

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588b.3
(432 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
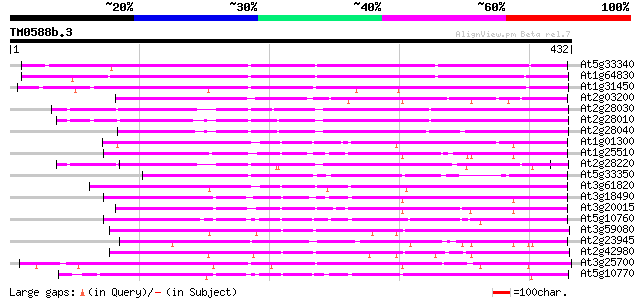
Score E
Sequences producing significant alignments: (bits) Value
At5g33340 putative protein 318 3e-87
At1g64830 hypothetical protein 310 1e-84
At1g31450 hypothetical protein 278 3e-75
At2g03200 putative chloroplast nucleoid DNA binding protein 220 1e-57
At2g28030 putative chloroplast nucleoid DNA binding protein 216 1e-56
At2g28010 putative chloroplast nucleoid DNA binding protein 211 6e-55
At2g28040 chloroplast nucleoid DNA binding like protein 206 2e-53
At1g01300 chloroplast nucleoid DNA binding protein like 206 3e-53
At1g25510 unknown protein 205 3e-53
At2g28220 putative chloroplast nucleoid DNA binding protein 204 6e-53
At5g33350 putative protein 201 6e-52
At3g61820 unknown protein 190 1e-48
At3g18490 chloroplast nucleoid DNA-binding protein like 189 3e-48
At3g20015 putative nucleoid chloroplast DNA-binding protein 186 3e-47
At5g10760 nucleoid DNA-binding protein cnd41 - like protein 181 9e-46
At3g59080 unknown protein 173 2e-43
At2g23945 putative protein 169 2e-42
At2g42980 hypothetical protein 168 5e-42
At3g25700 unknown protein 156 2e-38
At5g10770 nucleoid DNA-binding protein cnd41 - like protein 153 2e-37
>At5g33340 putative protein
Length = 437
Score = 318 bits (816), Expect = 3e-87
Identities = 176/430 (40%), Positives = 257/430 (58%), Gaps = 17/430 (3%)
Query: 10 LCFLVVLYLLSCQTPIEAQDAGFSVQLTRQNSPHSPFYKPDNLHRHKLPS-FHQVPKKAF 68
LC L L+L + + GF+ L ++SP SPFY P +L + H+ + F
Sbjct: 12 LCLLSSLFLSNANAKPKL---GFTADLIHRDSPKSPFYNPMETSSQRLRNAIHRSVNRVF 68
Query: 69 APNGPFSTR-----VTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQK 123
+T +TSN+G+YLM +++G+PP I + DTGSDL+W QC+PC CY Q
Sbjct: 69 HFTEKDNTPQPQIDLTSNSGEYLMNVSIGTPPFPIMAIADTGSDLLWTQCAPCDDCYTQV 128
Query: 124 SPMFEPLSSKTFNPIPCDSEQCGSLFSH-SCSP-QKLCAYSYSYADSSVTKGVLARETIT 181
P+F+P +S T+ + C S QC +L + SCS C+YS SY D+S TKG +A +T+T
Sbjct: 129 DPLFDPKTSSTYKDVSCSSSQCTALENQASCSTNDNTCSYSLSYGDNSYTKGNIAVDTLT 188
Query: 182 FSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQC 241
S ++ + + +II GCGH+N+G FN+ G++GLGGGP+SL+ Q+G +FS C
Sbjct: 189 LGS-SDTRPMQLKNIIIGCGHNNAGTFNKKGSGIVGLGGGPVSLIKQLGDSIDG-KFSYC 246
Query: 242 LVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQ-TPYLVTLEGISVGDTFVSFNS 300
LVP + + I+FG + VSG GVV+TPL+++ Q T Y +TL+ ISVG + ++
Sbjct: 247 LVPLTSKKDQTSKINFGTNAIVSGSGVVSTPLIAKASQETFYYLTLKSISVGSKQIQYSG 306
Query: 301 SE-KLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNL 359
S+ + S+GN++IDSGT T LP EFY L E+ S DP G LCY + +L
Sbjct: 307 SDSESSEGNIIIDSGTTLTLLPTEFYSEL-EDAVASSIDAEKKQDPQSGLSLCYSATGDL 365
Query: 360 EGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDR 419
+ P++T HF+GADV+L F+ + + CFA G+ I+GN AQ N L+G+D
Sbjct: 366 KVPVITMHFDGADVKLDSSNAFVQVSEDLVCFAFRGSPSFS-IYGNVAQMNFLVGYDTVS 424
Query: 420 KTISFKPTDC 429
KT+SFKPTDC
Sbjct: 425 KTVSFKPTDC 434
>At1g64830 hypothetical protein
Length = 431
Score = 310 bits (794), Expect = 1e-84
Identities = 175/431 (40%), Positives = 247/431 (56%), Gaps = 15/431 (3%)
Query: 10 LCFLVVLYLLSCQTPIEAQDAGFSVQLTRQNSPHSPFY--------KPDNLHRHKLPSFH 61
L F +L LL GF++ L ++SP SPFY + N R S
Sbjct: 4 LIFATLLSLLLLSNVNAYPKDGFTIDLIHRDSPKSPFYNSAETSSQRMRNAIRRSARSTL 63
Query: 62 QVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYK 121
Q +PN P S +TSN G+YLM +++G+PPV I + DTGSDL+W QC+PC CY+
Sbjct: 64 QFSNDDASPNSPQSF-ITSNRGEYLMNISIGTPPVPILAIADTGSDLIWTQCNPCEDCYQ 122
Query: 122 QKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCS-PQKLCAYSYSYADSSVTKGVLARETI 180
Q SP+F+P S T+ + C S QC +L SCS + C+Y+ +Y D+S TKG +A +T+
Sbjct: 123 QTSPLFDPKESSTYRKVSCSSSQCRALEDASCSTDENTCSYTITYGDNSYTKGDVAVDTV 182
Query: 181 TFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQ 240
T S + + + ++I GCGH N+G F+ G+IGLGGG SLVSQ+ +FS
Sbjct: 183 TMGS-SGRRPVSLRNMIIGCGHENTGTFDPAGSGIIGLGGGSTSLVSQLRKSING-KFSY 240
Query: 241 CLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNS 300
CLVPF +++ + I+FG VSG+GVV+T +V ++ T Y + LE ISVG + F S
Sbjct: 241 CLVPFTSETGLTSKINFGTNGIVSGDGVVSTSMVKKDPATYYFLNLEAISVGSKKIQFTS 300
