 402 ACAYFVTEQQAL----AVHSQVIKRGFQKDTV-LSNALIHAYARSGSLALSEQVFDEMCC 456
Y + E+ L VH VI G V + N L++ YA+ GS+A + +VF M
Sbjct: 318 FPEYSLAEEVGLKKGREVHGHVITTGLVDFMVGIGNGLVNMYAKCGSIADARRVFYFMTD 377
Query: 457 HDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEI 513
D VSWNSM+ +G +A+E +K + D+ P S T ++ LS+C+ + G +I
Sbjct: 378 KDSVSWNSMITGLDQNGCFIEAVERYKSMRRHDILPGSFTLISSLSSCASLKWAKLGQQI 437
Query: 514 FNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHG 573
+ S GI + ++ LY G ++E + +MP + D V W+S++G+ +
Sbjct: 438 -HGESLKLGIDLNVSVSNALMTLYAETGYLNECRKIFSSMP-EHDQVSWNSIIGALAR-S 494
Query: 574 ETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWV 633
E L L + S++ S+ S LS+
Sbjct: 495 ERSLPEAVVCFLNAQRAGQKLNRITFSSVLSAVSS---------------------LSFG 533
Query: 634 EVGKQVH 640
E+GKQ+H
Sbjct: 534 ELGKQIH 540
Score = 65.5 bits (158), Expect = 1e-10
Identities = 48/173 (27%), Positives = 86/173 (48%), Gaps = 16/173 (9%)
Query: 416 HSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKA 475
HS++ K KD L N LI+AY +G + +VFDEM + VSW ++ Y+ +G+
Sbjct: 24 HSRLYKNRLDKDVYLCNNLINAYLETGDSVSARKVFDEMPLRNCVSWACIVSGYSRNGEH 83
Query: 476 KDALELFK---KLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYA- 531
K+AL + K + + FV++L AC G V G+ + HG++ +L +
Sbjct: 84 KEALVFLRDMVKEGIFSNQYAFVSVLRACQEIGSV--GILFGRQI---HGLMFKLSYAVD 138
Query: 532 -----CMVDLYGR-VGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLA 578
++ +Y + +G + A + +K +SV W+S++ + G+ R A
Sbjct: 139 AVVSNVLISMYWKCIGSVGYALCAFGDIEVK-NSVSWNSIISVYSQAGDQRSA 190
>At2g27610 putative selenium-binding protein
Length = 868
Score = 416 bits (1068), Expect = e-116
Identities = 242/701 (34%), Positives = 386/701 (54%), Gaps = 37/701 (5%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T+A+ A+ +G+ +H ++ + + ++N L+N+Y KCG++ AR +FD
Sbjct: 196 TFAAALGVLAEEGVGGRGLQVHTVVVKNG--LDKTIPVSNSLINLYLKCGNVRKARILFD 253
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHY-RPNEFAFASLLSACEE-HDIKC 192
+ +++V+W ++ISGYA GL E +F + +Y R +E +FAS++ C +++
Sbjct: 254 KTEVKSVVTWNSMISGYAANGLDLEALGMFYSMRLNYVRLSESSFASVIKLCANLKELRF 313
Query: 193 GMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSME-FRNLIS 251
Q+H +K + AL+ YSKC+ DA +FK + N++S
Sbjct: 314 TEQLHCSVVKYGFLFDQNIRTALMVAYSKCTAML---------DALRLFKEIGCVGNVVS 364
Query: 252 WNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCF 311
W +MI+GF ++A+ LF+ M G+ + T + ++L P+ +
Sbjct: 365 WTAMISGFLQNDGKEEAVDLFSEMKRKGVRPNEFTYSVILTAL-----------PVISPS 413
Query: 312 VLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLAD 371
+H KT V TAL+ +Y LG ++ + ++F K DIV+W+A++ A
Sbjct: 414 EVHAQVVKTNYERSSTVGTALLDAYVKLG-KVEEAAKVFSGIDDK-DIVAWSAMLAGYAQ 471
Query: 372 Q-DPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQ-QALAVHSQVIKRGFQKDTV 429
+ E A +F +L + P+ TFS L CA Q H IK
Sbjct: 472 TGETEAAIKMFGELTKGGIKPNEFTFSSILNVCAATNASMGQGKQFHGFAIKSRLDSSLC 531
Query: 430 LSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLD--- 486
+S+AL+ YA+ G++ +E+VF DLVSWNSM+ YA HG+A AL++FK++
Sbjct: 532 VSSALLTMYAKKGNIESAEEVFKRQREKDLVSWNSMISGYAQHGQAMKALDVFKEMKKRK 591
Query: 487 VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEA 546
V D TF+ + +AC+HAGLVEEG + F+ M + I P +H +CMVDLY R G++ +A
Sbjct: 592 VKMDGVTFIGVFAACTHAGLVEEGEKYFDIMVRDCKIAPTKEHNSCMVDLYSRAGQLEKA 651
Query: 547 EDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSE 606
+I MP S IW ++L +CR H +T L R+AAEK + P++S YV +SN+Y+
Sbjct: 652 MKVIENMPNPAGSTIWRTILAACRVHKKTELGRLAAEKIIAMKPEDSAAYVLLSNMYAES 711
Query: 607 GSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQL 666
G + E +RK M + VKK+PG SW+EV + + F +G HP K+ I +LE L +L
Sbjct: 712 GDWQERAKVRKLMNERNVKKEPGYSWIEVKNKTYSFLAGDRSHPLKDQIYMKLEDLSTRL 771
Query: 667 KEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRIC 726
K++GY P+ + L D + EHKE L HSE++A+ F ++ G+ + I+KN+R+C
Sbjct: 772 KDLGYEPDTSYVLQDIDDEHKEAVLAQHSERLAIAFGLIATPK----GSPLLIIKNLRVC 827
Query: 727 ADCHNFMKLASNLFQKEIVVRDSNRFHHF-KNATCSCNDYW 766
DCH +KL + + ++EIVVRDSNRFHHF + CSC D+W
Sbjct: 828 GDCHLVIKLIAKIEEREIVVRDSNRFHHFSSDGVCSCGDFW 868
Score = 211 bits (537), Expect = 1e-54
Identities = 143/487 (29%), Positives = 250/487 (50%), Gaps = 31/487 (6%)
Query: 108 NDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGL 167
+D+ + LV+ Y K + + R VFD+M RN+V+WT LISGYA+ + E LF +
Sbjct: 126 DDVSVGTSLVDTYMKGSNFKDGRKVFDEMKERNVVTWTTLISGYARNSMNDEVLTLFMRM 185
Query: 168 LAH-YRPNEFAFASLLSACEEHDI-KCGMQVHAVALKISLDASVYVANALITMYSKCSGG 225
+PN F FA+ L E + G+QVH V +K LD ++ V+N+LI +Y KC
Sbjct: 186 QNEGTQPNSFTFAAALGVLAEEGVGGRGLQVHTVVVKNGLDKTIPVSNSLINLYLKCG-- 243
Query: 226 FHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRA 285
A +F E +++++WNSMI+G+ GL +A+ +F Y + + R
Sbjct: 244 -------NVRKARILFDKTEVKSVVTWNSMISGYAANGLDLEALGMF---YSMRLNYVRL 293
Query: 286 TLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISD 345
+ S S + C+ EL + LHC K G + + + TAL+ +Y+ + D
Sbjct: 294 SESSFASVIKLCANLKELRFTEQ----LHCSVVKYGFLFDQNIRTALMVAYSKCTAML-D 348
Query: 346 CYRLFLDTSGKQDIVSWTAIIT-VLADQDPEQAFLLFCQLHRENFVPDWHTFSIALKACA 404
RLF + ++VSWTA+I+ L + E+A LF ++ R+ P+ T+S+ L A
Sbjct: 349 ALRLFKEIGCVGNVVSWTAMISGFLQNDGKEEAVDLFSEMKRKGVRPNEFTYSVILTALP 408
Query: 405 YFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNS 464
+ VH+QV+K +++ + + AL+ AY + G + + +VF + D+V+W++
Sbjct: 409 VISPSE----VHAQVVKTNYERSSTVGTALLDAYVKLGKVEEAAKVFSGIDDKDIVAWSA 464
Query: 465 MLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNH 521
ML YA G+ + A+++F +L + P+ TF ++L+ C+ + F+ +
Sbjct: 465 MLAGYAQTGETEAAIKMFGELTKGGIKPNEFTFSSILNVCAATNASMGQGKQFHGFAIKS 524
Query: 522 GIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIA 581
+ L + ++ +Y + G I AE++ K D V W+S++ +HG+ A A
Sbjct: 525 RLDSSLCVSSALLTMYAKKGNIESAEEVFKRQREK-DLVSWNSMISGYAQHGQ---AMKA 580
Query: 582 AEKFKEL 588
+ FKE+
Sbjct: 581 LDVFKEM 587
Score = 120 bits (300), Expect = 4e-27
Identities = 113/462 (24%), Positives = 205/462 (43%), Gaps = 41/462 (8%)
Query: 129 ARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHYRPNEF---AFASLLSA- 184
A +FD+ P R+ S+ +L+ G+++ G +E LF + H E F+S+L
Sbjct: 46 AHNLFDKSPGRDRESYISLLFGFSRDGRTQEAKRLFLNI--HRLGMEMDCSIFSSVLKVS 103
Query: 185 ---CEEHDIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMF 241
C+E G Q+H +K V V +L+ Y K S D +F
Sbjct: 104 ATLCDE---LFGRQLHCQCIKFGFLDDVSVGTSLVDTYMKGS---------NFKDGRKVF 151
Query: 242 KSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFD 301
M+ RN+++W ++I+G+ + D+ + LF M G T + F+
Sbjct: 152 DEMKERNVVTWTTLISGYARNSMNDEVLTLFMRMQNEG------TQPNSFTFAAALGVLA 205
Query: 302 ELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVS 361
E + R V H + K GL + V +L+ Y G R+ D + + +V+
Sbjct: 206 EEGVGGRGLQV-HTVVVKNGLDKTIPVSNSLINLYLKCGNVRK--ARILFDKTEVKSVVT 262
Query: 362 WTAIITVLADQDPEQAFLLFCQLHRENFVP-DWHTFSIALKACAYFVTEQQALAVHSQVI 420
W ++I+ A + L R N+V +F+ +K CA + +H V+
Sbjct: 263 WNSMISGYAANGLDLEALGMFYSMRLNYVRLSESSFASVIKLCANLKELRFTEQLHCSVV 322
Query: 421 KRGFQKDTVLSNALIHAYARSGSLALSEQVFDEM-CCHDLVSWNSMLKSYALHGKAKDAL 479
K GF D + AL+ AY++ ++ + ++F E+ C ++VSW +M+ + + ++A+
Sbjct: 323 KYGFLFDQNIRTALMVAYSKCTAMLDALRLFKEIGCVGNVVSWTAMISGFLQNDGKEEAV 382
Query: 480 ELF---KKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDL 536
+LF K+ V P+ T+ +L+A E ++ + + V ++D
Sbjct: 383 DLFSEMKRKGVRPNEFTYSVILTALPVISPSEVHAQVVKTNYERSSTVG-----TALLDA 437
Query: 537 YGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLA 578
Y ++GK+ EA + + K D V WS++L + GET A
Sbjct: 438 YVKLGKVEEAAKVFSGIDDK-DIVAWSAMLAGYAQTGETEAA 478
Score = 108 bits (269), Expect = 1e-23
Identities = 81/274 (29%), Positives = 140/274 (50%), Gaps = 17/274 (6%)
Query: 236 DAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLN 295
+A +F R+ S+ S++ GF G +A RLF +++ G+ D S+FSS+
Sbjct: 45 NAHNLFDKSPGRDRESYISLLFGFSRDGRTQEAKRLFLNIHRLGMEMD----CSIFSSVL 100
Query: 296 ECSAF--DELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDT 353
+ SA DEL LHC K G + +V V T+LV +Y G D ++F D
Sbjct: 101 KVSATLCDELFGRQ-----LHCQCIKFGFLDDVSVGTSLVDTYMK-GSNFKDGRKVF-DE 153
Query: 354 SGKQDIVSWTAIITVLADQDP-EQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQA 412
++++V+WT +I+ A ++ LF ++ E P+ TF+ AL A +
Sbjct: 154 MKERNVVTWTTLISGYARNSMNDEVLTLFMRMQNEGTQPNSFTFAAALGVLAEEGVGGRG 213
Query: 413 LAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALH 472
L VH+ V+K G K +SN+LI+ Y + G++ + +FD+ +V+WNSM+ YA +
Sbjct: 214 LQVHTVVVKNGLDKTIPVSNSLINLYLKCGNVRKARILFDKTEVKSVVTWNSMISGYAAN 273
Query: 473 GKAKDALELFKKLD---VHPDSTTFVALLSACSH 503
G +AL +F + V ++F +++ C++
Sbjct: 274 GLDLEALGMFYSMRLNYVRLSESSFASVIKLCAN 307
>At1g11290 hypothetical protein
Length = 809
Score = 409 bits (1052), Expect = e-114
Identities = 240/697 (34%), Positives = 372/697 (52%), Gaps = 31/697 (4%)
Query: 76 YASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQ 135
+ L C L+ G +H ++ ++ DLF L NMY+KC + AR VFD+
Sbjct: 138 FTYLLKVCGDEAELRVGKEIHGLLVKSGFSL--DLFAMTGLENMYAKCRQVNEARKVFDR 195
Query: 136 MPRRNIVSWTALISGYAQCGLIRECFYLFSGLLA-HYRPNEFAFASLLSACEE-HDIKCG 193
MP R++VSW +++GY+Q G+ R + + + +P+ S+L A I G
Sbjct: 196 MPERDLVSWNTIVAGYSQNGMARMALEMVKSMCEENLKPSFITIVSVLPAVSALRLISVG 255
Query: 194 MQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWN 253
++H A++ D+ V ++ AL+ MY+KC + + A +F M RN++SWN
Sbjct: 256 KEIHGYAMRSGFDSLVNISTALVDMYAKCG---------SLETARQLFDGMLERNVVSWN 306
Query: 254 SMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVL 313
SMI + +A+ +F M G+ + T +SV +L+ C+ +L R F+
Sbjct: 307 SMIDAYVQNENPKEAMLIFQKMLDEGV---KPTDVSVMGALHACADLGDLE---RGRFI- 359
Query: 314 HCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQD 373
H L+ + GL V VV +L+ Y ++ +F + +VSW A+I A
Sbjct: 360 HKLSVELGLDRNVSVVNSLISMYCKCK-EVDTAASMFGKLQSRT-LVSWNAMILGFAQNG 417
Query: 374 -PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSN 432
P A F Q+ PD T+ + A A A +H V++ K+ ++
Sbjct: 418 RPIDALNYFSQMRSRTVKPDTFTYVSVITAIAELSITHHAKWIHGVVMRSCLDKNVFVTT 477
Query: 433 ALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLD---VHP 489
AL+ YA+ G++ ++ +FD M + +WN+M+ Y HG K ALELF+++ + P
Sbjct: 478 ALVDMYAKCGAIMIARLIFDMMSERHVTTWNAMIDGYGTHGFGKAALELFEEMQKGTIKP 537
Query: 490 DSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDL 549
+ TF++++SACSH+GLVE G++ F M +N+ I +DHY MVDL GR G+++EA D
Sbjct: 538 NGVTFLSVISACSHSGLVEAGLKCFYMMKENYSIELSMDHYGAMVDLLGRAGRLNEAWDF 597
Query: 550 IHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSF 609
I MP+KP ++ ++LG+C+ H A AAE+ EL+P + +V ++NIY + +
Sbjct: 598 IMQMPVKPAVNVYGAMLGACQIHKNVNFAEKAAERLFELNPDDGGYHVLLANIYRAASMW 657
Query: 610 IEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEM 669
+ G +R M ++K PG S VE+ +VH F SG HP + I + LE LI +KE
Sbjct: 658 EKVGQVRVSMLRQGLRKTPGCSMVEIKNEVHSFFSGSTAHPDSKKIYAFLEKLICHIKEA 717
Query: 670 GYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADC 729
GYVP+ L L E + KE L HSEK+A+ F ++N G I + KN+R+CADC
Sbjct: 718 GYVPDTNLVL-GVENDVKEQLLSTHSEKLAISFGLLN----TTAGTTIHVRKNLRVCADC 772
