

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0587.7
(319 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
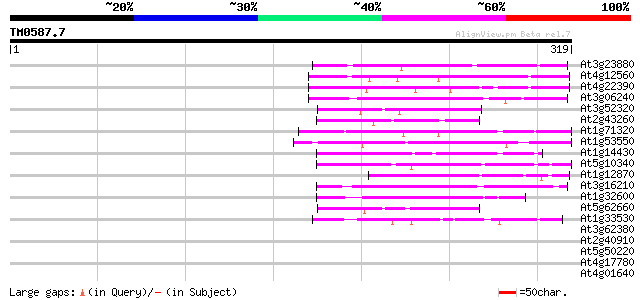
Score E
Sequences producing significant alignments: (bits) Value
At3g23880 unknown protein 79 2e-15
At4g12560 putative protein 58 6e-09
At4g22390 unknown protein 51 7e-07
At3g06240 unknown protein 51 7e-07
At3g52320 putative protein 51 1e-06
At2g43260 unknown protein 47 1e-05
At1g71320 hypothetical protein 47 2e-05
At1g53550 hypothetical protein 46 3e-05
At1g14430 hypothetical protein 45 4e-05
At5g10340 putative protein 45 5e-05
At1g12870 hypothetical protein 45 5e-05
At3g16210 hypothetical protein 45 7e-05
At1g32600 hypothetical protein 43 2e-04
At5g62660 putative protein 42 4e-04
At1g33530 hypothetical protein 41 8e-04
At3g62380 putative protein 40 0.002
At2g40910 unknown protein 40 0.002
At5g50220 putative protein 40 0.002
At4g17780 MYB transcription factor like protein 40 0.002
At4g01640 hypothetical protein 40 0.002
>At3g23880 unknown protein
Length = 364
Score = 79.3 bits (194), Expect = 2e-15
Identities = 51/147 (34%), Positives = 79/147 (53%), Gaps = 8/147 (5%)
Query: 173 SFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFP--VSLSHISQL 230
++GFGYD D YKVVA+ + H+ + K+Y WR+ SFP V ++ S+
Sbjct: 158 TYGFGYDESEDDYKVVALLQ---QRHQVKIETKIYSTRQKLWRSNTSFPSGVVVADKSRS 214
Query: 231 GVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCI 290
G+YI+GTLNW + + ++ I+S D+ +E+ + LP G L L+ CL +
Sbjct: 215 GIYINGTLNWAATSSSSSWTIISYDMSRDEFKE--LPGPVCCGRGCFTMTLGDLRGCLSM 272
Query: 291 LENNEGDNRFLVWQMKKFGVHNSWIQL 317
+ +G N VW MK+FG SW +L
Sbjct: 273 VCYCKGANAD-VWVMKEFGEVYSWSKL 298
>At4g12560 putative protein
Length = 408
Score = 58.2 bits (139), Expect = 6e-09
Identities = 46/168 (27%), Positives = 78/168 (46%), Gaps = 23/168 (13%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEERV-------VKVYDMGGSFWRNIQS---- 219
Y +G GYD ++D YKVV + F+ E+ + VKV+ + + W+ I+S
Sbjct: 137 YVFYGLGYDSVSDDYKVVRMVQ--FKIDSEDELGCSFPYEVKVFSLKKNSWKRIESVASS 194
Query: 220 ------FPVSLSHISQLGVYISGTLNWEV--KTDNYTF-IIVSLDLGTEEYSQLSLPYSP 270
F L + GV +L+W + + F +IV DL EE+ + P +
Sbjct: 195 IQLLFYFYYHLLYRRGYGVLAGNSLHWVLPRRPGLIAFNLIVRFDLALEEFEIVRFPEAV 254
Query: 271 LYGEDFEFPFLAVLKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLY 318
G + VL CLC++ N + + VW MK++ V +SW +++
Sbjct: 255 ANGNVDIQMDIGVLDGCLCLMCNYD-QSYVDVWMMKEYNVRDSWTKVF 301
>At4g22390 unknown protein
Length = 401
Score = 51.2 bits (121), Expect = 7e-07
Identities = 44/165 (26%), Positives = 76/165 (45%), Gaps = 21/165 (12%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEE----RVVKVYDMGGSFWRNI-QSFPVSLS 225
Y +G GYD + D +KVV + +E K++ VKV+ + + W+ + F +
Sbjct: 137 YVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFPCPVEVKVFSLKKNSWKRVCLMFEFQIL 196
Query: 226 HISQ---------LGVYISGTLNWEVKTDNYTF---IIVSLDLGTEEYSQLSLPYSPLYG 273
IS GV ++ L+W + I+ DL +++ LS P LY
Sbjct: 197 WISYYYHLLPRRGYGVVVNNHLHWILPRRQGVIAFNAIIKYDLASDDIGVLSFP-QELYI 255
Query: 274 EDFEFPFLAVLKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLY 318