Query: 301 S-EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNL 359
+ +GN++IDSGT T LP FY L E + + DPD LCYR ++
Sbjct: 301 TIFGTGEGNIVIDSGTTLTLLPSNFYYEL-ESVVASTIKAERVQDPDGILSLCYRDSSSF 359
Query: 360 EGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDR 419
+ P +T HF+G DV+L + TF+ + V CFA A + IFGN AQ N L+G+D
Sbjct: 360 KVPDITVHFKGGDVKLGNLNTFVAVSEDVSCFAFAAN-EQLTIFGNLAQMNFLVGYDTVS 418
Query: 420 KTISFKPTDCT 430
T+SFK TDC+
Sbjct: 419 GTVSFKKTDCS 429
>At1g31450 hypothetical protein
Length = 445
Score = 278 bits (712), Expect = 3e-75
Identities = 159/444 (35%), Positives = 238/444 (52%), Gaps = 27/444 (6%)
Query: 7 FFHLCFLVVLYLLSCQTPIEAQDAGFSVQLTRQNSPHSPFYKP-----DNLHRHKLPSFH 61
F + L + + + + A +V+L ++SPHSP Y P D L+ L S
Sbjct: 6 FLYCSLLAISFFFASNS--SANRENLTVELIHRDSPHSPLYNPHHTVSDRLNAAFLRSIS 63
Query: 62 QVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYK 121
+ + F + + SN G+Y M +++G+PP ++ + DTGSDL W QC PC CYK
Sbjct: 64 R--SRRFTTKTDLQSGLISNGGEYFMSISIGTPPSKVFAIADTGSDLTWVQCKPCQQCYK 121
Query: 122 QKSPMFEPLSSKTFNPIPCDSEQCGSLFSH--SCSPQK-LCAYSYSYADSSVTKGVLARE 178
Q SP+F+ S T+ CDS+ C +L H C K +C Y YSY D+S TKG +A E
Sbjct: 122 QNSPLFDKKKSSTYKTESCDSKTCQALSEHEEGCDESKDICKYRYSYGDNSFTKGDVATE 181
Query: 179 TITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRF 238
TI+ S ++G + +FGCG++N G F E G+IGLGGGPLSLVSQ+G+ G ++F
Sbjct: 182 TISIDS-SSGSSVSFPGTVFGCGYNNGGTFEETGSGIIGLGGGPLSLVSQLGSSIG-KKF 239
Query: 239 SQCLVPFHADSRTSGTISFGDASDVSG----EGVVTTPLVSEEGQTPYLVTLEGISVGDT 294
S CL A + + I+ G S S +TTPL+ ++ +T Y +TLE ++VG T
Sbjct: 240 SYCLSHTAATTNGTSVINLGTNSIPSNPSKDSATLTTPLIQKDPETYYFLTLEAVTVGKT 299
Query: 295 FVSF-------NSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDL 347
+ + N GN++IDSGT T L FYD ++ + +DP
Sbjct: 300 KLPYTGGGYGLNGKSSKRTGNIIIDSGTTLTLLDSGFYDDFGTAVEESVTGAKRVSDPQG 359
Query: 348 GTQLCYRS-ETNLEGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNF 406
C++S + + P +T HF ADV+L PI F+ + C +M T + I+GN
Sbjct: 360 LLTHCFKSGDKEIGLPAITMHFTNADVKLSPINAFVKLNEDTVCLSMIPTTE-VAIYGNM 418
Query: 407 AQSNILIGFDLDRKTISFKPTDCT 430
Q + L+G+DL+ KT+SF+ DC+
Sbjct: 419 VQMDFLVGYDLETKTVSFQRMDCS 442
>At2g03200 putative chloroplast nucleoid DNA binding protein
Length = 461
Score = 220 bits (561), Expect = 1e-57
Identities = 137/368 (37%), Positives = 201/368 (54%), Gaps = 33/368 (8%)
Query: 82 NGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCD 141
+G++LM+L++G+P V +VDTGSDL+W QC PC C+ Q +P+F+P S +++ + C
Sbjct: 104 SGEFLMELSIGNPAVKYSAIVDTGSDLIWTQCKPCTECFDQPTPIFDPEKSSSYSKVGCS 163
Query: 142 SEQCGSLFSHSCSPQK-LCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGC 200
S C +L +C+ K C Y Y+Y D S T+G+LA ET TF DE + I FGC
Sbjct: 164 SGLCNALPRSNCNEDKDACEYLYTYGDYSSTRGLLATETFTFE-----DENSISGIGFGC 218
Query: 201 GHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGD- 259
G N G G++GLG GPLSL+SQ+ +FS CL DS S ++ G
Sbjct: 219 GVENEGDGFSQGSGLVGLGRGPLSLISQL----KETKFSYCLTSIE-DSEASSSLFIGSL 273
Query: 260 --------ASDVSGEGVVTTPLVSEEGQTP-YLVTLEGISVGDTFVSFNSS----EKLSK 306
+ + GE T L+ Q Y + L+GI+VG +S S +
Sbjct: 274 ASGIVNKTGASLDGEVTKTMSLLRNPDQPSFYYLELQGITVGAKRLSVEKSTFELAEDGT 333
Query: 307 GNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYR---SETNLEGPI 363
G M+IDSGT TYL + + L EE + S LPVD+ G LC++ + N+ P
Sbjct: 334 GGMIIDSGTTITYLEETAFKVLKEEFTSRMS-LPVDDSGSTGLDLCFKLPDAAKNIAVPK 392
Query: 364 LTAHFEGADVQLMPIQTFI--PPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKT 421
+ HF+GAD++L P + ++ GV C AM G+++G IFGN Q N + DL+++T
Sbjct: 393 MIFHFKGADLEL-PGENYMVADSSTGVLCLAM-GSSNGMSIFGNVQQQNFNVLHDLEKET 450
Query: 422 ISFKPTDC 429
+SF PT+C
Sbjct: 451 VSFVPTEC 458
>At2g28030 putative chloroplast nucleoid DNA binding protein
Length = 392
Score = 216 bits (551), Expect = 1e-56
Identities = 144/403 (35%), Positives = 207/403 (50%), Gaps = 31/403 (7%)
Query: 33 SVQLTRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLG 92
S+ T +SPH + D + R S ++ K P++ + N YLMKL +G
Sbjct: 12 SLFTTTASSPHG--FTIDLIQRRSNSSSSRLSKNQLQGASPYADTLFDYN-IYLMKLQVG 68
Query: 93 SPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHS 152
+PP +I +DTGSDL+W QC PC CY Q +P+F+P +S TF C+