Query: 730 HNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
HN K S + +EIVVRD RFHHFKN CSC DYW
Sbjct: 773 HNATKYISLVTGREIVVRDMQRFHHFKNGACSCGDYW 809
Score = 158 bits (400), Expect = 9e-39
Identities = 135/554 (24%), Positives = 260/554 (46%), Gaps = 51/554 (9%)
Query: 77 ASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQM 136
A L C+ K L+Q + L + K+ Q F T LV+++ + G ++ A VF+ +
Sbjct: 41 ALLLERCSSLKELRQILPL----VFKNGLYQEHFFQTK-LVSLFCRYGSVDEAARVFEPI 95
Query: 137 PRRNIVSWTALISGYAQCGLIRECFYLFSGL-LAHYRPNEFAFASLLSAC-EEHDIKCGM 194
+ V + ++ G+A+ + + F + P + F LL C +E +++ G
Sbjct: 96 DSKLNVLYHTMLKGFAKVSDLDKALQFFVRMRYDDVEPVVYNFTYLLKVCGDEAELRVGK 155
Query: 195 QVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNS 254
++H + +K ++ L MY+KC ++A +F M R+L+SWN+
Sbjct: 156 EIHGLLVKSGFSLDLFAMTGLENMYAKCR---------QVNEARKVFDRMPERDLVSWNT 206
Query: 255 MIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLH 314
++AG+ G+ A+ + M + + + +++ S L SA +++ +H
Sbjct: 207 IVAGYSQNGMARMALEMVKSMCEENL---KPSFITIVSVLPAVSALRLISVGKE----IH 259
Query: 315 CLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAII-TVLADQD 373
A ++G S V + TALV YA G + R D ++++VSW ++I + +++
Sbjct: 260 GYAMRSGFDSLVNISTALVDMYAKCGS--LETARQLFDGMLERNVVSWNSMIDAYVQNEN 317
Query: 374 PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNA 433
P++A L+F ++ E P + AL ACA ++ +H ++ G ++ + N+
Sbjct: 318 PKEAMLIFQKMLDEGVKPTDVSVMGALHACADLGDLERGRFIHKLSVELGLDRNVSVVNS 377
Query: 434 LIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPD 490
LI Y + + + +F ++ LVSWN+M+ +A +G+ DAL F ++ V PD
Sbjct: 378 LISMYCKCKEVDTAASMFGKLQSRTLVSWNAMILGFAQNGRPIDALNYFSQMRSRTVKPD 437
Query: 491 STTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQ--LDH----YACMVDLYGRVGKIS 544
+ T+V++++A + + I HG+V + LD +VD+Y + G I
Sbjct: 438 TFTYVSVITAIAELSITHHAKWI-------HGVVMRSCLDKNVFVTTALVDMYAKCGAIM 490
Query: 545 EAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELD----PKNSLGYVQMS 600
A LI M + W++++ HG + A E F+E+ N + ++ +
Sbjct: 491 IAR-LIFDMMSERHVTTWNAMIDGYGTHG---FGKAALELFEEMQKGTIKPNGVTFLSVI 546
Query: 601 NIYSSEGSFIEAGL 614
+ S G +EAGL
Sbjct: 547 SACSHSG-LVEAGL 559
>At3g15130 hypothetical protein
Length = 689
Score = 407 bits (1047), Expect = e-114
Identities = 246/703 (34%), Positives = 382/703 (53%), Gaps = 30/703 (4%)
Query: 74 QTYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVF 133
Q S+ C + QG +H Y+L + +L +N+L++MY KC A VF
Sbjct: 7 QNLVSILRVCTRKGLSDQGGQVHCYLLKSGSGL--NLITSNYLIDMYCKCREPLMAYKVF 64
Query: 134 DQMPRRNIVSWTALISGYAQCGLIRECFYLFS--GLLAHYRPNEFAFASLLSACEE-HDI 190
D MP RN+VSW+AL+SG+ G ++ LFS G Y PNEF F++ L AC + +
Sbjct: 65 DSMPERNVVSWSALMSGHVLNGDLKGSLSLFSEMGRQGIY-PNEFTFSTNLKACGLLNAL 123
Query: 191 KCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLI 250
+ G+Q+H LKI + V V N+L+ MYSKC ++A +F+ + R+LI
Sbjct: 124 EKGLQIHGFCLKIGFEMMVEVGNSLVDMYSKCG---------RINEAEKVFRRIVDRSLI 174
Query: 251 SWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNC 310
SWN+MIAGF G G KA+ F M + I +R ++ S L CS+ + +
Sbjct: 175 SWNAMIAGFVHAGYGSKALDTFGMMQEANIK-ERPDEFTLTSLLKACSSTGMIYAGKQ-- 231
Query: 311 FVLHCLATKTGLI--SEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITV 368
+H ++G S + +LV Y G S R D ++ ++SW+++I
Sbjct: 232 --IHGFLVRSGFHCPSSATITGSLVDLYVKCGYLFSA--RKAFDQIKEKTMISWSSLILG 287
Query: 369 LADQDP-EQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKD 427
A + +A LF +L N D S + A F +Q + + +K +
Sbjct: 288 YAQEGEFVEAMGLFKRLQELNSQIDSFALSSIIGVFADFALLRQGKQMQALAVKLPSGLE 347
Query: 428 TVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL-- 485
T + N+++ Y + G + +E+ F EM D++SW ++ Y HG K ++ +F ++
Sbjct: 348 TSVLNSVVDMYLKCGLVDEAEKCFAEMQLKDVISWTVVITGYGKHGLGKKSVRIFYEMLR 407
Query: 486 -DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKIS 544
++ PD ++A+LSACSH+G+++EG E+F+ + + HGI P+++HYAC+VDL GR G++
Sbjct: 408 HNIEPDEVCYLAVLSACSHSGMIKEGEELFSKLLETHGIKPRVEHYACVVDLLGRAGRLK 467
Query: 545 EAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYS 604
EA+ LI TMP+KP+ IW +LL CR HG+ L + + +D KN YV MSN+Y
Sbjct: 468 EAKHLIDTMPIKPNVGIWQTLLSLCRVHGDIELGKEVGKILLRIDAKNPANYVMMSNLYG 527
Query: 605 SEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIG 664
G + E G R+ +KK+ G+SWVE+ ++VH F SG HP IQ L+
Sbjct: 528 QAGYWNEQGNARELGNIKGLKKEAGMSWVEIEREVHFFRSGEDSHPLTPVIQETLKEAER 587
Query: 665 QLK-EMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNI 723
+L+ E+GYV + L+D + E KE+ L HSEK+A+ A+ G L G I++ KN+
Sbjct: 588 RLREELGYVYGLKHELHDIDDESKEENLRAHSEKLAIGLALAT-GGLNQKGKTIRVFKNL 646
Query: 724 RICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
R+C DCH F+K S + + VVRD+ RFH F++ CSC DYW
Sbjct: 647 RVCVDCHEFIKGLSKITKIAYVVRDAVRFHSFEDGCCSCGDYW 689
>At4g13650 unknown protein
Length = 1024
Score = 407 bits (1045), Expect = e-113
Identities = 238/732 (32%), Positives = 385/732 (52%), Gaps = 35/732 (4%)
Query: 42 IRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILH 101
I LS G E+A+ L L S T ASL AC+ + L +G LH Y
Sbjct: 321 INGLSQCGYGEKAMELFKRMHLDGLEPDS---NTLASLVVACSADGTLFRGQQLHAYTTK 377
Query: 102 KDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECF 161
N + L+N+Y+KC +E A F + N+V W ++ Y +R F
Sbjct: 378 LGFASNNKI--EGALLNLYAKCADIETALDYFLETEVENVVLWNVMLVAYGLLDDLRNSF 435
Query: 162 YLFSGL-LAHYRPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMY 219
+F + + PN++ + S+L C D++ G Q+H+ +K + + YV + LI MY
Sbjct: 436 RIFRQMQIEEIVPNQYTYPSILKTCIRLGDLELGEQIHSQIIKTNFQLNAYVCSVLIDMY 495
Query: 220 SKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSG 279
+K D AW + ++++SW +MIAG+ DKA+ F M G
Sbjct: 496 AKLG---------KLDTAWDILIRFAGKDVVSWTTMIAGYTQYNFDDKALTTFRQMLDRG 546
Query: 280 IGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANL 339
I D L + S+ A E +H A +G S++ ALV Y+
Sbjct: 547 IRSDEVGLTNAVSACAGLQALKEGQ-------QIHAQACVSGFSSDLPFQNALVTLYSRC 599
Query: 340 GGQISDCYRLFLDTSGKQDIVSWTAIITVLADQ-DPEQAFLLFCQLHRENFVPDWHTFSI 398
G +I + Y F T D ++W A+++ + E+A +F +++RE + TF
Sbjct: 600 G-KIEESYLAFEQTEAG-DNIAWNALVSGFQQSGNNEEALRVFVRMNREGIDNNNFTFGS 657
Query: 399 ALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHD 458
A+KA + +Q VH+ + K G+ +T + NALI YA+ GS++ +E+ F E+ +
Sbjct: 658 AVKAASETANMKQGKQVHAVITKTGYDSETEVCNALISMYAKCGSISDAEKQFLEVSTKN 717
Query: 459 LVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFN 515
VSWN+++ +Y+ HG +AL+ F ++ +V P+ T V +LSACSH GLV++G+ F
Sbjct: 718 EVSWNAIINAYSKHGFGSEALDSFDQMIHSNVRPNHVTLVGVLSACSHIGLVDKGIAYFE 777
Query: 516 SMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGET 575
SM+ +G+ P+ +HY C+VD+ R G +S A++ I MP+KPD+++W +LL +C H
Sbjct: 778 SMNSEYGLSPKPEHYVCVVDMLTRAGLLSRAKEFIQEMPIKPDALVWRTLLSACVVHKNM 837
Query: 576 RLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEV 635
+ AA EL+P++S YV +SN+Y+ + L R++M++ VKK+PG SW+EV
Sbjct: 838 EIGEFAAHHLLELEPEDSATYVLLSNLYAVSKKWDARDLTRQKMKEKGVKKEPGQSWIEV 897
Query: 636 GKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHS 695
+H F G +HP + I + L + E+GYV + L + + E K+ +F HS
Sbjct: 898 KNSIHSFYVGDQNHPLADEIHEYFQDLTKRASEIGYVQDCFSLLNELQHEQKDPIIFIHS 957
Query: 696 EKMALVFAIMN-EGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHH 754
EK+A+ F +++ +P I +MKN+R+C DCH ++K S + +EI+VRD+ RFHH
Sbjct: 958 EKLAISFGLLSLPATVP-----INVMKNLRVCNDCHAWIKFVSKVSNREIIVRDAYRFHH 1012
Query: 755 FKNATCSCNDYW 766
F+ CSC DYW
Sbjct: 1013 FEGGACSCKDYW 1024
Score = 187 bits (476), Expect = 1e-47
Identities = 153/557 (27%), Positives = 265/557 (47%), Gaps = 35/557 (6%)
Query: 30 PETTISTNIDAQIRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCL 89
PE TI T + I+ L+ + + E L +T + T++ + AC
Sbjct: 107 PERTIFT-WNKMIKELASRNLIGEVFGLFVRMVSENVTPNE---GTFSGVLEACRGGSVA 162
Query: 90 QQGMA-LHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALI 148
+ +H IL++ +++ + N L+++YS+ G ++ AR VFD + ++ SW A+I
Sbjct: 163 FDVVEQIHARILYQG--LRDSTVVCNPLIDLYSRNGFVDLARRVFDGLRLKDHSSWVAMI 220
Query: 149 SGYAQCGLIRECFYLFSGL-LAHYRPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLD 206
SG ++ E LF + + P +AF+S+LSAC++ ++ G Q+H + LK+
Sbjct: 221 SGLSKNECEAEAIRLFCDMYVLGIMPTPYAFSSVLSACKKIESLEIGEQLHGLVLKLGFS 280
Query: 207 ASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGD 266
+ YV NAL+++Y FH G + + +F +M R+ +++N++I G G G+
Sbjct: 281 SDTYVCNALVSLY------FHLGNLISAEH---IFSNMSQRDAVTYNTLINGLSQCGYGE 331
Query: 267 KAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEV 326
KA+ LF M+ G+ D TL S+ + CSA L + LH TK G S
Sbjct: 332 KAMELFKRMHLDGLEPDSNTLASLVVA---CSADGTLFRGQQ----LHAYTTKLGFASNN 384
Query: 327 EVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITV--LADQDPEQAFLLFCQL 384
++ AL+ YA I FL+T +++V W ++ L D D +F +F Q+
Sbjct: 385 KIEGALLNLYAKC-ADIETALDYFLETE-VENVVLWNVMLVAYGLLD-DLRNSFRIFRQM 441
Query: 385 HRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSL 444
E VP+ +T+ LK C + +HSQ+IK FQ + + + LI YA+ G L
Sbjct: 442 QIEEIVPNQYTYPSILKTCIRLGDLELGEQIHSQIIKTNFQLNAYVCSVLIDMYAKLGKL 501
Query: 445 ALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSAC 501
+ + D+VSW +M+ Y + AL F+++ + D +SAC
Sbjct: 502 DTAWDILIRFAGKDVVSWTTMIAGYTQYNFDDKALTTFRQMLDRGIRSDEVGLTNAVSAC 561
Query: 502 SHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVI 561
+ ++EG +I ++ + G L +V LY R GKI E+ L D++
Sbjct: 562 AGLQALKEGQQI-HAQACVSGFSSDLPFQNALVTLYSRCGKIEESY-LAFEQTEAGDNIA 619
Query: 562 WSSLLGSCRKHGETRLA 578
W++L+ ++ G A
Sbjct: 620 WNALVSGFQQSGNNEEA 636
Score = 181 bits (458), Expect = 2e-45
Identities = 150/503 (29%), Positives = 243/503 (47%), Gaps = 34/503 (6%)
Query: 74 QTYASLFHACAK-NKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYV 132
QT L C K N L +G LH+ IL + ++ L+ L + Y G L A V
Sbjct: 45 QTLKWLLEGCLKTNGSLDEGRKLHSQILKLG--LDSNGCLSEKLFDFYLFKGDLYGAFKV 102
Query: 133 FDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLA-HYRPNEFAFASLLSACEEHDIK 191
FD+MP R I +W +I A LI E F LF +++ + PNE F+ +L AC +
Sbjct: 103 FDEMPERTIFTWNKMIKELASRNLIGEVFGLFVRMVSENVTPNEGTFSGVLEACRGGSVA 162
Query: 192 CGM--QVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNL 249
+ Q+HA L L S V N LI +YS+ GF D A +F + ++
Sbjct: 163 FDVVEQIHARILYQGLRDSTVVCNPLIDLYSR--NGF-------VDLARRVFDGLRLKDH 213
Query: 250 ISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRN 309
SW +MI+G +AIRLF MY GI T + S L+ C + L I +
Sbjct: 214 SSWVAMISGLSKNECEAEAIRLFCDMYVLGI---MPTPYAFSSVLSACKKIESLEIGEQ- 269
Query: 310 CFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVL 369
LH L K G S+ V ALV Y +LG IS +F + S ++D V++ +I L
Sbjct: 270 ---LHGLVLKLGFSSDTYVCNALVSLYFHLGNLIS-AEHIFSNMS-QRDAVTYNTLINGL 324
Query: 370 ADQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDT 428
+ E+A LF ++H + PD +T + + AC+ T + +H+ K GF +
Sbjct: 325 SQCGYGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTLFRGQQLHAYTTKLGFASNN 384
Query: 429 VLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLDVH 488
+ AL++ YA+ + + F E ++V WN ML +Y L +++ +F+++ +
Sbjct: 385 KIEGALLNLYAKCADIETALDYFLETEVENVVLWNVMLVAYGLLDDLRNSFRIFRQMQIE 444
