

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588b.6
(142 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
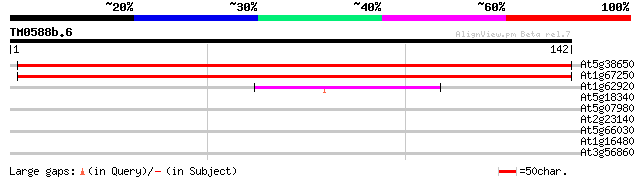
Score E
Sequences producing significant alignments: (bits) Value
At5g38650 unknown protein 214 9e-57
At1g67250 unknown protein 214 2e-56
At1g62920 hypothetical protein 57 3e-09
At5g18340 putative protein 27 4.8
At5g07980 putative protein 27 4.8
At2g23140 hypothetical protein 27 4.8
At5g66030 Golgi-localized protein GRIP 26 6.2
At1g16480 hypothetical protein 26 6.2
At3g56860 unknown protein 26 8.1
>At5g38650 unknown protein
Length = 141
Score = 214 bits (546), Expect = 9e-57
Identities = 102/140 (72%), Positives = 120/140 (84%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKSDI+ SHPL++A ES + E MKR+ + + YGA
Sbjct: 2 ESQKKIAHEIGGMKNDALRFGLHGVKSDILRSHPLETAYESGKQSQEEMKRRVITHTYGA 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMDLDRQILSRFQRP G IPSSMLGLE +TG+LD+FGFEDYL+DPRESET++P+D
Sbjct: 62 ALPLKMDLDRQILSRFQRPPGPIPSSMLGLEVYTGALDNFGFEDYLNDPRESETLKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLGLSKGP PSFM
Sbjct: 122 HHGMEVRLGLSKGPASPSFM 141
>At1g67250 unknown protein
Length = 141
Score = 214 bits (544), Expect = 2e-56
Identities = 98/140 (70%), Positives = 119/140 (85%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKS+I+GSHPL+S+ ES K E +KR + + YG
Sbjct: 2 ESEKKIAHEIGGVKNDALRFGLHGVKSNIIGSHPLESSYESEKKSKEALKRTVIAHAYGT 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMD+DRQILSRFQRP G IPSSMLGLE +TG++DDFGFEDYL+DPR+SET +P+D
Sbjct: 62 ALPLKMDMDRQILSRFQRPPGPIPSSMLGLEVYTGAVDDFGFEDYLNDPRDSETFKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLG+SKGP+ PSFM
Sbjct: 122 HHGMEVRLGISKGPIYPSFM 141
>At1g62920 hypothetical protein
Length = 260
Score = 57.0 bits (136), Expect = 3e-09
Identities = 32/71 (45%), Positives = 38/71 (53%), Gaps = 24/71 (33%)
Query: 63 AFPLKMDLDRQILSRF------------------------QRPSGAIPSSMLGLEAFTGS 98
A PLKMDLDR IL R+ RP+G IPS ML LE +TG+
Sbjct: 4 ALPLKMDLDRPILPRYGMLDQYHYEIIPLNSDAYSAIRYENRPTGPIPSLMLDLEVYTGA 63
Query: 99 LDDFGFEDYLS 109
+DDF FEDYL+
Sbjct: 64 IDDFDFEDYLN 74
>At5g18340 putative protein
Length = 456
Score = 26.6 bits (57), Expect = 4.8
Identities = 14/49 (28%), Positives = 21/49 (42%), Gaps = 10/49 (20%)
Query: 47 INEVMKRQCMVNLYGAAFPLKMDLD----------RQILSRFQRPSGAI 85
INE++ R C+ N Y P D+D +L R PS ++
Sbjct: 132 INELITRWCLANKYDRPAPKPSDIDYVTELFTDGIESLLQRISSPSSSV 180
>At5g07980 putative protein
Length = 1501
Score = 26.6 bits (57), Expect = 4.8
Identities = 17/73 (23%), Positives = 31/73 (42%), Gaps = 1/73 (1%)
Query: 65 PLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHH 124
P++ + R +L Q+P+ LGL+ + G + S + SE D+H
Sbjct: 84 PMRSEYSRSVLQEPQQPTNGYMHGNLGLQTMPNEANVLGMDVESSRDKLSERGFTPDLHK 143
Query: 125 GMEVRLGLSKGPV 137
+ R + + PV
Sbjct: 144 -IPTRFEMGESPV 155
>At2g23140 hypothetical protein
Length = 924
Score = 26.6 bits (57), Expect = 4.8
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Query: 82 SGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRP 119
S I + GL+A GSL+D FED +D RE T P
Sbjct: 459 SPGISGNGYGLDARRGSLND--FEDRSNDSRELRTDAP 494
>At5g66030 Golgi-localized protein GRIP
Length = 788
Score = 26.2 bits (56), Expect = 6.2
Identities = 14/59 (23%), Positives = 32/59 (53%), Gaps = 4/59 (6%)
Query: 15 IQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQ 73
++ND LR +G+K ++ LQ A++ + ++ + Q V A+ ++D+++Q
Sbjct: 49 LENDFLRSQFEGLKDEVAQGRSLQKAEQVEADSAQLKQLQEQV----ASLSREIDVEKQ 103
>At1g16480 hypothetical protein
Length = 905
Score = 26.2 bits (56), Expect = 6.2
Identities = 13/40 (32%), Positives = 20/40 (49%)
Query: 92 LEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLG 131
+ +F SLD F F + LS + + HG+ V+LG
Sbjct: 524 MRSFGVSLDQFSFSEGLSAAAKLAVLEEGQQLHGLAVKLG 563
>At3g56860 unknown protein
Length = 478
Score = 25.8 bits (55), Expect = 8.1
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 9/73 (12%)
Query: 28 KSDIVGSHPLQSAQESAS---KINEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPSGA 84
K + G P+ +A SA +E +++ V+ GA +LD Q L F G
Sbjct: 217 KGPVFGGAPIAAAAVSAPAQHSNSEHTQKKIYVSNVGA------ELDPQKLLMFFSKFGE 270
Query: 85 IPSSMLGLEAFTG 97
I LGL+ +TG
Sbjct: 271 IEEGPLGLDKYTG 283
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,930,518
Number of Sequences: 26719
Number of extensions: 111271
Number of successful extensions: 238
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 233
Number of HSP's gapped (non-prelim): 9
length of query: 142
length of database: 11,318,596
effective HSP length: 89
effective length of query: 53
effective length of database: 8,940,605
effective search space: 473852065
effective search space used: 473852065
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0588b.6