Sbjct: 69 TPPFEIEAEIDTGSDLIWTQCMPCTNCYSQYAPIFDPSNSSTFKEKRCNGNS-------- 120
Query: 153 CSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNEND 212
C Y YAD++ +KG LA ET+T S T+G+ V+ + GCGH NS F
Sbjct: 121 ------CHYKIIYADTTYSKGTLATETVTIHS-TSGEPFVMPETTIGCGH-NSSWFKPTF 172
Query: 213 MGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVV-TT 271
G++GL GP SL++QMG Y L+ + S+ + I+FG + V+G+GVV TT
Sbjct: 173 SGMVGLSWGPSSLITQMGGEYPG------LMSYCFASQGTSKINFGTNAIVAGDGVVSTT 226
Query: 272 PLVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVE 330
++ Y + L+ +SVGDT V + ++ +GN++IDSGT TY P Y LV
Sbjct: 227 MFLTTAKPGLYYLNLDAVSVGDTHVETMGTTFHALEGNIIIDSGTTLTYFPVS-YCNLVR 285
Query: 331 ELKVQSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPP-KDGV 388
E DP LCY ++T P++T HF GAD+ L +I G
Sbjct: 286 EAVDHYVTAVRTADPTGNDMLCYYTDTIDIFPVITMHFSGGADLVLDKYNMYIETITRGT 345
Query: 389 FCFA-MAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
FC A + D IFGN AQ+N L+G+D +SF PT+C+
Sbjct: 346 FCLAIICNNPPQDAIFGNRAQNNFLVGYDSSSLLVSFSPTNCS 388
>At2g28010 putative chloroplast nucleoid DNA binding protein
Length = 396
Score = 211 bits (537), Expect = 6e-55
Identities = 139/399 (34%), Positives = 208/399 (51%), Gaps = 33/399 (8%)
Query: 37 TRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPV 96
T + PH + D +HR S + + P++ V N+ YLMKL +G+PP
Sbjct: 22 TTASPPHG--FTMDLIHRRSNASSRV--SNTQSGSSPYANTVFDNSV-YLMKLQVGTPPF 76
Query: 97 DIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSPQ 156
+I ++DTGS++ W QC PC CY+Q +P+F+P S TF CD HS
Sbjct: 77 EIQAIIDTGSEITWTQCLPCVHCYEQNAPIFDPSKSSTFKEKRCD--------GHS---- 124
Query: 157 KLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVI 216
C Y Y D + T G LA ETIT S T+G+ V+ + I GCGH+NS F + G++
Sbjct: 125 --CPYEVDYFDHTYTMGTLATETITLHS-TSGEPFVMPETIIGCGHNNSW-FKPSFSGMV 180
Query: 217 GLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSE 276
GL GP SL++QMG Y L+ + + + I+FG + V+G+GVV+T +
Sbjct: 181 GLNWGPSSLITQMGGEYPG------LMSYCFSGQGTSKINFGANAIVAGDGVVSTTMFMT 234
Query: 277 EGQTP-YLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKV 334
+ Y + L+ +SVG+T + + ++ +GN++IDSGT TY P Y LV +
Sbjct: 235 TAKPGFYYLNLDAVSVGNTRIETMGTTFHALEGNIVIDSGTTLTYFPVS-YCNLVRQAVE 293
Query: 335 QSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPPKD-GVFCFA 392
DP LCY S+T P++T HF G D+ L ++ + GVFC A
Sbjct: 294 HVVTAVRAADPTGNDMLCYNSDTIDIFPVITMHFSGGVDLVLDKYNMYMESNNGGVFCLA 353
Query: 393 -MAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
+ + + IFGN AQ+N L+G+D +SF PT+C+
Sbjct: 354 IICNSPTQEAIFGNRAQNNFLVGYDSSSLLVSFSPTNCS 392
>At2g28040 chloroplast nucleoid DNA binding like protein
Length = 395
Score = 206 bits (524), Expect = 2e-53
Identities = 135/352 (38%), Positives = 190/352 (53%), Gaps = 29/352 (8%)
Query: 84 DYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSE 143
+YLMKL +G+PP +I ++DTGS+ +W QC PC CY Q +P+F+P S TF I CD+
Sbjct: 64 EYLMKLQIGTPPFEIEAVLDTGSEHIWTQCLPCVHCYNQTAPIFDPSKSSTFKEIRCDTH 123
Query: 144 QCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHS 203
HS C Y Y S TKG L ET+T S T+G V+ + I GCG +
Sbjct: 124 ------DHS------CPYELVYGGKSYTKGTLVTETVTIHS-TSGQPFVMPETIIGCGRN 170
Query: 204 NSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDV 263
NSG F GV+GL GP SL++QMG Y L+ + + + I+FG + V
Sbjct: 171 NSG-FKPGFAGVVGLDRGPKSLITQMGGEYPG------LMSYCFAGKGTSKINFGANAIV 223
Query: 264 SGEGVV-TTPLVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLP 321
+G+GVV TT V Y + L+ +SVG+T + + + KGN++IDSG+ TY P
Sbjct: 224 AGDGVVSTTVFVKTAKPGFYYLNLDAVSVGNTRIETVGTPFHALKGNIVIDSGSTLTYFP 283
Query: 322 QEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQT 380
E Y LV + Q D+ LCY S+T P++T HF GAD+ L
Sbjct: 284 -ESYCNLVRKAVEQVVTAVRFPRSDI---LCYYSKTIDIFPVITMHFSGGADLVLDKYNM 339
Query: 381 FIPPK-DGVFCFA-MAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
++ GVFC A + + + IFGN AQ+N L+G+D +SFKPT+C+
Sbjct: 340 YVASNTGGVFCLAIICNSPIEEAIFGNRAQNNFLVGYDSSSLLVSFKPTNCS 391
>At1g01300 chloroplast nucleoid DNA binding protein like
Length = 485
Score = 206 bits (523), Expect = 3e-53
Identities = 140/371 (37%), Positives = 189/371 (50%), Gaps = 24/371 (6%)