Query: 489 ---PDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYAC--MVDLYGRVGKI 543
P+ T+ ++L C G +E G +I + + + QL+ Y C ++D+Y ++GK+
Sbjct: 445 EIVPNQYTYPSILKTCIRLGDLELGEQIHSQIIKTNF---QLNAYVCSVLIDMYAKLGKL 501
Query: 544 SEAEDLIHTMPMKPDSVIWSSLL 566
A D++ K D V W++++
Sbjct: 502 DTAWDILIRFAGK-DVVSWTTMI 523
Score = 171 bits (432), Expect = 2e-42
Identities = 125/504 (24%), Positives = 238/504 (46%), Gaps = 28/504 (5%)
Query: 76 YASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQ 135
++S+ AC K + L+ G LH +L +D ++ N LV++Y G+L A ++F
Sbjct: 251 FSSVLSACKKIESLEIGEQLHGLVLKLG--FSSDTYVCNALVSLYFHLGNLISAEHIFSN 308
Query: 136 MPRRNIVSWTALISGYAQCGLIRECFYLFSGL-LAHYRPNEFAFASLLSACE-EHDIKCG 193
M +R+ V++ LI+G +QCG + LF + L P+ ASL+ AC + + G
Sbjct: 309 MSQRDAVTYNTLINGLSQCGYGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTLFRG 368
Query: 194 MQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWN 253
Q+HA K+ ++ + AL+ +Y+KC+ + A F E N++ WN
Sbjct: 369 QQLHAYTTKLGFASNNKIEGALLNLYAKCA---------DIETALDYFLETEVENVVLWN 419
Query: 254 SMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVL 313
M+ + + R+F M I ++ T S+ L C +L + + +
Sbjct: 420 VMLVAYGLLDDLRNSFRIFRQMQIEEIVPNQYTYPSI---LKTCIRLGDLELGEQ----I 472
Query: 314 HCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQD 373
H KT V + L+ YA L G++ + + + +GK D+VSWT +I +
Sbjct: 473 HSQIIKTNFQLNAYVCSVLIDMYAKL-GKLDTAWDILIRFAGK-DVVSWTTMIAGYTQYN 530
Query: 374 -PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSN 432
++A F Q+ D + A+ ACA ++ +H+Q GF D N
Sbjct: 531 FDDKALTTFRQMLDRGIRSDEVGLTNAVSACAGLQALKEGQQIHAQACVSGFSSDLPFQN 590
Query: 433 ALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLD---VHP 489
AL+ Y+R G + S F++ D ++WN+++ + G ++AL +F +++ +
Sbjct: 591 ALVTLYSRCGKIEESYLAFEQTEAGDNIAWNALVSGFQQSGNNEEALRVFVRMNREGIDN 650
Query: 490 DSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDL 549
++ TF + + A S +++G ++ +++ G + + ++ +Y + G IS+AE
Sbjct: 651 NNFTFGSAVKAASETANMKQGKQV-HAVITKTGYDSETEVCNALISMYAKCGSISDAEKQ 709
Query: 550 IHTMPMKPDSVIWSSLLGSCRKHG 573
+ K + V W++++ + KHG
Sbjct: 710 FLEVSTK-NEVSWNAIINAYSKHG 732
Score = 128 bits (322), Expect = 1e-29
Identities = 106/402 (26%), Positives = 178/402 (43%), Gaps = 27/402 (6%)
Query: 172 RPNEFAFASLLSACEEHD--IKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGG 229
RPN LL C + + + G ++H+ LK+ LD++ ++ L Y G +G
Sbjct: 41 RPNHQTLKWLLEGCLKTNGSLDEGRKLHSQILKLGLDSNGCLSEKLFDFYL-FKGDLYG- 98
Query: 230 YDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLS 289
A+ +F M R + +WN MI R L + LF M + + T
Sbjct: 99 -------AFKVFDEMPERTIFTWNKMIKELASRNLIGEVFGLFVRMVSENVTPNEGTFSG 151
Query: 290 VFSSLNECS-AFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYR 348
V + S AFD + +H GL V L+ Y+ G D R
Sbjct: 152 VLEACRGGSVAFDVVE-------QIHARILYQGLRDSTVVCNPLIDLYSRNG--FVDLAR 202
Query: 349 LFLDTSGKQDIVSWTAIITVLADQDPE-QAFLLFCQLHRENFVPDWHTFSIALKACAYFV 407
D +D SW A+I+ L+ + E +A LFC ++ +P + FS L AC
Sbjct: 203 RVFDGLRLKDHSSWVAMISGLSKNECEAEAIRLFCDMYVLGIMPTPYAFSSVLSACKKIE 262
Query: 408 TEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLK 467
+ + +H V+K GF DT + NAL+ Y G+L +E +F M D V++N+++
Sbjct: 263 SLEIGEQLHGLVLKLGFSSDTYVCNALVSLYFHLGNLISAEHIFSNMSQRDAVTYNTLIN 322
Query: 468 SYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIV 524
+ G + A+ELFK++ + PDS T +L+ ACS G + G ++ ++ + G
Sbjct: 323 GLSQCGYGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTLFRGQQL-HAYTTKLGFA 381
Query: 525 PQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLL 566
+++LY + I A D ++ + V+W+ +L
Sbjct: 382 SNNKIEGALLNLYAKCADIETALDYFLETEVE-NVVLWNVML 422
Score = 61.6 bits (148), Expect = 2e-09
Identities = 44/174 (25%), Positives = 76/174 (43%), Gaps = 3/174 (1%)
Query: 415 VHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGK 474
+HSQ++K G + LS L Y G L + +VFDEM + +WN M+K A
Sbjct: 67 LHSQILKLGLDSNGCLSEKLFDFYLFKGDLYGAFKVFDEMPERTIFTWNKMIKELASRNL 126
Query: 475 AKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYA 531
+ LF ++ +V P+ TF +L AC + + VE ++ G+
Sbjct: 127 IGEVFGLFVRMVSENVTPNEGTFSGVLEACRGGSVAFDVVEQIHARILYQGLRDSTVVCN 186
Query: 532 CMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKF 585
++DLY R G + A + + +K S + + G + E R+ + +
Sbjct: 187 PLIDLYSRNGFVDLARRVFDGLRLKDHSSWVAMISGLSKNECEAEAIRLFCDMY 240
>At3g03580 unknown protein (At3g03580)
Length = 882
Score = 406 bits (1044), Expect = e-113
Identities = 245/733 (33%), Positives = 385/733 (52%), Gaps = 37/733 (5%)
Query: 42 IRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILH 101
I S G EEAL + + + S T +S+ A ++QG LH + L
Sbjct: 179 ISGYSSHGYYEEALEIYHELKNSWIVPDSF---TVSSVLPAFGNLLVVKQGQGLHGFALK 235
Query: 102 KDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECF 161
+ + + + N LV MY K AR VFD+M R+ VS+ +I GY + ++ E
Sbjct: 236 SG--VNSVVVVNNGLVAMYLKFRRPTDARRVFDEMDVRDSVSYNTMICGYLKLEMVEESV 293
Query: 162 YLFSGLLAHYRPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMYS 220
+F L ++P+ +S+L AC D+ ++ LK V N LI +Y+
Sbjct: 294 RMFLENLDQFKPDLLTVSSVLRACGHLRDLSLAKYIYNYMLKAGFVLESTVRNILIDVYA 353
Query: 221 KCSGGFHGGYDPTGD--DAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCS 278
KC GD A +F SME ++ +SWNS+I+G+ G +A++LF M
Sbjct: 354 KC-----------GDMITARDVFNSMECKDTVSWNSIISGYIQSGDLMEAMKLFKMMMIM 402
Query: 279 GIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYAN 338
D T L + S + L+ LH K+G+ ++ V AL+ YA
Sbjct: 403 EEQADHITYLMLISVSTRLA-------DLKFGKGLHSNGIKSGICIDLSVSNALIDMYAK 455
Query: 339 LGGQISDCYRLFLDTSGKQDIVSWTAIITVLAD-QDPEQAFLLFCQLHRENFVPDWHTFS 397
G ++ D ++F + G D V+W +I+ D + Q+ + VPD TF
Sbjct: 456 CG-EVGDSLKIF-SSMGTGDTVTWNTVISACVRFGDFATGLQVTTQMRKSEVVPDMATFL 513
Query: 398 IALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCH 457
+ L CA ++ +H +++ G++ + + NALI Y++ G L S +VF+ M
Sbjct: 514 VTLPMCASLAAKRLGKEIHCCLLRFGYESELQIGNALIEMYSKCGCLENSSRVFERMSRR 573
Query: 458 DLVSWNSMLKSYALHGKAKDALELF---KKLDVHPDSTTFVALLSACSHAGLVEEGVEIF 514
D+V+W M+ +Y ++G+ + ALE F +K + PDS F+A++ ACSH+GLV+EG+ F
Sbjct: 574 DVVTWTGMIYAYGMYGEGEKALETFADMEKSGIVPDSVVFIAIIYACSHSGLVDEGLACF 633
Query: 515 NSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGE 574
M ++ I P ++HYAC+VDL R KIS+AE+ I MP+KPD+ IW+S+L +CR G+
Sbjct: 634 EKMKTHYKIDPMIEHYACVVDLLSRSQKISKAEEFIQAMPIKPDASIWASVLRACRTSGD 693
Query: 575 TRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVE 634
A + + EL+P + + SN Y++ + + LIRK ++D + K PG SW+E
Sbjct: 694 METAERVSRRIIELNPDDPGYSILASNAYAALRKWDKVSLIRKSLKDKHITKNPGYSWIE 753
Query: 635 VGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLF-H 693
VGK VH F+SG P EAI LEIL + + GY+P+ + E E ++ +L
Sbjct: 754 VGKNVHVFSSGDDSAPQSEAIYKSLEILYSLMAKEGYIPDPREVSQNLEEEEEKRRLICG 813
Query: 694 HSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFH 753
HSE++A+ F ++N G +++MKN+R+C DCH KL S + +EI+VRD+NRFH
Sbjct: 814 HSERLAIAFGLLNTE----PGTPLQVMKNLRVCGDCHEVTKLISKIVGREILVRDANRFH 869
Query: 754 HFKNATCSCNDYW 766
FK+ TCSC D W
Sbjct: 870 LFKDGTCSCKDRW 882
Score = 183 bits (464), Expect = 4e-46
Identities = 166/596 (27%), Positives = 274/596 (45%), Gaps = 38/596 (6%)
Query: 42 IRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILH 101
IRA S G EAL + K + S T+ S+ ACA + G ++ IL
Sbjct: 78 IRAFSKNGLFPEALEF---YGKLRESKVSPDKYTFPSVIKACAGLFDAEMGDLVYEQIL- 133
Query: 102 KDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECF 161
D ++DLF+ N LV+MYS+ G L AR VFD+MP R++VSW +LISGY+ G E
Sbjct: 134 -DMGFESDLFVGNALVDMYSRMGLLTRARQVFDEMPVRDLVSWNSLISGYSSHGYYEEAL 192
Query: 162 YLFSGLLAHY-RPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMY 219
++ L + P+ F +S+L A +K G +H ALK +++ V V N L+ MY
Sbjct: 193 EIYHELKNSWIVPDSFTVSSVLPAFGNLLVVKQGQGLHGFALKSGVNSVVVVNNGLVAMY 252
Query: 220 SKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSG 279
K PT DA +F M+ R+ +S+N+MI G+ + ++++R M+
Sbjct: 253 LKFR-------RPT--DARRVFDEMDVRDSVSYNTMICGYLKLEMVEESVR----MFLEN 299
Query: 280 IGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANL 339
+ + LL+V S L C +L++ ++ K G + E V L+ YA
Sbjct: 300 LDQFKPDLLTVSSVLRACGHLRDLSL----AKYIYNYMLKAGFVLESTVRNILIDVYAKC 355
Query: 340 GGQISDCYRLFLDTSGKQDIVSWTAIIT-VLADQDPEQAFLLFCQLHRENFVPDWHTFSI 398
G I+ R ++ +D VSW +II+ + D +A LF + D T+ +
Sbjct: 356 GDMIT--ARDVFNSMECKDTVSWNSIISGYIQSGDLMEAMKLFKMMMIMEEQADHITYLM 413
Query: 399 ALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHD 458
+ + +HS IK G D +SNALI YA+ G + S ++F M D
Sbjct: 414 LISVSTRLADLKFGKGLHSNGIKSGICIDLSVSNALIDMYAKCGEVGDSLKIFSSMGTGD 473
Query: 459 LVSWNSMLKSYALHGKAKDALEL---FKKLDVHPDSTTFVALLSACSHAGLVEEGVEIFN 515
V+WN+++ + G L++ +K +V PD TF+ L C+ G EI
Sbjct: 474 TVTWNTVISACVRFGDFATGLQVTTQMRKSEVVPDMATFLVTLPMCASLAAKRLGKEIHC 533
Query: 516 SMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGET 575
+ G +L ++++Y + G + + + M + D V W+ ++ + +GE
Sbjct: 534 CLL-RFGYESELQIGNALIEMYSKCGCLENSSRVFERMSRR-DVVTWTGMIYAYGMYGE- 590
Query: 576 RLARIAAEKFKELDPKNSL--GYVQMSNIYS-SEGSFIEAGLIRKEMRDSRVKKQP 628
A E F +++ + V ++ IY+ S ++ GL E + K P
Sbjct: 591 --GEKALETFADMEKSGIVPDSVVFIAIIYACSHSGLVDEGLACFEKMKTHYKIDP 644
Score = 165 bits (417), Expect = 1e-40
Identities = 127/471 (26%), Positives = 233/471 (48%), Gaps = 28/471 (5%)
Query: 111 FLTNHLVNMYSKCGHLEYARYVFDQM-PRRNIVSWTALISGYAQCGLIRECFYLFSGLL- 168
F + L++ YS + VF ++ P +N+ W ++I +++ GL E + L
Sbjct: 40 FFSGKLIDKYSHFREPASSLSVFRRVSPAKNVYLWNSIIRAFSKNGLFPEALEFYGKLRE 99
Query: 169 AHYRPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFH 227
+ P+++ F S++ AC D + G V+ L + ++ ++V NAL+ MYS+
Sbjct: 100 SKVSPDKYTFPSVIKACAGLFDAEMGDLVYEQILDMGFESDLFVGNALVDMYSRMG---- 155
Query: 228 GGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATL 287
A +F M R+L+SWNS+I+G+ G ++A+ ++ + S I D T+
Sbjct: 156 -----LLTRARQVFDEMPVRDLVSWNSLISGYSSHGYYEEALEIYHELKNSWIVPDSFTV 210
Query: 288 LSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCY 347
SV AF L + ++ LH A K+G+ S V V LV Y + +D
Sbjct: 211 SSVL------PAFGNL-LVVKQGQGLHGFALKSGVNSVVVVNNGLVAMYLKF-RRPTDAR 262
Query: 348 RLFLDTSGKQDIVSW-TAIITVLADQDPEQAFLLFCQLHRENFVPDWHTFSIALKACAYF 406
R+F D +D VS+ T I L + E++ +F + + + F PD T S L+AC +
Sbjct: 263 RVF-DEMDVRDSVSYNTMICGYLKLEMVEESVRMFLE-NLDQFKPDLLTVSSVLRACGHL 320
Query: 407 VTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSML 466
A +++ ++K GF ++ + N LI YA+ G + + VF+ M C D VSWNS++
Sbjct: 321 RDLSLAKYIYNYMLKAGFVLESTVRNILIDVYAKCGDMITARDVFNSMECKDTVSWNSII 380
Query: 467 KSYALHGKAKDALELFKK---LDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGI 523
Y G +A++LFK ++ D T++ L+S + ++ G + +S GI
Sbjct: 381 SGYIQSGDLMEAMKLFKMMMIMEEQADHITYLMLISVSTRLADLKFGKGL-HSNGIKSGI 439
Query: 524 VPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGE 574
L ++D+Y + G++ ++ + +M D+V W++++ +C + G+
Sbjct: 440 CIDLSVSNALIDMYAKCGEVGDSLKIFSSMG-TGDTVTWNTVISACVRFGD 489
Score = 112 bits (281), Expect = 6e-25
Identities = 97/384 (25%), Positives = 180/384 (46%), Gaps = 24/384 (6%)
Query: 195 QVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSME-FRNLISWN 253
++HA+ + + LD+S + + LI YS +P + ++F+ + +N+ WN
Sbjct: 25 RIHALVISLGLDSSDFFSGKLIDKYSHFR-------EPA--SSLSVFRRVSPAKNVYLWN 75
Query: 254 SMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVL 313
S+I F GL +A+ + + S + D+ T SV + C+ + + ++
Sbjct: 76 SIIRAFSKNGLFPEALEFYGKLRESKVSPDKYTFPSV---IKACAGLFDAEMG----DLV 128
Query: 314 HCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQD 373
+ G S++ V ALV Y+ +G + R D +D+VSW ++I+ +
Sbjct: 129 YEQILDMGFESDLFVGNALVDMYSRMG--LLTRARQVFDEMPVRDLVSWNSLISGYSSHG 186
Query: 374 -PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSN 432
E+A ++ +L VPD T S L A + +Q +H +K G V++N
Sbjct: 187 YYEEALEIYHELKNSWIVPDSFTVSSVLPAFGNLLVVKQGQGLHGFALKSGVNSVVVVNN 246
Query: 433 ALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELF-KKLD-VHPD 490
L+ Y + + +VFDEM D VS+N+M+ Y ++++ +F + LD PD
Sbjct: 247 GLVAMYLKFRRPTDARRVFDEMDVRDSVSYNTMICGYLKLEMVEESVRMFLENLDQFKPD 306
Query: 491 STTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLI 550
T ++L AC H + I+N M G V + ++D+Y + G + A D+
Sbjct: 307 LLTVSSVLRACGHLRDLSLAKYIYNYML-KAGFVLESTVRNILIDVYAKCGDMITARDVF 365
Query: 551 HTMPMKPDSVIWSSLLGSCRKHGE 574
++M K D+V W+S++ + G+
Sbjct: 366 NSMECK-DTVSWNSIISGYIQSGD 388
>At4g33170 putative protein
Length = 990
Score = 402 bits (1034), Expect = e-112
Identities = 228/667 (34%), Positives = 368/667 (54%), Gaps = 32/667 (4%)
Query: 109 DLFLT--NHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSG 166
DL LT N L+NMY K +AR VFD M R+++SW ++I+G AQ GL E LF
Sbjct: 347 DLMLTVSNSLINMYCKLRKFGFARTVFDNMSERDLISWNSVIAGIAQNGLEVEAVCLFMQ 406
Query: 167 LL-AHYRPNEFAFASLLSACEE--HDIKCGMQVHAVALKISLDASVYVANALITMYSKCS 223
LL +P+++ S+L A + QVH A+KI+ + +V+ ALI YS+
Sbjct: 407 LLRCGLKPDQYTMTSVLKAASSLPEGLSLSKQVHVHAIKINNVSDSFVSTALIDAYSR-- 464
Query: 224 GGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFD 283
+ +A +F+ F +L++WN+M+AG+ G K ++LFA M+ G D
Sbjct: 465 -------NRCMKEAEILFERHNF-DLVAWNAMMAGYTQSHDGHKTLKLFALMHKQGERSD 516
Query: 284 RATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQI 343
TL +VF + A ++ +H A K+G ++ V + ++ Y G
Sbjct: 517 DFTLATVFKTCGFLFAINQGK-------QVHAYAIKSGYDLDLWVSSGILDMYVKCGDM- 568
Query: 344 SDCYRLFLDTSGKQDIVSWTAIIT-VLADQDPEQAFLLFCQLHRENFVPDWHTFSIALKA 402
+ D+ D V+WT +I+ + + + E+AF +F Q+ +PD T + KA
Sbjct: 569 -SAAQFAFDSIPVPDDVAWTTMISGCIENGEEERAFHVFSQMRLMGVLPDEFTIATLAKA 627
Query: 403 CAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSW 462
+ +Q +H+ +K D + +L+ YA+ GS+ + +F + ++ +W
Sbjct: 628 SSCLTALEQGRQIHANALKLNCTNDPFVGTSLVDMYAKCGSIDDAYCLFKRIEMMNITAW 687
Query: 463 NSMLKSYALHGKAKDALELFKK---LDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSD 519
N+ML A HG+ K+ L+LFK+ L + PD TF+ +LSACSH+GLV E + SM
Sbjct: 688 NAMLVGLAQHGEGKETLQLFKQMKSLGIKPDKVTFIGVLSACSHSGLVSEAYKHMRSMHG 747
Query: 520 NHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLAR 579
++GI P+++HY+C+ D GR G + +AE+LI +M M+ + ++ +LL +CR G+T +
Sbjct: 748 DYGIKPEIEHYSCLADALGRAGLVKQAENLIESMSMEASASMYRTLLAACRVQGDTETGK 807
Query: 580 IAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQV 639
A K EL+P +S YV +SN+Y++ + E L R M+ +VKK PG SW+EV ++
Sbjct: 808 RVATKLLELEPLDSSAYVLLSNMYAAASKWDEMKLARTMMKGHKVKKDPGFSWIEVKNKI 867
Query: 640 HEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMA 699
H F + E I +++ +I +K+ GYVPE L D E E KE L++HSEK+A
Sbjct: 868 HIFVVDDRSNRQTELIYRKVKDMIRDIKQEGYVPETDFTLVDVEEEEKERALYYHSEKLA 927
Query: 700 LVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNAT 759
+ F +++ I+++KN+R+C DCHN MK + ++ +EIV+RD+NRFH FK+
Sbjct: 928 VAFGLLSTP----PSTPIRVIKNLRVCGDCHNAMKYIAKVYNREIVLRDANRFHRFKDGI 983
Query: 760 CSCNDYW 766
CSC DYW
Sbjct: 984 CSCGDYW 990
Score = 171 bits (434), Expect = 1e-42
Identities = 157/585 (26%), Positives = 255/585 (42%), Gaps = 93/585 (15%)
Query: 69 SSLSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEY 128
SS S Q + L +A + L G H IL + + FL N+L++MYSKCG L Y
Sbjct: 36 SSSSSQWFGFLRNAITSSD-LMLGKCTHARILTFEENPER--FLINNLISMYSKCGSLTY 92
Query: 129 ARYVFDQMPRRNIVSWTALISGYAQCG-----LIRECFYLFSGLLAH-YRPNEFAFASLL 182
AR VFD+MP R++VSW ++++ YAQ I++ F LF L + + +L
Sbjct: 93 ARRVFDKMPDRDLVSWNSILAAYAQSSECVVENIQQAFLLFRILRQDVVYTSRMTLSPML 152
Query: 183 SAC-EEHDIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMF 241
C + H A KI LD +VA AL+ +Y K G +F
Sbjct: 153 KLCLHSGYVWASESFHGYACKIGLDGDEFVAGALVNIYLKFGKVKEGK---------VLF 203
Query: 242 KSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATL-------------- 287
+ M +R+++ WN M+ + G ++AI L + + SG+ + TL
Sbjct: 204 EEMPYRDVVLWNLMLKAYLEMGFKEEAIDLSSAFHSSGLNPNEITLRLLARISGDDSDAG 263
Query: 288 -LSVFSSLNECSAFDELNI---------------PLRNCFV------------------- 312
+ F++ N+ S+ E+ L CF
Sbjct: 264 QVKSFANGNDASSVSEIIFRNKGLSEYLHSGQYSALLKCFADMVESDVECDQVTFILMLA 323
Query: 313 -------------LHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDI 359
+HC+A K GL + V +L+ Y L + R D ++D+
Sbjct: 324 TAVKVDSLALGQQVHCMALKLGLDLMLTVSNSLINMYCKL--RKFGFARTVFDNMSERDL 381
Query: 360 VSWTAIITVLADQDPE-QAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALA--VH 416
+SW ++I +A E +A LF QL R PD +T + LKA A + E +L+ VH
Sbjct: 382 ISWNSVIAGIAQNGLEVEAVCLFMQLLRCGLKPDQYTMTSVLKA-ASSLPEGLSLSKQVH 440
Query: 417 SQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAK 476
IK D+ +S ALI AY+R+ + +E +F E DLV+WN+M+ Y
Sbjct: 441 VHAIKINNVSDSFVSTALIDAYSRNRCMKEAEILF-ERHNFDLVAWNAMMAGYTQSHDGH 499
Query: 477 DALELF---KKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACM 533
L+LF K D T + C + +G ++ ++ + G L + +
Sbjct: 500 KTLKLFALMHKQGERSDDFTLATVFKTCGFLFAINQGKQV-HAYAIKSGYDLDLWVSSGI 558
Query: 534 VDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLA 578
+D+Y + G +S A+ ++P+ PD V W++++ C ++GE A
Sbjct: 559 LDMYVKCGDMSAAQFAFDSIPV-PDDVAWTTMISGCIENGEEERA 602
Score = 124 bits (311), Expect = 2e-28
Identities = 91/326 (27%), Positives = 157/326 (47%), Gaps = 28/326 (8%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T A++F C + QG +H Y + + DL++++ +++MY KCG + A++ FD
Sbjct: 519 TLATVFKTCGFLFAINQGKQVHAYAIKSGYDL--DLWVSSGILDMYVKCGDMSAAQFAFD 576
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGL-LAHYRPNEFAFASLLSACE-EHDIKC 192
+P + V+WT +ISG + G F++FS + L P+EF A+L A ++
Sbjct: 577 SIPVPDDVAWTTMISGCIENGEEERAFHVFSQMRLMGVLPDEFTIATLAKASSCLTALEQ 636
Query: 193 GMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISW 252
G Q+HA ALK++ +V +L+ MY+KC + DDA+ +FK +E N+ +W
Sbjct: 637 GRQIHANALKLNCTNDPFVGTSLVDMYAKCG---------SIDDAYCLFKRIEMMNITAW 687
Query: 253 NSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCF- 311
N+M+ G G G + ++LF M GI D+ T + V S+ + E +R+
Sbjct: 688 NAMLVGLAQHGEGKETLQLFKQMKSLGIKPDKVTFIGVLSACSHSGLVSEAYKHMRSMHG 747
Query: 312 -------VLH--CLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQ-DIVS 361
+ H CLA G V+ L++S + + YR L Q D +
Sbjct: 748 DYGIKPEIEHYSCLADALGRAGLVKQAENLIESMSMEAS--ASMYRTLLAACRVQGDTET 805
Query: 362 WTAIITVLADQDP--EQAFLLFCQLH 385
+ T L + +P A++L ++
Sbjct: 806 GKRVATKLLELEPLDSSAYVLLSNMY 831
>At2g29760 hypothetical protein
Length = 738
Score = 400 bits (1028), Expect = e-111
Identities = 241/726 (33%), Positives = 391/726 (53%), Gaps = 55/726 (7%)
Query: 76 YASLFHACAKNKCLQQGMALHNYILH----KDPTIQNDLFLTNHLVNMYSKCGHLEYARY 131
+ SL C + L+Q H +++ DP + LF L + S LEYAR
Sbjct: 33 HISLIERCVSLRQLKQ---THGHMIRTGTFSDPYSASKLFAMAALSSFAS----LEYARK 85
Query: 132 VFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHYR--PNEFAFASLLSACEE-H 188
VFD++P+ N +W LI YA + F +++ + PN++ F L+ A E
Sbjct: 86 VFDEIPKPNSFAWNTLIRAYASGPDPVLSIWAFLDMVSESQCYPNKYTFPFLIKAAAEVS 145
Query: 189 DIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRN 248
+ G +H +A+K ++ + V+VAN+LI Y C D A +F +++ ++
Sbjct: 146 SLSLGQSLHGMAVKSAVGSDVFVANSLIHCYFSCGDL---------DSACKVFTTIKEKD 196
Query: 249 LISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNE----------CS 298
++SWNSMI GF +G DKA+ LF M + T++ V S+ + CS
Sbjct: 197 VVSWNSMINGFVQKGSPDKALELFKKMESEDVKASHVTMVGVLSACAKIRNLEFGRQVCS 256
Query: 299 AFDE--LNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANL-------GGQISDCY-- 347
+E +N+ L + + TK G I + + + ++ N+ G IS+ Y
Sbjct: 257 YIEENRVNVNLTLANAMLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGYAISEDYEA 316
Query: 348 -RLFLDTSGKQDIVSWTAIITVLADQD-PEQAFLLFCQLH-RENFVPDWHTFSIALKACA 404
R L++ ++DIV+W A+I+ P +A ++F +L ++N + T L ACA
Sbjct: 317 AREVLNSMPQKDIVAWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITLVSTLSACA 376
Query: 405 YFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNS 464
+ +HS + K G + + +++ALIH Y++ G L S +VF+ + D+ W++
Sbjct: 377 QVGALELGRWIHSYIKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEKRDVFVWSA 436
Query: 465 MLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNH 521
M+ A+HG +A+++F K+ +V P+ TF + ACSH GLV+E +F+ M N+
Sbjct: 437 MIGGLAMHGCGNEAVDMFYKMQEANVKPNGVTFTNVFCACSHTGLVDEAESLFHQMESNY 496
Query: 522 GIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIA 581
GIVP+ HYAC+VD+ GR G + +A I MP+ P + +W +LLG+C+ H LA +A
Sbjct: 497 GIVPEEKHYACIVDVLGRSGYLEKAVKFIEAMPIPPSTSVWGALLGACKIHANLNLAEMA 556
Query: 582 AEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHE 641
+ EL+P+N +V +SNIY+ G + +RK MR + +KK+PG S +E+ +HE
Sbjct: 557 CTRLLELEPRNDGAHVLLSNIYAKLGKWENVSELRKHMRVTGLKKEPGCSSIEIDGMIHE 616
Query: 642 FTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEH-KEDQLFHHSEKMAL 700
F SG + HP E + +L ++ +LK GY PEI+ L E E KE L HSEK+A+
Sbjct: 617 FLSGDNAHPMSEKVYGKLHEVMEKLKSNGYEPEISQVLQIIEEEEMKEQSLNLHSEKLAI 676
Query: 701 VFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATC 760
+ +++ VI+++KN+R+C DCH+ KL S L+ +EI+VRD RFHHF+N C
Sbjct: 677 CYGLISTE----APKVIRVIKNLRVCGDCHSVAKLISQLYDREIIVRDRYRFHHFRNGQC 732
Query: 761 SCNDYW 766
SCND+W
Sbjct: 733 SCNDFW 738
Score = 77.4 bits (189), Expect = 3e-14
Identities = 49/148 (33%), Positives = 79/148 (53%), Gaps = 5/148 (3%)
Query: 39 DAQIRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNY 98
+A I A G EAL + H+ + + L+ T S ACA+ L+ G +H+Y
Sbjct: 333 NALISAYEQNGKPNEALIVF--HELQLQKNMKLNQITLVSTLSACAQVGALELGRWIHSY 390
Query: 99 ILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIR 158
I K I+ + +T+ L++MYSKCG LE +R VF+ + +R++ W+A+I G A G
Sbjct: 391 I--KKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEKRDVFVWSAMIGGLAMHGCGN 448
Query: 159 ECFYLFSGLL-AHYRPNEFAFASLLSAC 185
E +F + A+ +PN F ++ AC
Sbjct: 449 EAVDMFYKMQEANVKPNGVTFTNVFCAC 476
>At3g22690 hypothetical protein
Length = 978
Score = 399 bits (1024), Expect = e-111
Identities = 248/761 (32%), Positives = 396/761 (51%), Gaps = 62/761 (8%)
Query: 42 IRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLF--HACAKNKCLQQGMALHNYI 99
IR + G EA+ L + +S +S Y F ACAK++ G+ +H I
Sbjct: 106 IRGYASSGLCNEAILLFLR-----MMNSGISPDKYTFPFGLSACAKSRAKGNGIQIHGLI 160
Query: 100 LHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRE 159
+ DLF+ N LV+ Y++CG L+ AR VFD+M RN+VSWT++I GYA+ ++
Sbjct: 161 VKMG--YAKDLFVQNSLVHFYAECGELDSARKVFDEMSERNVVSWTSMICGYARRDFAKD 218
Query: 160 CFYLFSGLLA--HYRPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALI 216