Query: 72 GPFSTRVTSN----NGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMF 127
G FS+ V S +G+Y +L +G+P +Y ++DTGSD+VW QC+PC CY Q P+F
Sbjct: 125 GGFSSSVVSGLSQGSGEYFTRLGVGTPARYVYMVLDTGSDIVWLQCAPCRRCYSQSDPIF 184
Query: 128 EPLSSKTFNPIPCDSEQCGSLFSHSCSP-QKLCAYSYSYADSSVTKGVLARETITFSSPT 186
+P SKT+ IPC S C L S C+ +K C Y SY D S T G + ET+TF
Sbjct: 185 DPRKSKTYATIPCSSPHCRRLDSAGCNTRRKTCLYQVSYGDGSFTVGDFSTETLTFRRNR 244
Query: 187 NGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFH 246
V + GCGH N G F G++GLG G LS Q G + +++FS CLV
Sbjct: 245 ------VKGVALGCGHDNEGLF-VGAAGLLGLGKGKLSFPGQTGHRF-NQKFSYCLVDRS 296
Query: 247 ADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFV-----SFNSS 301
A S+ S + FG+A+ VS T L + + T Y V L GISVG T V S
Sbjct: 297 ASSKPSSVV-FGNAA-VSRIARFTPLLSNPKLDTFYYVGLLGISVGGTRVPGVTASLFKL 354
Query: 302 EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETN-LE 360
+++ G ++IDSGT T L + Y + + +V + L D L S N ++
Sbjct: 355 DQIGNGGVIIDSGTSVTRLIRPAYIAMRDAFRVGAKTLKRAPDFSLFDTCFDLSNMNEVK 414
Query: 361 GPILTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLD 418
P + HF GADV L P ++ P D G FCFA AGT G I GN Q + +DL
Sbjct: 415 VPTVVLHFRGADVSL-PATNYLIPVDTNGKFCFAFAGTMGGLSIIGNIQQQGFRVVYDLA 473
Query: 419 RKTISFKPTDC 429
+ F P C
Sbjct: 474 SSRVGFAPGGC 484
>At1g25510 unknown protein
Length = 483
Score = 205 bits (522), Expect = 3e-53
Identities = 131/368 (35%), Positives = 188/368 (50%), Gaps = 31/368 (8%)
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSS 132
P + T +G+Y ++ +G P ++Y ++DTGSD+ W QC+PC CY Q P+FEP SS
Sbjct: 136 PLISGTTQGSGEYFTRVGIGKPAREVYMVLDTGSDVNWLQCTPCADCYHQTEPIFEPSSS 195
Query: 133 KTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELV 192
++ P+ CD+ QC +L C C Y SY D S T G A ET+T S +
Sbjct: 196 SSYEPLSCDTPQCNALEVSECR-NATCLYEVSYGDGSYTVGDFATETLTIGS------TL 248
Query: 193 VGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTS 252
V ++ GCGHSN G F G++GLGGG L+L SQ+ + FS CLV DS ++
Sbjct: 249 VQNVAVGCGHSNEGLF-VGAAGLLGLGGGLLALPSQL----NTTSFSYCLV--DRDSDSA 301
Query: 253 GTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKGN 308
T+ FG + +S + VV L + + T Y + L GISVG + S ++ G
Sbjct: 302 STVDFG--TSLSPDAVVAPLLRNHQLDTFYYLGLTGISVGGELLQIPQSSFEMDESGSGG 359
Query: 309 MMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQL---CYR--SETNLEGPI 363
++IDSGT T L E Y+ L + V+ +L D + G + CY ++T +E P
Sbjct: 360 IIIDSGTAVTRLQTEIYNSLRDSF-VKGTL---DLEKAAGVAMFDTCYNLSAKTTVEVPT 415
Query: 364 LTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKT 421
+ HF G + +P + ++ P D G FC A A TA I GN Q + FDL
Sbjct: 416 VAFHFPGGKMLALPAKNYMIPVDSVGTFCLAFAPTASSLAIIGNVQQQGTRVTFDLANSL 475
Query: 422 ISFKPTDC 429
I F C
Sbjct: 476 IGFSSNKC 483
>At2g28220 putative chloroplast nucleoid DNA binding protein
Length = 756
Score = 204 bits (520), Expect = 6e-53
Identities = 124/357 (34%), Positives = 188/357 (51%), Gaps = 36/357 (10%)
Query: 85 YLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQ 144
YLMKL +G+PP +I +DTGSD++W QC PC CY Q +P+F+P S TF C+
Sbjct: 421 YLMKLQVGTPPFEIVAEIDTGSDIIWTQCMPCPNCYSQFAPIFDPSKSSTFREQRCNGNS 480
Query: 145 CGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSN 204
C Y YAD + +KG+LA ET+T S T+G+ V+ + GCG N
Sbjct: 481 --------------CHYEIIYADKTYSKGILATETVTIPS-TSGEPFVMAETKIGCGLDN 525
Query: 205 S----GAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDA 260
+ F + G++GL GPLSL+SQM Y L+ + + + I+FG
Sbjct: 526 TNLQYSGFASSSSGIVGLNMGPLSLISQMDLPYPG------LISYCFSGQGTSKINFGTN 579
Query: 261 SDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATY 319
+ V+G+G V + ++ Y + L+ +SV D + + + GN+ IDSGT TY
Sbjct: 580 AIVAGDGTVAADMFIKKDNPFYYLNLDAVSVEDNLIATLGTPFHAEDGNIFIDSGTTLTY 639
Query: 320 LPQEFYDRLVEELKVQSSLLPVDNDPDLGTQ--LCYRSETNLEGPILTAHFE-GADVQLM 376
P + + + E ++ + + V PD+G+ LCY S+T P++T HF GAD+ L
Sbjct: 640 FPMSYCNLVREAVEQVVTAVKV---PDMGSDNLLCYYSDTIDIFPVITMHFSGGADLVLD 696
Query: 377 PIQTFIPP-KDGVFCFAMAGTADGDY--IFGNFAQSNILIGFDLDRKTISFKPTDCT 430
++ G+FC A+ G D +FGN AQ+N L+G+D ISF PT+C+
Sbjct: 697 KYNMYLETITGGIFCLAI-GCNDPSMPAVFGNRAQNNFLVGYDPSSNVISFSPTNCS 752
Score = 193 bits (491), Expect = 1e-49
Identities = 129/388 (33%), Positives = 191/388 (48%), Gaps = 33/388 (8%)