LF ++ PN ++SAC + D++ G +V+A ++ + + +AL+
Sbjct: 219 AVDLFFRMVRDEEVTPNSVTMVCVISACAKLEDLETGEKVYAFIRNSGIEVNDLMVSALV 278
Query: 217 TMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMY 276
MY KC+ D A +F NL N+M + + +GL +A+ +F M
Sbjct: 279 DMYMKCNAI---------DVAKRLFDEYGASNLDLCNAMASNYVRQGLTREALGVFNLMM 329
Query: 277 CSGIGFDRATLLSVFSSLNECSAFDEL-------NIPLRNCF-------------VLHCL 316
SG+ DR ++LS SS CS + LRN F + C
Sbjct: 330 DSGVRPDRISMLSAISS---CSQLRNILWGKSCHGYVLRNGFESWDNICNALIDMYMKCH 386
Query: 317 ATKTGL-----ISEVEVVT--ALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVL 369
T +S VVT ++V Y G++ + F +T +++IVSW II+ L
Sbjct: 387 RQDTAFRIFDRMSNKTVVTWNSIVAGYVE-NGEVDAAWETF-ETMPEKNIVSWNTIISGL 444
Query: 370 ADQDP-EQAFLLFCQLHRENFV-PDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKD 427
E+A +FC + + V D T AC + A ++ + K G Q D
Sbjct: 445 VQGSLFEEAIEVFCSMQSQEGVNADGVTMMSIASACGHLGALDLAKWIYYYIEKNGIQLD 504
Query: 428 TVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL-- 485
L L+ ++R G + +F+ + D+ +W + + + A+ G A+ A+ELF +
Sbjct: 505 VRLGTTLVDMFSRCGDPESAMSIFNSLTNRDVSAWTAAIGAMAMAGNAERAIELFDDMIE 564
Query: 486 -DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKIS 544
+ PD FV L+ACSH GLV++G EIF SM HG+ P+ HY CMVDL GR G +
Sbjct: 565 QGLKPDGVAFVGALTACSHGGLVQQGKEIFYSMLKLHGVSPEDVHYGCMVDLLGRAGLLE 624
Query: 545 EAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYS 604
EA LI MPM+P+ VIW+SLL +CR G +A AAEK + L P+ + YV +SN+Y+
Sbjct: 625 EAVQLIEDMPMEPNDVIWNSLLAACRVQGNVEMAAYAAEKIQVLAPERTGSYVLLSNVYA 684
Query: 605 SEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIG 664
S G + + +R M++ ++K PG S +++ + HEFTSG HP I++ L+ +
Sbjct: 685 SAGRWNDMAKVRLSMKEKGLRKPPGTSSIQIRGKTHEFTSGDESHPEMPNIEAMLDEVSQ 744
Query: 665 QLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIR 724
+ +G+VP+++ L D + + K L HSEK+A+ + +++ G I+I+KN+R
Sbjct: 745 RASHLGHVPDLSNVLMDVDEKEKIFMLSRHSEKLAMAYGLISSNK----GTTIRIVKNLR 800
Query: 725 ICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDY 765
+C+DCH+F K AS ++ +EI++RD+NRFH+ + CSC D+
Sbjct: 801 VCSDCHSFAKFASKVYNREIILRDNNRFHYIRQGKCSCGDF 841
Score = 98.2 bits (243), Expect = 2e-20
Identities = 88/370 (23%), Positives = 164/370 (43%), Gaps = 41/370 (11%)
Query: 243 SMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDE 302
S + +NS+I G+ GL ++AI LF M SGI D+ T S+ + A
Sbjct: 93 SESYGTCFMYNSLIRGYASSGLCNEAILLFLRMMNSGISPDKYTFPFGLSACAKSRA--- 149
Query: 303 LNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSW 362
N +H L K G ++ V +LV YA G D R D ++++VSW
Sbjct: 150 ----KGNGIQIHGLIVKMGYAKDLFVQNSLVHFYAECGE--LDSARKVFDEMSERNVVSW 203
Query: 363 TAIITVLADQD--PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVI 420
T++I A +D + L F + E P+ T + ACA + V++ +
Sbjct: 204 TSMICGYARRDFAKDAVDLFFRMVRDEEVTPNSVTMVCVISACAKLEDLETGEKVYAFIR 263
Query: 421 KRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALE 480
G + + ++ +AL+ Y + ++ +++++FDE +L N+M +Y G ++AL
Sbjct: 264 NSGIEVNDLMVSALVDMYMKCNAIDVAKRLFDEYGASNLDLCNAMASNYVRQGLTREALG 323
Query: 481 LFKKL---DVHPDSTTFVALLSACS-----------HAGLVEEGVEIFNSMSDNHGIVPQ 526
+F + V PD + ++ +S+CS H ++ G E ++++ +
Sbjct: 324 VFNLMMDSGVRPDRISMLSAISSCSQLRNILWGKSCHGYVLRNGFESWDNICN------- 376
Query: 527 LDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFK 586
++D+Y + + A + M K V W+S++ ++GE A E F+
Sbjct: 377 -----ALIDMYMKCHRQDTAFRIFDRMSNK-TVVTWNSIVAGYVENGE---VDAAWETFE 427
Query: 587 ELDPKNSLGY 596
+ KN + +
Sbjct: 428 TMPEKNIVSW 437
>At4g18750 putative protein
Length = 871
Score = 398 bits (1022), Expect = e-111
Identities = 239/699 (34%), Positives = 374/699 (53%), Gaps = 31/699 (4%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T++ + + + + + G LH +IL +N + N LV Y K ++ AR VFD
Sbjct: 197 TFSCVSKSFSSLRSVHGGEQLHGFILKSGFGERNSV--GNSLVAFYLKNQRVDSARKVFD 254
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHYRPNEFA-FASLLSACEEHD-IKC 192
+M R+++SW ++I+GY GL + +F +L + A S+ + C + I
Sbjct: 255 EMTERDVISWNSIINGYVSNGLAEKGLSVFVQMLVSGIEIDLATIVSVFAGCADSRLISL 314
Query: 193 GMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISW 252
G VH++ +K N L+ MYSKC D A +F+ M R+++S+
Sbjct: 315 GRAVHSIGVKACFSREDRFCNTLLDMYSKCGDL---------DSAKAVFREMSDRSVVSY 365
Query: 253 NSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFV 312
SMIAG+ GL +A++LF M GI D T+ +V LN C+ + L+ R
Sbjct: 366 TSMIAGYAREGLAGEAVKLFEEMEEEGISPDVYTVTAV---LNCCARYRLLDEGKR---- 418
Query: 313 LHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQ 372
+H + L ++ V AL+ YA G + + +F + K DI+SW II +
Sbjct: 419 VHEWIKENDLGFDIFVSNALMDMYAKCGS-MQEAELVFSEMRVK-DIISWNTIIGGYSKN 476
Query: 373 D-PEQAFLLF-CQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVL 430
+A LF L + F PD T + L ACA + +H +++ G+ D +
Sbjct: 477 CYANEALSLFNLLLEEKRFSPDERTVACVLPACASLSAFDKGREIHGYIMRNGYFSDRHV 536
Query: 431 SNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DV 487
+N+L+ YA+ G+L L+ +FD++ DLVSW M+ Y +HG K+A+ LF ++ +
Sbjct: 537 ANSLVDMYAKCGALLLAHMLFDDIASKDLVSWTVMIAGYGMHGFGKEAIALFNQMRQAGI 596
Query: 488 HPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAE 547
D +FV+LL ACSH+GLV+EG FN M I P ++HYAC+VD+ R G + +A
Sbjct: 597 EADEISFVSLLYACSHSGLVDEGWRFFNIMRHECKIEPTVEHYACIVDMLARTGDLIKAY 656
Query: 548 DLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEG 607
I MP+ PD+ IW +LL CR H + +LA AEK EL+P+N+ YV M+NIY+
Sbjct: 657 RFIENMPIPPDATIWGALLCGCRIHHDVKLAEKVAEKVFELEPENTGYYVLMANIYAEAE 716
Query: 608 SFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLK 667
+ + +RK + ++K PG SW+E+ +V+ F +G +P E I++ L + ++
Sbjct: 717 KWEQVKRLRKRIGQRGLRKNPGCSWIEIKGRVNIFVAGDSSNPETENIEAFLRKVRARMI 776
Query: 668 EMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICA 727
E GY P AL D E KE+ L HSEK+A+ I++ G+ G +I++ KN+R+C
Sbjct: 777 EEGYSPLTKYALIDAEEMEKEEALCGHSEKLAMALGIISSGH----GKIIRVTKNLRVCG 832
Query: 728 DCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
DCH K S L ++EIV+RDSNRFH FK+ CSC +W
Sbjct: 833 DCHEMAKFMSKLTRREIVLRDSNRFHQFKDGHCSCRGFW 871
Score = 188 bits (477), Expect = 1e-47
Identities = 152/545 (27%), Positives = 247/545 (44%), Gaps = 34/545 (6%)
Query: 36 TNIDAQIRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMAL 95
T+ + Q+R GNLE A+ LL K + +T S+ CA +K L+ G +
Sbjct: 62 TDANTQLRRFCESGNLENAVKLLCVSGKW-----DIDPRTLCSVLQLCADSKSLKDGKEV 116
Query: 96 HNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCG 155
N+I I ++L + L MY+ CG L+ A VFD++ + W L++ A+ G
Sbjct: 117 DNFIRGNGFVIDSNL--GSKLSLMYTNCGDLKEASRVFDEVKIEKALFWNILMNELAKSG 174
Query: 156 LIRECFYLFSGLLAH-YRPNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVAN 213
LF +++ + + F+ + + + G Q+H LK V N
Sbjct: 175 DFSGSIGLFKKMMSSGVEMDSYTFSCVSKSFSSLRSVHGGEQLHGFILKSGFGERNSVGN 234
Query: 214 ALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFA 273
+L+ Y K + D A +F M R++ISWNS+I G+ GL +K + +F
Sbjct: 235 SLVAFYLK---------NQRVDSARKVFDEMTERDVISWNSIINGYVSNGLAEKGLSVFV 285
Query: 274 HMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALV 333
M SGI D AT++SVF+ C+ D I L +H + K E L+
Sbjct: 286 QMLVSGIEIDLATIVSVFAG---CA--DSRLISLGR--AVHSIGVKACFSREDRFCNTLL 338
Query: 334 KSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQD-PEQAFLLFCQLHRENFVPD 392
Y+ G + +F + S + +VS+T++I A + +A LF ++ E PD
Sbjct: 339 DMYSKC-GDLDSAKAVFREMSDR-SVVSYTSMIAGYAREGLAGEAVKLFEEMEEEGISPD 396
Query: 393 WHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFD 452
+T + L CA + + VH + + D +SNAL+ YA+ GS+ +E VF
Sbjct: 397 VYTVTAVLNCCARYRLLDEGKRVHEWIKENDLGFDIFVSNALMDMYAKCGSMQEAELVFS 456
Query: 453 EMCCHDLVSWNSMLKSYALHGKAKDALELFKKL----DVHPDSTTFVALLSACSHAGLVE 508
EM D++SWN+++ Y+ + A +AL LF L PD T +L AC+ +
Sbjct: 457 EMRVKDIISWNTIIGGYSKNCYANEALSLFNLLLEEKRFSPDERTVACVLPACASLSAFD 516
Query: 509 EGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGS 568
+G EI + N G +VD+Y + G + A L + K D V W+ ++
Sbjct: 517 KGREIHGYIMRN-GYFSDRHVANSLVDMYAKCGALLLAHMLFDDIASK-DLVSWTVMIAG 574
Query: 569 CRKHG 573
HG
Sbjct: 575 YGMHG 579
Score = 80.5 bits (197), Expect = 3e-15
Identities = 81/342 (23%), Positives = 145/342 (41%), Gaps = 48/342 (14%)
Query: 321 GLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQ-DPEQAFL 379
G + + + + L Y N G + + R+F D + + W ++ LA D +
Sbjct: 124 GFVIDSNLGSKLSLMYTNCG-DLKEASRVF-DEVKIEKALFWNILMNELAKSGDFSGSIG 181
Query: 380 LFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYA 439
LF ++ D +TFS K+ + + +H ++K GF + + N+L+ Y
Sbjct: 182 LFKKMMSSGVEMDSYTFSCVSKSFSSLRSVHGGEQLHGFILKSGFGERNSVGNSLVAFYL 241
Query: 440 RSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVA 496
++ + + +VFDEM D++SWNS++ Y +G A+ L +F ++ + D T V+
Sbjct: 242 KNQRVDSARKVFDEMTERDVISWNSIINGYVSNGLAEKGLSVFVQMLVSGIEIDLATIVS 301
Query: 497 LLSACSHAGLVEEGVEIF---------------NSMSDNHGIVPQLD------------- 528
+ + C+ + L+ G + N++ D + LD
Sbjct: 302 VFAGCADSRLISLGRAVHSIGVKACFSREDRFCNTLLDMYSKCGDLDSAKAVFREMSDRS 361
Query: 529 --HYACMVDLYGRVGKISEAEDLIHTMP---MKPDSVIWSSLLGSCRKHGETRLARIAAE 583
Y M+ Y R G EA L M + PD +++L C ++ + E
Sbjct: 362 VVSYTSMIAGYAREGLAGEAVKLFEEMEEEGISPDVYTVTAVLNCCARYRLLDEGKRVHE 421
Query: 584 KFKELDPKNSLGY-VQMSN----IYSSEGSFIEAGLIRKEMR 620
KE N LG+ + +SN +Y+ GS EA L+ EMR
Sbjct: 422 WIKE----NDLGFDIFVSNALMDMYAKCGSMQEAELVFSEMR 459
>At5g16860 putative protein
Length = 850
Score = 397 bits (1019), Expect = e-110
Identities = 247/740 (33%), Positives = 383/740 (51%), Gaps = 66/740 (8%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T+ +F AC + ++ G + H L +++F+ N LV MYS+C L AR VFD
Sbjct: 129 TFPFVFKACGEISSVRCGESAHALSLVTG--FISNVFVGNALVAMYSRCRSLSDARKVFD 186
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHY--RPNEFAFASLLSACEE---HD 189
+M ++VSW ++I YA+ G + +FS + + RP+ ++L C H
Sbjct: 187 EMSVWDVVSWNSIIESYAKLGKPKVALEMFSRMTNEFGCRPDNITLVNVLPPCASLGTHS 246
Query: 190 IKCGMQVHAVALKISLDASVYVANALITMYSKCS--------------------GGFHGG 229
+ G Q+H A+ + +++V N L+ MY+KC G
Sbjct: 247 L--GKQLHCFAVTSEMIQNMFVGNCLVDMYAKCGMMDEANTVFSNMSVKDVVSWNAMVAG 304
Query: 230 YDPTG--DDAWTMFKSMEFRNL----ISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFD 283
Y G +DA +F+ M+ + ++W++ I+G+ RGLG +A+ + M SGI +
Sbjct: 305 YSQIGRFEDAVRLFEKMQEEKIKMDVVTWSAAISGYAQRGLGYEALGVCRQMLSSGIKPN 364
Query: 284 RATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATK-------TGLISEVEVVTALVKSY 336
TL+SV S A L + +HC A K G E V+ L+ Y
Sbjct: 365 EVTLISVLSGCASVGA-------LMHGKEIHCYAIKYPIDLRKNGHGDENMVINQLIDMY 417
Query: 337 ANLGGQISDCYRLFLDTSGKQ-DIVSWTAIITVLADQ-DPEQAFLLFCQLHRENFV--PD 392
A ++ +F S K+ D+V+WT +I + D +A L ++ E+ P+
Sbjct: 418 AKCK-KVDTARAMFDSLSPKERDVVTWTVMIGGYSQHGDANKALELLSEMFEEDCQTRPN 476
Query: 393 WHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTV---LSNALIHAYARSGSLALSEQ 449
T S AL ACA + +H+ ++ Q++ V +SN LI YA+ GS++ +
Sbjct: 477 AFTISCALVACASLAALRIGKQIHAYALRN--QQNAVPLFVSNCLIDMYAKCGSISDARL 534
Query: 450 VFDEMCCHDLVSWNSMLKSYALHGKAKDALELF---KKLDVHPDSTTFVALLSACSHAGL 506
VFD M + V+W S++ Y +HG ++AL +F +++ D T + +L ACSH+G+
Sbjct: 535 VFDNMMAKNEVTWTSLMTGYGMHGYGEEALGIFDEMRRIGFKLDGVTLLVVLYACSHSGM 594
Query: 507 VEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLL 566
+++G+E FN M G+ P +HYAC+VDL GR G+++ A LI MPM+P V+W + L
Sbjct: 595 IDQGMEYFNRMKTVFGVSPGPEHYACLVDLLGRAGRLNAALRLIEEMPMEPPPVVWVAFL 654
Query: 567 GSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKK 626
CR HG+ L AAEK EL + Y +SN+Y++ G + + IR MR VKK
Sbjct: 655 SCCRIHGKVELGEYAAEKITELASNHDGSYTLLSNLYANAGRWKDVTRIRSLMRHKGVKK 714
Query: 627 QPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEH 686
+PG SWVE K F G HPH + I L + ++K++GYVPE AL+D + E
Sbjct: 715 RPGCSWVEGIKGTTTFFVGDKTHPHAKEIYQVLLDHMQRIKDIGYVPETGFALHDVDDEE 774
Query: 687 KEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVV 746
K+D LF HSEK+AL + I+ G I+I KN+R+C DCH S + +I++
Sbjct: 775 KDDLLFEHSEKLALAYGILTTPQ----GAAIRITKNLRVCGDCHTAFTYMSRIIDHDIIL 830
Query: 747 RDSNRFHHFKNATCSCNDYW 766
RDS+RFHHFKN +CSC YW
Sbjct: 831 RDSSRFHHFKNGSCSCKGYW 850
Score = 165 bits (418), Expect = 8e-41
Identities = 146/537 (27%), Positives = 234/537 (43%), Gaps = 63/537 (11%)
Query: 85 KNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRN--IV 142
K K + Q +H +L N LT+HL++ Y G L +A + + P + +
Sbjct: 37 KCKTISQVKLIHQKLLSFGILTLN---LTSHLISTYISVGCLSHAVSLLRRFPPSDAGVY 93
Query: 143 SWTALISGYAQCGLIRECFYLFSGLL--AHYRPNEFAFASLLSACEE-HDIKCGMQVHAV 199
W +LI Y G +C YLF GL+ + P+ + F + AC E ++CG HA+
Sbjct: 94 HWNSLIRSYGDNGCANKCLYLF-GLMHSLSWTPDNYTFPFVFKACGEISSVRCGESAHAL 152
Query: 200 ALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGF 259
+L ++V+V NAL+ MYS+C + DA +F M +++SWNS+I +
Sbjct: 153 SLVTGFISNVFVGNALVAMYSRCR---------SLSDARKVFDEMSVWDVVSWNSIIESY 203
Query: 260 QFRGLGDKAIRLFAHMYCS-GIGFDRATLLSVF---SSLNECSAFDELNI------PLRN 309
G A+ +F+ M G D TL++V +SL S +L+ ++N
Sbjct: 204 AKLGKPKVALEMFSRMTNEFGCRPDNITLVNVLPPCASLGTHSLGKQLHCFAVTSEMIQN 263
Query: 310 CFVLHCLA---TKTGLISEVEVV------------TALVKSYANLGGQISDCYRLF---L 351
FV +CL K G++ E V A+V Y+ + G+ D RLF
Sbjct: 264 MFVGNCLVDMYAKCGMMDEANTVFSNMSVKDVVSWNAMVAGYSQI-GRFEDAVRLFEKMQ 322
Query: 352 DTSGKQDIVSWTAIITVLADQDPEQAFLLFC-QLHRENFVPDWHTFSIALKACAYFVTEQ 410
+ K D+V+W+A I+ A + L C Q+ P+ T L CA
Sbjct: 323 EEKIKMDVVTWSAAISGYAQRGLGYEALGVCRQMLSSGIKPNEVTLISVLSGCASVGALM 382
Query: 411 QALAVHSQVI-------KRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCC--HDLVS 461
+H I K G + ++ N LI YA+ + + +FD + D+V+
Sbjct: 383 HGKEIHCYAIKYPIDLRKNGHGDENMVINQLIDMYAKCKKVDTARAMFDSLSPKERDVVT 442
Query: 462 WNSMLKSYALHGKAKDALELFKKL-----DVHPDSTTFVALLSACSHAGLVEEGVEIFNS 516
W M+ Y+ HG A ALEL ++ P++ T L AC+ + G +I
Sbjct: 443 WTVMIGGYSQHGDANKALELLSEMFEEDCQTRPNAFTISCALVACASLAALRIGKQIHAY 502
Query: 517 MSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHG 573
N L C++D+Y + G IS+A L+ M + V W+SL+ HG
Sbjct: 503 ALRNQQNAVPLFVSNCLIDMYAKCGSISDAR-LVFDNMMAKNEVTWTSLMTGYGMHG 558
Score = 110 bits (276), Expect = 2e-24
Identities = 77/273 (28%), Positives = 135/273 (49%), Gaps = 8/273 (2%)
Query: 359 IVSWTAIITVLADQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHS 417
+ W ++I D + LF +H ++ PD +TF KAC + + + H+
Sbjct: 92 VYHWNSLIRSYGDNGCANKCLYLFGLMHSLSWTPDNYTFPFVFKACGEISSVRCGESAHA 151
Query: 418 QVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKD 477
+ GF + + NAL+ Y+R SL+ + +VFDEM D+VSWNS+++SYA GK K
Sbjct: 152 LSLVTGFISNVFVGNALVAMYSRCRSLSDARKVFDEMSVWDVVSWNSIIESYAKLGKPKV 211
Query: 478 ALELFKKL----DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACM 533
ALE+F ++ PD+ T V +L C+ G G ++ + + ++ + C+
Sbjct: 212 ALEMFSRMTNEFGCRPDNITLVNVLPPCASLGTHSLGKQL-HCFAVTSEMIQNMFVGNCL 270
Query: 534 VDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPK-N 592
VD+Y + G + EA + M +K D V W++++ + G A EK +E K +
Sbjct: 271 VDMYAKCGMMDEANTVFSNMSVK-DVVSWNAMVAGYSQIGRFEDAVRLFEKMQEEKIKMD 329
Query: 593 SLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVK 625
+ + + Y+ G EA + ++M S +K
Sbjct: 330 VVTWSAAISGYAQRGLGYEALGVCRQMLSSGIK 362
>At3g57430 putative protein
Length = 803
Score = 396 bits (1018), Expect = e-110
Identities = 239/715 (33%), Positives = 379/715 (52%), Gaps = 48/715 (6%)
Query: 75 TYASLFHACAK---NKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARY 131
T S+ AC+ + L G +H Y L K + + F+ N LV MY K G L ++
Sbjct: 114 TLVSVVTACSNLPMPEGLMMGKQVHAYGLRKG---ELNSFIINTLVAMYGKLGKLASSKV 170
Query: 132 VFDQMPRRNIVSWTALISGYAQCGLIRECF-YLFSGLLAHYRPNEFAFASLLSACEEHD- 189
+ R++V+W ++S Q + E YL +L P+EF +S+L AC +
Sbjct: 171 LLGSFGGRDLVTWNTVLSSLCQNEQLLEALEYLREMVLEGVEPDEFTISSVLPACSHLEM 230
Query: 190 IKCGMQVHAVALKI-SLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRN 248
++ G ++HA ALK SLD + +V +AL+ MY C G +F M R
Sbjct: 231 LRTGKELHAYALKNGSLDENSFVGSALVDMYCNCKQVLSGR---------RVFDGMFDRK 281
Query: 249 LISWNSMIAGFQFRGLGDKAIRLFAHMYCS-GIGFDRATLLSVFSSLNECSAFDELNIPL 307
+ WN+MIAG+ +A+ LF M S G+ + T+ V + AF
Sbjct: 282 IGLWNAMIAGYSQNEHDKEALLLFIGMEESAGLLANSTTMAGVVPACVRSGAFSRKE--- 338
Query: 308 RNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIIT 367
+H K GL + V L+ Y+ LG +I R+F + D+V+W +IT
Sbjct: 339 ----AIHGFVVKRGLDRDRFVQNTLMDMYSRLG-KIDIAMRIFGKMEDR-DLVTWNTMIT 392
Query: 368 -VLADQDPEQAFLLFCQLH-----------RENFVPDWHTFSIALKACAYFVTEQQALAV 415
+ + E A LL ++ R + P+ T L +CA + +
Sbjct: 393 GYVFSEHHEDALLLLHKMQNLERKVSKGASRVSLKPNSITLMTILPSCAALSALAKGKEI 452
Query: 416 HSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKA 475
H+ IK D + +AL+ YA+ G L +S +VFD++ ++++WN ++ +Y +HG
Sbjct: 453 HAYAIKNNLATDVAVGSALVDMYAKCGCLQMSRKVFDQIPQKNVITWNVIIMAYGMHGNG 512
Query: 476 KDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYAC 532
++A++L + + V P+ TF+++ +ACSH+G+V+EG+ IF M ++G+ P DHYAC
Sbjct: 513 QEAIDLLRMMMVQGVKPNEVTFISVFAACSHSGMVDEGLRIFYVMKPDYGVEPSSDHYAC 572
Query: 533 MVDLYGRVGKISEAEDLIHTMPMKPDSV-IWSSLLGSCRKHGETRLARIAAEKFKELDPK 591
+VDL GR G+I EA L++ MP + WSSLLG+ R H + IAA+ +L+P
Sbjct: 573 VVDLLGRAGRIKEAYQLMNMMPRDFNKAGAWSSLLGASRIHNNLEIGEIAAQNLIQLEPN 632
Query: 592 NSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPH 651
+ YV ++NIYSS G + +A +R+ M++ V+K+PG SW+E G +VH+F +G HP
Sbjct: 633 VASHYVLLANIYSSAGLWDKATEVRRNMKEQGVRKEPGCSWIEHGDEVHKFVAGDSSHPQ 692
Query: 652 KEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLP 711
E + LE L ++++ GYVP+ + L++ E + KE L HSEK+A+ F I+N
Sbjct: 693 SEKLSGYLETLWERMRKEGYVPDTSCVLHNVEEDEKEILLCGHSEKLAIAFGILNTS--- 749
Query: 712 CGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
G +I++ KN+R+C DCH K S + +EI++RD RFH FKN TCSC DYW
Sbjct: 750 -PGTIIRVAKNLRVCNDCHLATKFISKIVDREIILRDVRRFHRFKNGTCSCGDYW 803
Score = 149 bits (377), Expect = 4e-36
Identities = 129/520 (24%), Positives = 233/520 (44%), Gaps = 44/520 (8%)
Query: 76 YASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQ 135
+ +L A A + ++ G +H ++ +K + + + N LVN+Y KCG VFD+
Sbjct: 13 FPALLKAVADLQDMELGKQIHAHV-YKFGYGVDSVTVANTLVNLYRKCGDFGAVYKVFDR 71
Query: 136 MPRRNIVSWTALISGYAQCGLIRECFYLFSGLL-AHYRPNEFAFASLLSACEE----HDI 190
+ RN VSW +LIS F +L + P+ F S+++AC +
Sbjct: 72 ISERNQVSWNSLISSLCSFEKWEMALEAFRCMLDENVEPSSFTLVSVVTACSNLPMPEGL 131
Query: 191 KCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLI 250
G QVHA L+ + + ++ N L+ MY K + + S R+L+
Sbjct: 132 MMGKQVHAYGLRKG-ELNSFIINTLVAMYGKLG---------KLASSKVLLGSFGGRDLV 181
Query: 251 SWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNC 310
+WN++++ +A+ M G+ D T+ SV L CS + LR
Sbjct: 182 TWNTVLSSLCQNEQLLEALEYLREMVLEGVEPDEFTISSV---LPACSHLE----MLRTG 234
Query: 311 FVLHCLATKTGLISEVEVV-TALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVL 369
LH A K G + E V +ALV Y N +S R D + I W A+I
Sbjct: 235 KELHAYALKNGSLDENSFVGSALVDMYCNCKQVLSG--RRVFDGMFDRKIGLWNAMIAGY 292
Query: 370 A-DQDPEQAFLLFCQLHRE-NFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKD 427
+ ++ ++A LLF + + + T + + AC + A+H V+KRG +D
Sbjct: 293 SQNEHDKEALLLFIGMEESAGLLANSTTMAGVVPACVRSGAFSRKEAIHGFVVKRGLDRD 352
Query: 428 TVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL-- 485
+ N L+ Y+R G + ++ ++F +M DLV+WN+M+ Y +DAL L K+
Sbjct: 353 RFVQNTLMDMYSRLGKIDIAMRIFGKMEDRDLVTWNTMITGYVFSEHHEDALLLLHKMQN 412
Query: 486 ------------DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACM 533
+ P+S T + +L +C+ + +G EI ++ + + + + + +
Sbjct: 413 LERKVSKGASRVSLKPNSITLMTILPSCAALSALAKGKEI-HAYAIKNNLATDVAVGSAL 471
Query: 534 VDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHG 573
VD+Y + G + + + +P K + + W+ ++ + HG
Sbjct: 472 VDMYAKCGCLQMSRKVFDQIPQK-NVITWNVIIMAYGMHG 510
Score = 74.3 bits (181), Expect = 2e-13
Identities = 53/184 (28%), Positives = 99/184 (53%), Gaps = 8/184 (4%)
Query: 391 PDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTV-LSNALIHAYARSGSLALSEQ 449
PD + F LKA A + +H+ V K G+ D+V ++N L++ Y + G +
Sbjct: 8 PDNYAFPALLKAVADLQDMELGKQIHAHVYKFGYGVDSVTVANTLVNLYRKCGDFGAVYK 67
Query: 450 VFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGL 506
VFD + + VSWNS++ S K + ALE F+ + +V P S T V++++ACS+ +
Sbjct: 68 VFDRISERNQVSWNSLISSLCSFEKWEMALEAFRCMLDENVEPSSFTLVSVVTACSNLPM 127
Query: 507 VEEGVEIFNSMSDNHGIVPQLDHYA--CMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSS 564
EG+ + + +L+ + +V +YG++GK++ ++ L+ + + D V W++
Sbjct: 128 -PEGLMMGKQVHAYGLRKGELNSFIINTLVAMYGKLGKLASSKVLLGSFGGR-DLVTWNT 185
Query: 565 LLGS 568
+L S
Sbjct: 186 VLSS 189
>At3g12770 unknown protein
Length = 719
Score = 394 bits (1013), Expect = e-110
Identities = 239/720 (33%), Positives = 388/720 (53%), Gaps = 42/720 (5%)
Query: 54 ALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLT 113
A LLYT+ HS YASL + L+Q +H +L +Q FL
Sbjct: 8 ASPLLYTNSG---IHSD---SFYASLIDSATHKAQLKQ---IHARLLVLG--LQFSGFLI 56
Query: 114 NHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGL-LAHYR 172
L++ S G + +AR VFD +PR I W A+I GY++ ++ ++S + LA
Sbjct: 57 TKLIHASSSFGDITFARQVFDDLPRPQIFPWNAIIRGYSRNNHFQDALLMYSNMQLARVS 116
Query: 173 PNEFAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYD 231
P+ F F LL AC ++ G VHA ++ DA V+V N LI +Y+KC
Sbjct: 117 PDSFTFPHLLKACSGLSHLQMGRFVHAQVFRLGFDADVFVQNGLIALYAKCR-------- 168
Query: 232 PTGDDAWTMFKSMEF--RNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLS 289
A T+F+ + R ++SW ++++ + G +A+ +F+ M + D L+S
Sbjct: 169 -RLGSARTVFEGLPLPERTIVSWTAIVSAYAQNGEPMEALEIFSQMRKMDVKPDWVALVS 227
Query: 290 VFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRL 349
V LN + +L + +H K GL E +++ +L YA G Q++ L
Sbjct: 228 V---LNAFTCLQDL----KQGRSIHASVVKMGLEIEPDLLISLNTMYAKCG-QVATAKIL 279
Query: 350 FLDTSGKQDIVSWTAIITVLADQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVT 408
F D +++ W A+I+ A +A +F ++ ++ PD + + A+ ACA +
Sbjct: 280 F-DKMKSPNLILWNAMISGYAKNGYAREAIDMFHEMINKDVRPDTISITSAISACAQVGS 338
Query: 409 EQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKS 468
+QA +++ V + ++ D +S+ALI +A+ GS+ + VFD D+V W++M+
Sbjct: 339 LEQARSMYEYVGRSDYRDDVFISSALIDMFAKCGSVEGARLVFDRTLDRDVVVWSAMIVG 398
Query: 469 YALHGKAKDALELFKKLD---VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVP 525
Y LHG+A++A+ L++ ++ VHP+ TF+ LL AC+H+G+V EG FN M+D H I P