Query: 37 TRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPV 96
T +SPH + D + R S ++ K P++ + N YLMKL +G+PP
Sbjct: 37 TTVSSPHG--FTIDLIQRRSNSSSFRLSKNQLQGASPYADTLFDYN-IYLMKLQVGTPPF 93
Query: 97 DIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSPQ 156
+I +DTGSDL+W QC PC CY Q P+F+P S TFN C
Sbjct: 94 EIAAEIDTGSDLIWTQCMPCPDCYSQFDPIFDPSKSSTFNEQRCHG-------------- 139
Query: 157 KLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSN----SGAFNEND 212
K C Y Y D++ +KG+LA ET+T S T+G+ V+ + GCG N + F +
Sbjct: 140 KSCHYEIIYEDNTYSKGILATETVTIHS-TSGEPFVMAETTIGCGLHNTDLDNSGFASSS 198
Query: 213 MGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTP 272
G++GL GP SL+SQM Y L+ + + + I+FG + V+G+G V
Sbjct: 199 SGIVGLNMGPRSLISQMDLPYPG------LISYCFSGQGTSKINFGTNAIVAGDGTVAAD 252
Query: 273 LVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEE 331
+ ++ Y + L+ +SV D + + + GN++IDSG+ TY P Y LV +
Sbjct: 253 MFIKKDNPFYYLNLDAVSVEDNRIETLGTPFHAEDGNIVIDSGSTVTYFPVS-YCNLVRK 311
Query: 332 LKVQSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPPKD-GVF 389
Q DP LCY SET P++T HF GAD+ L ++ G+F
Sbjct: 312 AVEQVVTAVRVPDPSGNDMLCYFSETIDIFPVITMHFSGGADLVLDKYNMYMESNSGGLF 371
Query: 390 CFA-MAGTADGDYIFGNFAQSNILIGFD 416
C A + + + IFGN AQ+N L+G+D
Sbjct: 372 CLAIICNSPTQEAIFGNRAQNNFLVGYD 399
>At5g33350 putative protein
Length = 293
Score = 201 bits (511), Expect = 6e-52
Identities = 124/331 (37%), Positives = 180/331 (53%), Gaps = 49/331 (14%)
Query: 103 DTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSH-SCSPQ-KLCA 160
DTGSDL+W QC PC CY Q P+F+P +S T+ + C S QC +L SCS + C+
Sbjct: 5 DTGSDLIWTQCKPCDDCYTQVDPLFDPKASSTYKDVSCPSSQCRALKDDASCSKKDNTCS 64
Query: 161 YSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGG 220
YS +Y D+S ++G +A +T+T S T+ + V +II GCGH N+ F G++GLGG
Sbjct: 65 YSMNYGDNSYSRGNVAVDTLTLGS-TDNRPVQVKNIIIGCGHENAVTFRNKSSGIVGLGG 123
Query: 221 GPLSLVSQMG-ALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQ 279
G +SLV Q+G ++ G +FS CLVP ++ + ISFG + VSG G V+TPLV + +
Sbjct: 124 GAVSLVKQLGDSIEG--KFSYCLVP---ENDQTSKISFGTNAVVSGPGTVSTPLVVKSPE 178
Query: 280 TPYLVTLEGISVGDTFVSFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLL 339
T Y +TL+ I+VG + S+ KGNM+IDSGT T LP ++Y ++ + +SL+
Sbjct: 179 TFYFLTLKSITVGSKNMPTPGSD--IKGNMVIDSGTTLTLLPGKYYFQIESAV---ASLI 233
Query: 340 PVDNDPDLGTQLCYRSETNLEGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMA-GTAD 398
+ D + + D + CFA
Sbjct: 234 DAERSKD--------------------------------ERIV--SDDLVCFAFGLNLIT 259
Query: 399 GDYIFGNFAQSNILIGFDLDRKTISFKPTDC 429
D I+GN AQ N L+G+D K++SFK TDC
Sbjct: 260 RDGIYGNVAQKNFLVGYDTVSKSLSFKKTDC 290
>At3g61820 unknown protein
Length = 483
Score = 190 bits (483), Expect = 1e-48
Identities = 132/381 (34%), Positives = 182/381 (47%), Gaps = 22/381 (5%)
Query: 62 QVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYK 121
+ P+ A +G + ++ +G+Y M+L +G+P ++Y ++DTGSD+VW QCSPC CY
Sbjct: 112 RTPRTAGGFSGAVISGLSQGSGEYFMRLGVGTPATNVYMVLDTGSDVVWLQCSPCKACYN 171
Query: 122 QKSPMFEPLSSKTFNPIPCDSEQCGSLFSHS---CSPQKLCAYSYSYADSSVTKGVLARE 178
Q +F+P SKTF +PC S C L S K C Y SY D S T+G + E
Sbjct: 172 QTDAIFDPKKSKTFATVPCGSRLCRRLDDSSECVTRRSKTCLYQVSYGDGSFTEGDFSTE 231
Query: 179 TITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRF 238
T+TF V + GCGH N G F G++GLG G LS SQ Y +F
Sbjct: 232 TLTFHGAR------VDHVPLGCGHDNEGLF-VGAAGLLGLGRGGLSFPSQTKNRYNG-KF 283
Query: 239 SQCLV---PFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTF 295
S CLV + S+ TI FG+A+ V V T L + + T Y + L GISVG +
Sbjct: 284 SYCLVDRTSSGSSSKPPSTIVFGNAA-VPKTSVFTPLLTNPKLDTFYYLQLLGISVGGSR 342
Query: 296 VSFNSSEKL-----SKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDL-GT 349
V S + G ++IDSGT T L Q Y L + ++ ++ L L T
Sbjct: 343 VPGVSESQFKLDATGNGGVIIDSGTSVTRLTQPAYVALRDAFRLGATKLKRAPSYSLFDT 402
Query: 350 QLCYRSETNLEGPILTAHFEGADVQLMPIQTFIPPK-DGVFCFAMAGTADGDYIFGNFAQ 408
T ++ P + HF G +V L IP +G FCFA AGT I GN Q
Sbjct: 403 CFDLSGMTTVKVPTVVFHFGGGEVSLPASNYLIPVNTEGRFCFAFAGTMGSLSIIGNIQQ 462
Query: 409 SNILIGFDLDRKTISFKPTDC 429
+ +DL + F C
Sbjct: 463 QGFRVAYDLVGSRVGFLSRAC 483
>At3g18490 chloroplast nucleoid DNA-binding protein like