Sbjct: 399 YGLHGRAREAISLYRAMERGGVHPNDVTFLGLLMACNHSGMVREGWWFFNRMAD-HKINP 457
Query: 526 QLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKF 585
Q HYAC++DL GR G + +A ++I MP++P +W +LL +C+KH L AA++
Sbjct: 458 QQQHYACVIDLLGRAGHLDQAYEVIKCMPVQPGVTVWGALLSACKKHRHVELGEYAAQQL 517
Query: 586 KELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSG 645
+DP N+ YVQ+SN+Y++ + +R M++ + K G SWVEV ++ F G
Sbjct: 518 FSIDPSNTGHYVQLSNLYAAARLWDRVAEVRVRMKEKGLNKDVGCSWVEVRGRLEAFRVG 577
Query: 646 GHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIM 705
HP E I+ ++E + +LKE G+V +L+D E E+ L HSE++A+ + ++
Sbjct: 578 DKSHPRYEEIERQVEWIESRLKEGGFVANKDASLHDLNDEEAEETLCSHSERIAIAYGLI 637
Query: 706 NEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDY 765
+ G ++I KN+R C +CH KL S L +EIVVRD+NRFHHFK+ CSC DY
Sbjct: 638 STPQ----GTPLRITKNLRACVNCHAATKLISKLVDREIVVRDTNRFHHFKDGVCSCGDY 693
>At4g14850 unknown protein
Length = 684
Score = 394 bits (1012), Expect = e-109
Identities = 237/664 (35%), Positives = 358/664 (53%), Gaps = 30/664 (4%)
Query: 111 FLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH 170
FL N+L+NMYSK H E AR V P RN+VSWT+LISG AQ G F +
Sbjct: 43 FLANYLINMYSKLDHPESARLVLRLTPARNVVSWTSLISGLAQNGHFSTALVEFFEMRRE 102
Query: 171 -YRPNEFAFASLLSACEEHDIKC-GMQVHAVALKISLDASVYVANALITMYSKCSGGFHG 228
PN+F F A + G Q+HA+A+K V+V + MY K
Sbjct: 103 GVVPNDFTFPCAFKAVASLRLPVTGKQIHALAVKCGRILDVFVGCSAFDMYCKTR----- 157
Query: 229 GYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLL 288
DDA +F + RNL +WN+ I+ G +AI F G +
Sbjct: 158 ----LRDDARKLFDEIPERNLETWNAFISNSVTDGRPREAIEAFIEFRRID-GHPNSITF 212
Query: 289 SVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYR 348
F LN CS + LN+ ++ LH L ++G ++V V L+ Y QI
Sbjct: 213 CAF--LNACSDWLHLNLGMQ----LHGLVLRSGFDTDVSVCNGLIDFYGKCK-QIRSSEI 265
Query: 349 LFLDTSGKQDIVSWTAIITV-LADQDPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFV 407
+F + G ++ VSW +++ + + + E+A +L+ + ++ S L ACA
Sbjct: 266 IFTEM-GTKNAVSWCSLVAAYVQNHEDEKASVLYLRSRKDIVETSDFMISSVLSACAGMA 324
Query: 408 TEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLK 467
+ ++H+ +K ++ + +AL+ Y + G + SEQ FDEM +LV+ NS++
Sbjct: 325 GLELGRSIHAHAVKACVERTIFVGSALVDMYGKCGCIEDSEQAFDEMPEKNLVTRNSLIG 384
Query: 468 SYALHGKAKDALELFKKLDVH-----PDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHG 522
YA G+ AL LF+++ P+ TFV+LLSACS AG VE G++IF+SM +G
Sbjct: 385 GYAHQGQVDMALALFEEMAPRGCGPTPNYMTFVSLLSACSRAGAVENGMKIFDSMRSTYG 444
Query: 523 IVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAA 582
I P +HY+C+VD+ GR G + A + I MP++P +W +L +CR HG+ +L +AA
Sbjct: 445 IEPGAEHYSCIVDMLGRAGMVERAYEFIKKMPIQPTISVWGALQNACRMHGKPQLGLLAA 504
Query: 583 EKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEF 642
E +LDPK+S +V +SN +++ G + EA +R+E++ +KK G SW+ V QVH F
Sbjct: 505 ENLFKLDPKDSGNHVLLSNTFAAAGRWAEANTVREELKGVGIKKGAGYSWITVKNQVHAF 564
Query: 643 TSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVF 702
+ H + IQ+ L L +++ GY P++ L+LYD E E K ++ HHSEK+AL F
Sbjct: 565 QAKDRSHILNKEIQTTLAKLRNEMEAAGYKPDLKLSLYDLEEEEKAAEVSHHSEKLALAF 624
Query: 703 AIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSC 762
++ +LP I+I KN+RIC DCH+F K S ++EI+VRD+NRFH FK+ CSC
Sbjct: 625 GLL---SLPL-SVPIRITKNLRICGDCHSFFKFVSGSVKREIIVRDNNRFHRFKDGICSC 680
Query: 763 NDYW 766
DYW
Sbjct: 681 KDYW 684
Score = 98.6 bits (244), Expect = 1e-20
Identities = 64/226 (28%), Positives = 110/226 (48%), Gaps = 14/226 (6%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T+ + +AC+ L GM LH +L D+ + N L++ Y KC + + +F
Sbjct: 211 TFCAFLNACSDWLHLNLGMQLHGLVLRSG--FDTDVSVCNGLIDFYGKCKQIRSSEIIFT 268
Query: 135 QMPRRNIVSWTALISGYAQCGLIRECFYLF-SGLLAHYRPNEFAFASLLSACE-EHDIKC 192
+M +N VSW +L++ Y Q + L+ ++F +S+LSAC ++
Sbjct: 269 EMGTKNAVSWCSLVAAYVQNHEDEKASVLYLRSRKDIVETSDFMISSVLSACAGMAGLEL 328
Query: 193 GMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISW 252
G +HA A+K ++ +++V +AL+ MY KC +D+ F M +NL++
Sbjct: 329 GRSIHAHAVKACVERTIFVGSALVDMYGKCG---------CIEDSEQAFDEMPEKNLVTR 379
Query: 253 NSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECS 298
NS+I G+ +G D A+ LF M G G ++ S L+ CS
Sbjct: 380 NSLIGGYAHQGQVDMALALFEEMAPRGCG-PTPNYMTFVSLLSACS 424
Score = 95.5 bits (236), Expect = 1e-19
Identities = 109/443 (24%), Positives = 184/443 (40%), Gaps = 35/443 (7%)
Query: 75 TYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFD 134
T+ F A A + G +H + + D+F+ +MY K + AR +FD
Sbjct: 110 TFPCAFKAVASLRLPVTGKQIHALAVKCGRIL--DVFVGCSAFDMYCKTRLRDDARKLFD 167
Query: 135 QMPRRNIVSWTALISGYAQCGLIR---ECFYLFSGLLAHYRPNEFAFASLLSACEEH-DI 190
++P RN+ +W A IS G R E F F + H PN F + L+AC + +
Sbjct: 168 EIPERNLETWNAFISNSVTDGRPREAIEAFIEFRRIDGH--PNSITFCAFLNACSDWLHL 225
Query: 191 KCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLI 250
GMQ+H + L+ D V V N LI Y KC + +F M +N +
Sbjct: 226 NLGMQLHGLVLRSGFDTDVSVCNGLIDFYGKCK---------QIRSSEIIFTEMGTKNAV 276
Query: 251 SWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNC 310
SW S++A + +KA L+ S + + S L+ C+ L +
Sbjct: 277 SWCSLVAAYVQNHEDEKASVLYLR---SRKDIVETSDFMISSVLSACAGMAGLELGRS-- 331
Query: 311 FVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLA 370
+H A K + + V +ALV Y G I D + F D ++++V+ ++I A
Sbjct: 332 --IHAHAVKACVERTIFVGSALVDMYGKC-GCIEDSEQAF-DEMPEKNLVTRNSLIGGYA 387
Query: 371 DQ-DPEQAFLLFCQLHRENF--VPDWHTFSIALKACAYFVTEQQALAVHSQVIKR-GFQK 426
Q + A LF ++ P++ TF L AC+ + + + + G +
Sbjct: 388 HQGQVDMALALFEEMAPRGCGPTPNYMTFVSLLSACSRAGAVENGMKIFDSMRSTYGIEP 447
Query: 427 DTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVS-WNSMLKSYALHGKAKDAL---ELF 482
+ ++ R+G + + + +M +S W ++ + +HGK + L E
Sbjct: 448 GAEHYSCIVDMLGRAGMVERAYEFIKKMPIQPTISVWGALQNACRMHGKPQLGLLAAENL 507
Query: 483 KKLDVHPDSTTFVALLSACSHAG 505
KLD DS V L + + AG
Sbjct: 508 FKLD-PKDSGNHVLLSNTFAAAG 529
Score = 74.3 bits (181), Expect = 2e-13
Identities = 48/157 (30%), Positives = 82/157 (51%), Gaps = 8/157 (5%)
Query: 44 ALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKD 103
A +Q + +E S+LY + + +S + +S+ ACA L+ G ++H + +
Sbjct: 283 AAYVQNHEDEKASVLYLRSRKDIVETSDFM--ISSVLSACAGMAGLELGRSIHAHAVKA- 339
Query: 104 PTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYL 163
++ +F+ + LV+MY KCG +E + FD+MP +N+V+ +LI GYA G + L
Sbjct: 340 -CVERTIFVGSALVDMYGKCGCIEDSEQAFDEMPEKNLVTRNSLIGGYAHQGQVDMALAL 398
Query: 164 FSGLLAH---YRPNEFAFASLLSACEEHD-IKCGMQV 196
F + PN F SLLSAC ++ GM++
Sbjct: 399 FEEMAPRGCGPTPNYMTFVSLLSACSRAGAVENGMKI 435
>At5g39680 unknown protein
Length = 710
Score = 385 bits (989), Expect = e-107
Identities = 220/695 (31%), Positives = 365/695 (51%), Gaps = 28/695 (4%)
Query: 79 LFHACAKNKCLQQGMALHNYILHKDPTIQ-NDLFLTNHLVNMYSKCGHLEYARYVFDQMP 137
L CA + L+ G ++H +++ + + + D + N L+N+Y KC AR +FD MP
Sbjct: 37 LLKVCANSSYLRIGESIHAHLIVTNQSSRAEDAYQINSLINLYVKCRETVRARKLFDLMP 96
Query: 138 RRNIVSWTALISGYAQCGLIRECFYLFSGLL--AHYRPNEFAFASLLSACEEHD-IKCGM 194
RN+VSW A++ GY G E LF + RPNEF + +C I+ G
Sbjct: 97 ERNVVSWCAMMKGYQNSGFDFEVLKLFKSMFFSGESRPNEFVATVVFKSCSNSGRIEEGK 156
Query: 195 QVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNS 254
Q H LK L + +V N L+ MYS CSG +A + + + +L ++S
Sbjct: 157 QFHGCFLKYGLISHEFVRNTLVYMYSLCSGN---------GEAIRVLDDLPYCDLSVFSS 207
Query: 255 MIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFVLH 314
++G+ G + + + ++ T LS SL S +LN+ L+ +H
Sbjct: 208 ALSGYLECGAFKEGLDVLRKTANEDFVWNNLTYLS---SLRLFSNLRDLNLALQ----VH 260
Query: 315 CLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLADQDP 374
+ G +EVE AL+ Y G ++ R+F DT + ++ T + D+
Sbjct: 261 SRMVRFGFNAEVEACGALINMYGKCG-KVLYAQRVFDDTHAQNIFLNTTIMDAYFQDKSF 319
Query: 375 EQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNAL 434
E+A LF ++ + P+ +TF+I L + A +Q +H V+K G++ ++ NAL
Sbjct: 320 EEALNLFSKMDTKEVPPNEYTFAILLNSIAELSLLKQGDLLHGLVLKSGYRNHVMVGNAL 379
Query: 435 IHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLDVH---PDS 491
++ YA+SGS+ + + F M D+V+WN+M+ + HG ++ALE F ++ P+
Sbjct: 380 VNMYAKSGSIEDARKAFSGMTFRDIVTWNTMISGCSHHGLGREALEAFDRMIFTGEIPNR 439
Query: 492 TTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIH 551
TF+ +L ACSH G VE+G+ FN + + P + HY C+V L + G +AED +
Sbjct: 440 ITFIGVLQACSHIGFVEQGLHYFNQLMKKFDVQPDIQHYTCIVGLLSKAGMFKDAEDFMR 499
Query: 552 TMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIE 611
T P++ D V W +LL +C RL + AE E P +S YV +SNI++ +
Sbjct: 500 TAPIEWDVVAWRTLLNACYVRRNYRLGKKVAEYAIEKYPNDSGVYVLLSNIHAKSREWEG 559
Query: 612 AGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGY 671
+R M + VKK+PG+SW+ + Q H F + + HP I ++++ ++ ++K +GY
Sbjct: 560 VAKVRSLMNNRGVKKEPGVSWIGIRNQTHVFLAEDNQHPEITLIYAKVKEVMSKIKPLGY 619
Query: 672 VPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHN 731
P++ A +D + E +ED L +HSEK+A+ + ++ + + + KN+RIC DCH+
Sbjct: 620 SPDVAGAFHDVDEEQREDNLSYHSEKLAVAYGLIKTPE----KSPLYVTKNVRICDDCHS 675
Query: 732 FMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
+KL S + ++ IV+RDSNRFHHF + CSC DYW
Sbjct: 676 AIKLISKISKRYIVIRDSNRFHHFLDGQCSCCDYW 710
Score = 70.9 bits (172), Expect = 3e-12
Identities = 48/137 (35%), Positives = 70/137 (51%), Gaps = 6/137 (4%)
Query: 50 NLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQND 109
+ EEAL+L D + + T+A L ++ A+ L+QG LH +L +N
Sbjct: 318 SFEEALNLFSKMDTKEVPPNEY---TFAILLNSIAELSLLKQGDLLHGLVLKSG--YRNH 372
Query: 110 LFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLL- 168
+ + N LVNMY+K G +E AR F M R+IV+W +ISG + GL RE F ++
Sbjct: 373 VMVGNALVNMYAKSGSIEDARKAFSGMTFRDIVTWNTMISGCSHHGLGREALEAFDRMIF 432
Query: 169 AHYRPNEFAFASLLSAC 185
PN F +L AC
Sbjct: 433 TGEIPNRITFIGVLQAC 449
>At5g13270 putative protein
Length = 752
Score = 383 bits (983), Expect = e-106
Identities = 243/732 (33%), Positives = 376/732 (51%), Gaps = 45/732 (6%)
Query: 44 ALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILHKD 103
+LS L EA L DK ++ SS S Y LF AC + + L G LH+ +
Sbjct: 57 SLSKHRKLNEAFEFLQEMDKAGVSVSSYS---YQCLFEACRELRSLSHGRLLHDRMRMG- 112
Query: 104 PTIQN-DLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFY 162
I+N + L N ++ MY +C LE A +FD+M N VS T +IS YA+ G++ +
Sbjct: 113 --IENPSVLLQNCVLQMYCECRSLEDADKLFDEMSELNAVSRTTMISAYAEQGILDKAVG 170
Query: 163 LFSGLLAHY-RPNEFAFASLL-SACEEHDIKCGMQVHAVALKISLDASVYVANALITMYS 220
LFSG+LA +P + +LL S + G Q+HA ++ L ++ + ++ MY
Sbjct: 171 LFSGMLASGDKPPSSMYTTLLKSLVNPRALDFGRQIHAHVIRAGLCSNTSIETGIVNMYV 230
Query: 221 KCSGGFHGGYDPTGDDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGI 280
KC G+ G A +F M + ++ ++ G+ G A++LF + G+
Sbjct: 231 KC--GWLVG-------AKRVFDQMAVKKPVACTGLMVGYTQAGRARDALKLFVDLVTEGV 281
Query: 281 GFDRATLLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLG 340
+D V L C++ +ELN+ + +H K GL SEV V T LV Y
Sbjct: 282 EWDSFVFSVV---LKACASLEELNLGKQ----IHACVAKLGLESEVSVGTPLVDFYIKCS 334
Query: 341 GQISDCYRLFLDTSGKQDIVSWTAIITVLADQDP-EQAFLLFCQLHRENF-VPDWHTFSI 398
S C R F + D VSW+AII+ E+A F L +N + + T++
Sbjct: 335 SFESAC-RAFQEIREPND-VSWSAIISGYCQMSQFEEAVKTFKSLRSKNASILNSFTYTS 392
Query: 399 ALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHD 458