Length = 500
Score = 189 bits (479), Expect = 3e-48
Identities = 123/366 (33%), Positives = 182/366 (49%), Gaps = 24/366 (6%)
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSS 132
P + + +G+Y ++ +G+P ++Y ++DTGSD+ W QC PC CY+Q P+F P SS
Sbjct: 150 PVVSGASQGSGEYFSRIGVGTPAKEMYLVLDTGSDVNWIQCEPCADCYQQSDPVFNPTSS 209
Query: 133 KTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELV 192
T+ + C + QC L + +C K C Y SY D S T G LA +T+TF G+
Sbjct: 210 STYKSLTCSAPQCSLLETSACRSNK-CLYQVSYGDGSFTVGELATDTVTF-----GNSGK 263
Query: 193 VGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTS 252
+ ++ GCGH N G F G++GLGGG LS+ +QM A FS CLV DS S
Sbjct: 264 INNVALGCGHDNEGLFT-GAAGLLGLGGGVLSITNQMKA----TSFSYCLV--DRDSGKS 316
Query: 253 GTISFGDASDVSGEGVVTTPLV-SEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKG 307
++ F G G T PL+ +++ T Y V L G SVG V + + G
Sbjct: 317 SSLDFNSVQ--LGGGDATAPLLRNKKIDTFYYVGLSGFSVGGEKVVLPDAIFDVDASGSG 374
Query: 308 NMMIDSGTPATYLPQEFYDRLVEE-LKVQSSLLPVDNDPDL-GTQLCYRSETNLEGPILT 365
+++D GT T L + Y+ L + LK+ +L + L T + S + ++ P +
Sbjct: 375 GVILDCGTAVTRLQTQAYNSLRDAFLKLTVNLKKGSSSISLFDTCYDFSSLSTVKVPTVA 434
Query: 366 AHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKTIS 423
HF G +P + ++ P D G FCFA A T+ I GN Q I +DL + I
Sbjct: 435 FHFTGGKSLDLPAKNYLIPVDDSGTFCFAFAPTSSSLSIIGNVQQQGTRITYDLSKNVIG 494
Query: 424 FKPTDC 429
C
Sbjct: 495 LSGNKC 500
>At3g20015 putative nucleoid chloroplast DNA-binding protein
Length = 386
Score = 186 bits (471), Expect = 3e-47
Identities = 118/357 (33%), Positives = 176/357 (49%), Gaps = 23/357 (6%)
Query: 82 NGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCD 141
+G+Y +++ +GSPP D Y ++D+GSD+VW QC PC CYKQ P+F+P S ++ + C
Sbjct: 44 SGEYFVRIGVGSPPRDQYMVIDSGSDMVWVQCQPCKLCYKQSDPVFDPAKSGSYTGVSCG 103
Query: 142 SEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCG 201
S C + + C C Y Y D S TKG LA ET+TF+ + VV ++ GCG
Sbjct: 104 SSVCDRIENSGCHSGG-CRYEVMYGDGSYTKGTLALETLTFA------KTVVRNVAMGCG 156
Query: 202 HSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDAS 261
H N G F G++G+GGG +S V Q+ G F CLV DS +G++ FG
Sbjct: 157 HRNRGMF-IGAAGLLGIGGGSMSFVGQLSGQTGG-AFGYCLVSRGTDS--TGSLVFG--R 210
Query: 262 DVSGEGVVTTPLV-SEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKGNMMIDSGTP 316
+ G PLV + + Y V L+G+ VG + + G +++D+GT
Sbjct: 211 EALPVGASWVPLVRNPRAPSFYYVGLKGLGVGGVRIPLPDGVFDLTETGDGGVVMDTGTA 270
Query: 317 ATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCY--RSETNLEGPILTAHFEGADVQ 374
T LP Y + K Q++ LP + + CY ++ P ++ +F V
Sbjct: 271 VTRLPTAAYVAFRDGFKSQTANLPRASGVSI-FDTCYDLSGFVSVRVPTVSFYFTEGPVL 329
Query: 375 LMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDC 429
+P + F+ P D G +CFA A + G I GN Q I + FD + F P C
Sbjct: 330 TLPARNFLMPVDDSGTYCFAFAASPTGLSIIGNIQQEGIQVSFDGANGFVGFGPNVC 386
>At5g10760 nucleoid DNA-binding protein cnd41 - like protein
Length = 464
Score = 181 bits (458), Expect = 9e-46
Identities = 130/362 (35%), Positives = 181/362 (49%), Gaps = 22/362 (6%)
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHG-CYKQKSPMFEPLS 131
P + +T +G+Y++ + +G+P D+ + DTGSDL W QC PC G CY QK P F P S
Sbjct: 120 PAKSGITLGSGNYIVTIGIGTPKHDLSLVFDTGSDLTWTQCEPCLGSCYSQKEPKFNPSS 179
Query: 132 SKTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDEL 191
S T+ + C S C + SCS C YS Y D S T+G LA+E T TN D
Sbjct: 180 SSTYQNVSCSSPMCED--AESCSASN-CVYSIVYGDKSFTQGFLAKEKFTL---TNSD-- 231
Query: 192 VVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRT 251
V+ D+ FGCG +N G F + G++GLG G LSL +Q Y + FS CL F S +
Sbjct: 232 VLEDVYFGCGENNQGLF-DGVAGLLGLGPGKLSLPAQTTTTY-NNIFSYCLPSF--TSNS 287
Query: 252 SGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNSSEKLSKGNMMI 311
+G ++FG A E V TP+ S Y + + GISVGD ++ + ++G +I
Sbjct: 288 TGHLTFGSAG--ISESVKFTPISSFPSAFNYGIDIIGISVGDKELAITPNSFSTEG-AII 344
Query: 312 DSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNLEG---PILTAHF 368
DSGT T LP + Y L K + S + L CY T L+ P + F
Sbjct: 345 DSGTVFTRLPTKVYAELRSVFKEKMSSYKSTSGYGL-FDTCY-DFTGLDTVTYPTIAFSF 402
Query: 369 EGAD-VQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPT 427
G+ V+L +P K C A AG D IFGN Q+ + + +D+ + F P
Sbjct: 403 AGSTVVELDGSGISLPIKISQVCLAFAGNDDLPAIFGNVQQTTLDVVYDVAGGRVGFAPN 462
Query: 428 DC 429
C
Sbjct: 463 GC 464
>At3g59080 unknown protein
Length = 535
Score = 173 bits (438), Expect = 2e-43
Identities = 126/375 (33%), Positives = 190/375 (50%), Gaps = 24/375 (6%)
Query: 78 VTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNP 137
+T +G+Y M + +GSPP ++DTGSDL W QC PC+ C++Q ++P +S ++
Sbjct: 163 MTLGSGEYFMDVLVGSPPKHFSLILDTGSDLNWIQCLPCYDCFQQNGAFYDPKASASYKN 222
Query: 138 IPCDSEQCGSLFSHS----C-SPQKLCAYSYSYADSSVTKGVLARETITFSSPTNG--DE 190
I C+ ++C + S C S + C Y Y Y DSS T G A ET T + TNG E
Sbjct: 223 ITCNDQRCNLVSSPDPPMPCKSDNQSCPYYYWYGDSSNTTGDFAVETFTVNLTTNGGSSE 282
Query: 191 LV-VGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADS 249
L V +++FGCGH N G F+ G++GLG GPLS SQ+ +LYG FS CLV ++D+
Sbjct: 283 LYNVENMMFGCGHWNRGLFH-GAAGLLGLGRGPLSFSSQLQSLYG-HSFSYCLVDRNSDT 340
Query: 250 RTSGTISFGDASD-VSGEGVVTTPLVSEEG---QTPYLVTLEGISVGDTFV-----SFNS 300
S + FG+ D +S + T V+ + T Y V ++ I V + ++N
Sbjct: 341 NVSSKLIFGEDKDLLSHPNLNFTSFVAGKENLVDTFYYVQIKSILVAGEVLNIPEETWNI 400
Query: 301 SEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQS-SLLPVDND-PDLGTQLCYRSETN 358
S G +IDSGT +Y + Y+ + ++ ++ PV D P L N
Sbjct: 401 SSD-GAGGTIIDSGTTLSYFAEPAYEFIKNKIAEKAKGKYPVYRDFPILDPCFNVSGIHN 459
Query: 359 LEGPILTAHFEGADVQLMPIQ-TFIPPKDGVFCFAMAGTADGDY-IFGNFAQSNILIGFD 416
++ P L F V P + +FI + + C AM GT + I GN+ Q N I +D
Sbjct: 460 VQLPELGIAFADGAVWNFPTENSFIWLNEDLVCLAMLGTPKSAFSIIGNYQQQNFHILYD 519
Query: 417 LDRKTISFKPTDCTN 431
R + + PT C +
Sbjct: 520 TKRSRLGYAPTKCAD 534
>At2g23945 putative protein
Length = 458
Score = 169 bits (429), Expect = 2e-42
Identities = 121/369 (32%), Positives = 177/369 (47%), Gaps = 40/369 (10%)
Query: 85 YLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKS--PMFEPLSSKTFNPIPCDS 142
+L+ ++G PPV ++DTGS L+W QC PC C P+F P S TF CD
Sbjct: 96 FLVNFSVGQPPVPQLTIMDTGSSLLWIQCQPCKHCSSDHMIHPVFNPALSSTFVECSCDD 155
Query: 143 EQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGH 202
C + C C Y Y + +KGVLA+E +TF++P NG+ +V I FGCG+
Sbjct: 156 RFCRYAPNGHCGSSNKCVYEQVYISGTGSKGVLAKERLTFTTP-NGNTVVTQPIAFGCGY 214
Query: 203 SNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASD 262
N + G++GLG P SL Q+G+ +FS C+ + + G+ +D
Sbjct: 215 ENGEQLESHFTGILGLGAKPTSLAVQLGS-----KFSYCIGDLANKNYGYNQLVLGEDAD 269
Query: 263 VSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNSSEKLSKG---NMMIDSGTPATY 319
+ G+ TP+ E + Y + LEGISVGDT ++ +G +++DSGT T+
Sbjct: 270 ILGD---PTPIEFETENSIYYMNLEGISVGDTQLNIEPVVFKRRGPRTGVILDSGTLYTW 326
Query: 320 LPQEFYDRLVEELKVQSSLLPVDNDPDL-----GTQLCYR---SETNLEGPILTAHFE-G 370
L Y L E+K S+L DP L LCY SE + P++T HF G
Sbjct: 327 LADIAYRELYNEIK---SIL----DPKLERFWFRDFLCYHGRVSEELIGFPVVTFHFAGG 379
Query: 371 ADVQLMPIQTFIPPKD----GVFCFAMAGTAD--GDY----IFGNFAQSNILIGFDLDRK 420
A++ + F P + VFC ++ T + G+Y G AQ IG+DL K
Sbjct: 380 AELAMEATSMFYPLSEPNTFNVFCMSVKPTKEHGGEYKEFTAIGLMAQQYYNIGYDLKEK 439
Query: 421 TISFKPTDC 429
I + DC
Sbjct: 440 NIYLQRIDC 448
>At2g42980 hypothetical protein
Length = 481
Score = 168 bits (426), Expect = 5e-42
Identities = 124/382 (32%), Positives = 188/382 (48%), Gaps = 36/382 (9%)
Query: 78 VTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNP 137
+T +G+Y M + +G+PP ++DTGSDL W QC PC+ C+ Q ++P +S +F
Sbjct: 107 MTLGSGEYFMDVLVGTPPKHFSLILDTGSDLNWLQCLPCYDCFHQNGMFYDPKTSASFKN 166
Query: 138 IPCDSEQCGSLFSH----SC-SPQKLCAYSYSYADSSVTKGVLARETITFSSPT---NGD 189
I C+ +C + S C S + C Y Y Y D S T G A ET T + T
Sbjct: 167 ITCNDPRCSLISSPDPPVQCESDNQSCPYFYWYGDRSNTTGDFAVETFTVNLTTTEGGSS 226
Query: 190 ELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADS 249
E VG+++FGCGH N G F+ G++GLG GPLS SQ+ +LYG FS CLV ++++
Sbjct: 227 EYKVGNMMFGCGHWNRGLFS-GASGLLGLGRGPLSFSSQLQSLYG-HSFSYCLVDRNSNT 284
Query: 250 RTSGTISFGDASD-VSGEGVVTTPLVS---EEGQTPYLVTLEGISVGDTFV-----SFNS 300
S + FG+ D ++ + T V+ +T Y + ++ I VG + ++N
Sbjct: 285 NVSSKLIFGEDKDLLNHTNLNFTSFVNGKENSVETFYYIQIKSILVGGKALDIPEETWNI 344
Query: 301 SEKLSKGNMMIDSGTPATYLPQEFYD----RLVEELKVQSSLLPVDND-PDLGTQLCYR- 354
S G +IDSGT +Y + Y+ + E++K P+ D P L C+
Sbjct: 345 SSD-GDGGTIIDSGTTLSYFAEPAYEIIKNKFAEKMKEN---YPIFRDFPVLDP--CFNV 398