+AC+ VH+ IKR +ALI Y++ G L + +VF+ M D
Sbjct: 393 IFQACSVLADCNIGGQVHADAIKRSLIGSQYGESALITMYSKCGCLDDANEVFESMDNPD 452
Query: 459 LVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFN 515
+V+W + + +A +G A +AL LF+K+ + P+S TF+A+L+ACSHAGLVE+G +
Sbjct: 453 IVAWTAFISGHAYYGNASEALRLFEKMVSCGMKPNSVTFIAVLTACSHAGLVEQGKHCLD 512
Query: 516 SMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGET 575
+M + + P +DHY CM+D+Y R G + EA + MP +PD++ W L C H
Sbjct: 513 TMLRKYNVAPTIDHYDCMIDIYARSGLLDEALKFMKNMPFEPDAMSWKCFLSGCWTHKNL 572
Query: 576 RLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEV 635
L IA E+ ++LDP+++ GYV N+Y+ G + EA + K M + +KK+ SW++
Sbjct: 573 ELGEIAGEELRQLDPEDTAGYVLPFNLYTWAGKWEEAAEMMKLMNERMLKKELSCSWIQE 632
Query: 636 GKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHS 695
++H F G HHP + I +L+ G ++ ++ + + +QL HS
Sbjct: 633 KGKIHRFIVGDKHHPQTQEIYEKLKEFDGFMEG---------DMFQCNMTERREQLLDHS 683
Query: 696 EKMALVFAIMN-EGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHH 754
E++A+ F +++ GN P IK+ KN+R C DCH F K S + EIV+RDS RFHH
Sbjct: 684 ERLAIAFGLISVHGNAPA---PIKVFKNLRACPDCHEFAKHVSLVTGHEIVIRDSRRFHH 740
Query: 755 FKNATCSCNDYW 766
FK CSCNDYW
Sbjct: 741 FKEGKCSCNDYW 752
Score = 42.4 bits (98), Expect = 0.001
Identities = 48/239 (20%), Positives = 104/239 (43%), Gaps = 10/239 (4%)
Query: 354 SGKQDIVSWTAIITVLADQDPEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQAL 413
S KQ V ++++ + +AF ++ + +++ +AC +
Sbjct: 44 SHKQGQVENLHLVSLSKHRKLNEAFEFLQEMDKAGVSVSSYSYQCLFEACRELRSLSHGR 103
Query: 414 AVHSQVIKRGFQKDTVL-SNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALH 472
+H ++ + G + +VL N ++ Y SL ++++FDEM + VS +M+ +YA
Sbjct: 104 LLHDRM-RMGIENPSVLLQNCVLQMYCECRSLEDADKLFDEMSELNAVSRTTMISAYAEQ 162
Query: 473 GKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDH 529
G A+ LF + P S+ + LL + + ++ G +I ++ G+
Sbjct: 163 GILDKAVGLFSGMLASGDKPPSSMYTTLLKSLVNPRALDFGRQI-HAHVIRAGLCSNTSI 221
Query: 530 YACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKEL 588
+V++Y + G + A+ + M +K ++G + + AR A + F +L
Sbjct: 222 ETGIVNMYVKCGWLVGAKRVFDQMAVKKPVACTGLMVG----YTQAGRARDALKLFVDL 276
>At1g68930 hypothetical protein
Length = 743
Score = 380 bits (975), Expect = e-105
Identities = 221/711 (31%), Positives = 362/711 (50%), Gaps = 54/711 (7%)
Query: 95 LHNYILHKDPTI---------QNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWT 145
+H Y L K T Q +LF N+L+ YSK G + F+++P R+ V+W
Sbjct: 48 VHAYALMKSSTYARRVFDRIPQPNLFSWNNLLLAYSKAGLISEMESTFEKLPDRDGVTWN 107
Query: 146 ALISGYAQCGLIRECFYLFSGLLAHYRPNEFAFASL----LSACEEHDIKCGMQVHAVAL 201
LI GY+ GL+ ++ ++ + N + LS+ H + G Q+H +
Sbjct: 108 VLIEGYSLSGLVGAAVKAYNTMMRDFSANLTRVTLMTMLKLSSSNGH-VSLGKQIHGQVI 166
Query: 202 KISLDASVYVANALITMYSKCS--------------------GGFHGGYDPTG--DDAWT 239
K+ ++ + V + L+ MY+ GG G +DA
Sbjct: 167 KLGFESYLLVGSPLLYMYANVGCISDAKKVFYGLDDRNTVMYNSLMGGLLACGMIEDALQ 226
Query: 240 MFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSA 299
+F+ ME ++ +SW +MI G GL +AI F M G+ D+ SV + A
Sbjct: 227 LFRGME-KDSVSWAAMIKGLAQNGLAKEAIECFREMKVQGLKMDQYPFGSVLPACGGLGA 285
Query: 300 FDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDI 359
+E +H +T + V +AL+ Y + + D ++++
Sbjct: 286 INEGK-------QIHACIIRTNFQDHIYVGSALIDMYCKC--KCLHYAKTVFDRMKQKNV 336
Query: 360 VSWTAIITVLADQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQ 418
VSWTA++ E+A +F + R PD +T A+ ACA + ++ H +
Sbjct: 337 VSWTAMVVGYGQTGRAEEAVKIFLDMQRSGIDPDHYTLGQAISACANVSSLEEGSQFHGK 396
Query: 419 VIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDA 478
I G +SN+L+ Y + G + S ++F+EM D VSW +M+ +YA G+A +
Sbjct: 397 AITSGLIHYVTVSNSLVTLYGKCGDIDDSTRLFNEMNVRDAVSWTAMVSAYAQFGRAVET 456
Query: 479 LELFKKLDVH---PDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVD 535
++LF K+ H PD T ++SACS AGLVE+G F M+ +GIVP + HY+CM+D
Sbjct: 457 IQLFDKMVQHGLKPDGVTLTGVISACSRAGLVEKGQRYFKLMTSEYGIVPSIGHYSCMID 516
Query: 536 LYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLG 595
L+ R G++ EA I+ MP PD++ W++LL +CR G + + AAE ELDP + G
Sbjct: 517 LFSRSGRLEEAMRFINGMPFPPDAIGWTTLLSACRNKGNLEIGKWAAESLIELDPHHPAG 576
Query: 596 YVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAI 655
Y +S+IY+S+G + +R+ MR+ VKK+PG SW++ ++H F++ P+ + I
Sbjct: 577 YTLLSSIYASKGKWDSVAQLRRGMREKNVKKEPGQSWIKWKGKLHSFSADDESSPYLDQI 636
Query: 656 QSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGN 715
++LE L ++ + GY P+ + +D E K L +HSE++A+ F ++ G
Sbjct: 637 YAKLEELNNKIIDNGYKPDTSFVHHDVEEAVKVKMLNYHSERLAIAFGLI----FVPSGQ 692
Query: 716 VIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
I++ KN+R+C DCHN K S++ +EI+VRD+ RFH FK+ TCSC D+W
Sbjct: 693 PIRVGKNLRVCVDCHNATKHISSVTGREILVRDAVRFHRFKDGTCSCGDFW 743
Score = 65.9 bits (159), Expect = 8e-11
Identities = 73/294 (24%), Positives = 130/294 (43%), Gaps = 55/294 (18%)
Query: 396 FSIALKACAYFVTEQQALAV---HSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFD 452
+S+ +K C Q+ V H +I+ +T L N ++HAYA S + +VFD
Sbjct: 6 YSVQIKQCIGLGARNQSRYVKMIHGNIIRALPYPETFLYNNIVHAYALMKSSTYARRVFD 65
Query: 453 EMCCHDLVSWNSMLKSYALHGKAKDALELFKKLDVHPDSTTFVALLSACSHAGLVEEGVE 512
+ +L SWN++L +Y+ G + F+KL D T+ L+ S +GLV V+
Sbjct: 66 RIPQPNLFSWNNLLLAYSKAGLISEMESTFEKLP-DRDGVTWNVLIEGYSLSGLVGAAVK 124
Query: 513 IFNSM--------------------SDN---------HGIVPQL--DHY----ACMVDLY 537
+N+M S N HG V +L + Y + ++ +Y
Sbjct: 125 AYNTMMRDFSANLTRVTLMTMLKLSSSNGHVSLGKQIHGQVIKLGFESYLLVGSPLLYMY 184
Query: 538 GRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYV 597
VG IS+A+ + + + + ++V+++SL+G G + A + F+ ++ K+S+ +
Sbjct: 185 ANVGCISDAKKVFYGLDDR-NTVMYNSLMGGLLACG---MIEDALQLFRGME-KDSVSWA 239
Query: 598 QMSNIYSSEGSFIEAGLIRKEMRDSRVKKQP-----------GLSWVEVGKQVH 640
M + G EA +EM+ +K GL + GKQ+H
Sbjct: 240 AMIKGLAQNGLAKEAIECFREMKVQGLKMDQYPFGSVLPACGGLGAINEGKQIH 293
>At5g13230 putative protein
Length = 822
Score = 379 bits (972), Expect = e-105
Identities = 229/671 (34%), Positives = 357/671 (53%), Gaps = 39/671 (5%)
Query: 108 NDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGL 167
++ F+ L+N YS CG ++ AR VF+ + ++IV W ++S Y + G + L S +
Sbjct: 179 SNAFVGAALINAYSVCGSVDSARTVFEGILCKDIVVWAGIVSCYVENGYFEDSLKLLSCM 238
Query: 168 -LAHYRPNEFAFASLLSAC---EEHDIKCGMQVHAVALKISLDASVYVANALITMYSKCS 223
+A + PN + F + L A D G VH LK V L+ +Y++
Sbjct: 239 RMAGFMPNNYTFDTALKASIGLGAFDFAKG--VHGQILKTCYVLDPRVGVGLLQLYTQL- 295
Query: 224 GGFHGGYDPTGD--DAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIG 281
GD DA+ +F M +++ W+ MIA F G ++A+ LF M + +
Sbjct: 296 ----------GDMSDAFKVFNEMPKNDVVPWSFMIARFCQNGFCNEAVDLFIRMREAFVV 345
Query: 282 FDRATLLSVFS--SLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANL 339
+ TL S+ + ++ +CS E LH L K G ++ V AL+ YA
Sbjct: 346 PNEFTLSSILNGCAIGKCSGLGEQ---------LHGLVVKVGFDLDIYVSNALIDVYAKC 396
Query: 340 GGQISDCYRLFLDTSGKQDIVSWTAIITVLADQ-DPEQAFLLFCQLHRENFVPDWHTFSI 398
++ +LF + S K + VSW +I + + +AF +F + R TFS
Sbjct: 397 E-KMDTAVKLFAELSSKNE-VSWNTVIVGYENLGEGGKAFSMFREALRNQVSVTEVTFSS 454
Query: 399 ALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHD 458
AL ACA + + VH IK K +SN+LI YA+ G + ++ VF+EM D
Sbjct: 455 ALGACASLASMDLGVQVHGLAIKTNNAKKVAVSNSLIDMYAKCGDIKFAQSVFNEMETID 514
Query: 459 LVSWNSMLKSYALHGKAKDAL---ELFKKLDVHPDSTTFVALLSACSHAGLVEEGVEIFN 515
+ SWN+++ Y+ HG + AL ++ K D P+ TF+ +LS CS+AGL+++G E F
Sbjct: 515 VASWNALISGYSTHGLGRQALRILDIMKDRDCKPNGLTFLGVLSGCSNAGLIDQGQECFE 574
Query: 516 SMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGET 575
SM +HGI P L+HY CMV L GR G++ +A LI +P +P +IW ++L +
Sbjct: 575 SMIRDHGIEPCLEHYTCMVRLLGRSGQLDKAMKLIEGIPYEPSVMIWRAMLSASMNQNNE 634
Query: 576 RLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEV 635
AR +AE+ +++PK+ YV +SN+Y+ + IRK M++ VKK+PGLSW+E
Sbjct: 635 EFARRSAEEILKINPKDEATYVLVSNMYAGAKQWANVASIRKSMKEMGVKKEPGLSWIEH 694
Query: 636 GKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHS 695
VH F+ G HP + I LE L + GYVP+ L D + E K+ +L+ HS
Sbjct: 695 QGDVHYFSVGLSDHPDMKLINGMLEWLNMKATRAGYVPDRNAVLLDMDDEEKDKRLWVHS 754
Query: 696 EKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHF 755
E++AL + ++ +P N I IMKN+RIC+DCH+ MK+ S++ Q+++V+RD NRFHHF
Sbjct: 755 ERLALAYGLV---RMPSSRNRILIMKNLRICSDCHSAMKVISSIVQRDLVIRDMNRFHHF 811
Query: 756 KNATCSCNDYW 766
CSC D+W
Sbjct: 812 HAGVCSCGDHW 822
Score = 157 bits (396), Expect = 3e-38
Identities = 141/550 (25%), Positives = 245/550 (43%), Gaps = 45/550 (8%)
Query: 71 LSLQTYASLFHACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYAR 130
L Y ++ C + A+H IL K + DLF TN L+N Y K G + A
Sbjct: 47 LDSHAYGAMLRRCIQKNDPISAKAIHCDILKKGSCL--DLFATNILLNAYVKAGFDKDAL 104
Query: 131 YVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEEHD 189
+FD+MP RN VS+ L GYA ++ L+S L + N F S L D
Sbjct: 105 NLFDEMPERNNVSFVTLAQGYA----CQDPIGLYSRLHREGHELNPHVFTSFLKLFVSLD 160
Query: 190 -IKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRN 248
+ +H+ +K+ D++ +V ALI YS C + D A T+F+ + ++
Sbjct: 161 KAEICPWLHSPIVKLGYDSNAFVGAALINAYSVCG---------SVDSARTVFEGILCKD 211
Query: 249 LISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLR 308
++ W +++ + G + +++L + M +G + T + + AFD
Sbjct: 212 IVVWAGIVSCYVENGYFEDSLKLLSCMRMAGFMPNNYTFDTALKASIGLGAFDFAK---- 267
Query: 309 NCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITV 368
+H KT + + V L++ Y L G +SD +++F + K D+V W+ +I
Sbjct: 268 ---GVHGQILKTCYVLDPRVGVGLLQLYTQL-GDMSDAFKVF-NEMPKNDVVPWSFMIAR 322
Query: 369 LADQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKD 427
+A LF ++ VP+ T S L CA +H V+K GF D
Sbjct: 323 FCQNGFCNEAVDLFIRMREAFVVPNEFTLSSILNGCAIGKCSGLGEQLHGLVVKVGFDLD 382
Query: 428 TVLSNALIHAYARSGSLALSEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL-- 485
+SNALI YA+ + + ++F E+ + VSWN+++ Y G+ A +F++
Sbjct: 383 IYVSNALIDVYAKCEKMDTAVKLFAELSSKNEVSWNTVIVGYENLGEGGKAFSMFREALR 442
Query: 486 -DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYA------CMVDLYG 538
V TF + L AC+ ++ GV++ HG+ + ++ ++D+Y
Sbjct: 443 NQVSVTEVTFSSALGACASLASMDLGVQV-------HGLAIKTNNAKKVAVSNSLIDMYA 495
Query: 539 RVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPK-NSLGYV 597
+ G I A+ + + M D W++L+ HG R A + K+ D K N L ++
Sbjct: 496 KCGDIKFAQSVFNEME-TIDVASWNALISGYSTHGLGRQALRILDIMKDRDCKPNGLTFL 554
Query: 598 QMSNIYSSEG 607
+ + S+ G
Sbjct: 555 GVLSGCSNAG 564
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,016,763
Number of Sequences: 26719
Number of extensions: 720751
Number of successful extensions: 9360
Number of sequences better than 10.0: 438
Number of HSP's better than 10.0 without gapping: 355
Number of HSP's successfully gapped in prelim test: 83
Number of HSP's that attempted gapping in prelim test: 1725
Number of HSP's gapped (non-prelim): 1987
length of query: 766
length of database: 11,318,596
effective HSP length: 107
effective length of query: 659
effective length of database: 8,459,663
effective search space: 5574917917
effective search space used: 5574917917
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0588c.4