Query: 355 ---SETNLEGPILTAHFEGADVQLMPIQ-TFIPPKDGVFCFAMAGTADGDY-IFGNFAQS 409
E N+ P L F V P + +FI + + C A+ GT + I GN+ Q
Sbjct: 399 SGIEENNIHLPELGIAFVDGTVWNFPAENSFIWLSEDLVCLAILGTPKSTFSIIGNYQQQ 458
Query: 410 NILIGFDLDRKTISFKPTDCTN 431
N I +D R + F PT C +
Sbjct: 459 NFHILYDTKRSRLGFTPTKCAD 480
>At3g25700 unknown protein
Length = 452
Score = 156 bits (395), Expect = 2e-38
Identities = 129/458 (28%), Positives = 195/458 (42%), Gaps = 44/458 (9%)
Query: 8 FHLCFLVVLYLL--SCQTPIEAQDAGFSVQLTRQNSPHSPFYKPDN---LHRHKLPSFHQ 62
F LC + L+LL S + + + L R+ SPF P L +L
Sbjct: 6 FFLCSFLSLFLLPPSNIAAVSNHNKYLKLPLLRK----SPFPSPTQALALDTRRLHFLSL 61
Query: 63 VPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGC-YK 121
K P + S +G Y + L +G PP + + DTGSDLVW +CS C C +
Sbjct: 62 RRKPIPFVKSPVVSGAASGSGQYFVDLRIGQPPQSLLLIADTGSDLVWVKCSACRNCSHH 121
Query: 122 QKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSP-------QKLCAYSYSYADSSVTKGV 174
+ +F P S TF+P C C + +P C Y Y YAD S+T G+
Sbjct: 122 SPATVFFPRHSSTFSPAHCYDPVCRLVPKPDRAPICNHTRIHSTCHYEYGYADGSLTSGL 181
Query: 175 LARETITFSSPTNGDELVVGDIIFGC-----GHSNSGAFNENDMGVIGLGGGPLSLVSQM 229
ARET + + ++G E + + FGC G S SG GV+GLG GP+S SQ+
Sbjct: 182 FARETTSLKT-SSGKEARLKSVAFGCGFRISGQSVSGTSFNGANGVMGLGRGPISFASQL 240
Query: 230 GALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGI 289
G +G+ +FS CL+ + + + G+ D + T L + T Y V L+ +
Sbjct: 241 GRRFGN-KFSYCLMDYTLSPPPTSYLIIGNGGDGISKLFFTPLLTNPLSPTFYYVKLKSV 299
Query: 290 SVGDTFVSFNSS----EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDP 345
V + + S + G ++DSGT +L + Y ++ ++ + LP+ +
Sbjct: 300 FVNGAKLRIDPSIWEIDDSGNGGTVVDSGTTLAFLAEPAYRSVIAAVR-RRVKLPIADAL 358
Query: 346 DLGTQLCYRSETNLEG--------PILTAHFEGADVQLMPIQT-FIPPKDGVFCFAMAGT 396
G LC N+ G P L F G V + P + FI ++ + C A+
Sbjct: 359 TPGFDLC----VNVSGVTKPEKILPRLKFEFSGGAVFVPPPRNYFIETEEQIQCLAIQSV 414
Query: 397 AD--GDYIFGNFAQSNILIGFDLDRKTISFKPTDCTNP 432
G + GN Q L FD DR + F C P
Sbjct: 415 DPKVGFSVIGNLMQQGFLFEFDRDRSRLGFSRRGCALP 452
>At5g10770 nucleoid DNA-binding protein cnd41 - like protein
Length = 474
Score = 153 bits (386), Expect = 2e-37
Identities = 132/404 (32%), Positives = 182/404 (44%), Gaps = 34/404 (8%)
Query: 38 RQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVD 97
R NS HS K KL + H K+ + P T +G+Y++ + LG+P D
Sbjct: 94 RVNSIHSKLSK-------KLATDHVSESKS--TDLPAKDGSTLGSGNYIVTVGLGTPKND 144
Query: 98 IYGLVDTGSDLVWAQCSPC-HGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFS-----H 151
+ + DTGSDL W QC PC CY QK P+F P S ++ + C S CGSL S
Sbjct: 145 LSLIFDTGSDLTWTQCQPCVRTCYDQKEPIFNPSKSTSYYNVSCSSAACGSLSSATGNAG 204
Query: 152 SCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNEN 211
SCS C Y Y D S + G LA+E T TN D V + FGCG +N G F
Sbjct: 205 SCSASN-CIYGIQYGDQSFSVGFLAKEKFTL---TNSD--VFDGVYFGCGENNQGLFT-G 257
Query: 212 DMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTT 271
G++GLG LS SQ Y ++ FS CL + + +G ++FG A V T
Sbjct: 258 VAGLLGLGRDKLSFPSQTATAY-NKIFSYCL---PSSASYTGHLTFGSAG--ISRSVKFT 311
Query: 272 PLVS-EEGQTPYLVTLEGISVGDTFVSFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVE 330
P+ + +G + Y + + I+VG + S+ S +IDSGT T LP + Y L
Sbjct: 312 PISTITDGTSFYGLNIVAITVGGQKLPIPST-VFSTPGALIDSGTVITRLPPKAYAALRS 370
Query: 331 ELKVQSSLLPVDNDPD-LGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPPKDGV 388
K + S P + L T + P + F GA V+L F K
Sbjct: 371 SFKAKMSKYPTTSGVSILDTCFDLSGFKTVTIPKVAFSFSGGAVVELGSKGIFYVFKISQ 430
Query: 389 FCFAMAGTADGD--YIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
C A AG +D IFGN Q + + +D + F P C+
Sbjct: 431 VCLAFAGNSDDSNAAIFGNVQQQTLEVVYDGAGGRVGFAPNGCS 474
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,651,025
Number of Sequences: 26719
Number of extensions: 499401
Number of successful extensions: 1445
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 1118
Number of HSP's gapped (non-prelim): 90
length of query: 432
length of database: 11,318,596
effective HSP length: 102
effective length of query: 330
effective length of database: 8,593,258
effective search space: 2835775140
effective search space used: 2835775140
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0588b.3