ED + VL C+C++ +E + VW +K++ + SW +LY
Sbjct: 256 ED--NMDIGVLDGCVCLMCYDE-YSHVDVWVLKEYEDYKSWTKLY 297
>At3g06240 unknown protein
Length = 427
Score = 51.2 bits (121), Expect = 7e-07
Identities = 37/153 (24%), Positives = 72/153 (46%), Gaps = 14/153 (9%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQL 230
+ ++GFG+D + D YK+V + + ++ VY + WR I + + S
Sbjct: 211 FQTYGFGFDGLTDDYKLVKLVAT----SEDILDASVYSLKADSWRRICNLNYEHNDGSYT 266
Query: 231 -GVYISGTLNWEVKTDNYT-FIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPF----LAVL 284
GV+ +G ++W + ++V+ D+ TEE+ ++ +P ED F + L
Sbjct: 267 SGVHFNGAIHWVFTESRHNQRVVVAFDIQTEEFREMPVPDE---AEDCSHRFSNFVVGSL 323
Query: 285 KDCLCILENNEGDNRFLVWQMKKFGVHNSWIQL 317
LC++ N+ D +W M ++G SW ++
Sbjct: 324 NGRLCVV-NSCYDVHDDIWVMSEYGEAKSWSRI 355
>At3g52320 putative protein
Length = 390
Score = 50.8 bits (120), Expect = 1e-06
Identities = 32/98 (32%), Positives = 52/98 (52%), Gaps = 6/98 (6%)
Query: 176 FGYDCMNDTYKVVAVAVSCFREH---KEERVVKVYDMGGSFWRNIQSF--PVSLSHISQL 230
FG+D +D YKV+++ +E + E V V +G S WRN +S P H
Sbjct: 165 FGHDPFHDEYKVLSLFWEVTKEQTVVRSEHQVLVLGVGAS-WRNTKSHHTPHRPFHPYSR 223
Query: 231 GVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPY 268
G+ I G L + +TD +++S DL +EE++ + LP+
Sbjct: 224 GMTIDGVLYYSARTDANRCVLMSFDLSSEEFNLIELPF 261
>At2g43260 unknown protein
Length = 367
Score = 47.4 bits (111), Expect = 1e-05
Identities = 31/96 (32%), Positives = 56/96 (58%), Gaps = 7/96 (7%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVK--VYDMGGSFWRNIQSFPVSLSHISQLGV 232
GFG D +N YKVV ++ + +R +EE VV+ V D+ WR + S P + ++ V
Sbjct: 101 GFGRDKVNGRYKVVRMSFAFWRVRQEEPVVECGVLDVDTGEWRKL-SPPPYVVNVGSKSV 159
Query: 233 YISGTLNW-EVKTDNYTFIIVSLDLGTEEYSQLSLP 267
++G++ W ++T + I++LDL EE+ ++ +P
Sbjct: 160 CVNGSIYWLHIQT---VYRILALDLHKEEFHKVPVP 192
>At1g71320 hypothetical protein
Length = 392
Score = 46.6 bits (109), Expect = 2e-05
Identities = 40/160 (25%), Positives = 75/160 (46%), Gaps = 10/160 (6%)
Query: 165 VLVRFLYSSFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPV-- 222
+ V+ + S GFG D + +YKVV + + + H E KV + R++ + +
Sbjct: 181 ISVKRMPSLAGFGKDIVKKSYKVVLI-YTRYEIHDHEFKAKVLSLDNGEQRDVGFYDIHH 239
Query: 223 SLSHISQLGVYISGTLNWEV--KTDNYTFIIVSLDLGTEEYSQLSLP-YSPLYGEDFEFP 279
+ Q VY +G+L W K ++ ++++DL TEE+ + LP Y + E
Sbjct: 240 CIFCDEQTSVYANGSLFWLTLKKLSQTSYQLLAIDLHTEEFRWILLPECDTKYATNIE-- 297
Query: 280 FLAVLKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLYN 319
+ L + LC+ + E N +VW + + W ++Y+
Sbjct: 298 -MWNLNERLCLSDVLESSN-LVVWSLHQEYPTEKWEKIYS 335
>At1g53550 hypothetical protein
Length = 408
Score = 45.8 bits (107), Expect = 3e-05
Identities = 41/163 (25%), Positives = 67/163 (40%), Gaps = 15/163 (9%)
Query: 162 SEIVLVRFLYSSFGFGYDCMNDTYKVVAVAVSCFREHK--EERVVKVYDMGGSFWRNIQS 219
SE ++ + YS FGYD ++ +KV+ + H+ E V G WRN Q
Sbjct: 170 SETIIEKVRYS---FGYDPIDKQFKVLRITWLHRGSHEWSSEYQVLTLGFGNISWRNTQC 226
Query: 220 FPVSLSHISQLGVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFP 279
V + G+ I+G L + + DN + IV D+ TE++S S+ + F
Sbjct: 227 CVVHYL-LEDSGICINGVLYYPARLDNRKYTIVCFDVMTEKFSFTSIDKDMTIMTNLSFS 285
Query: 280 FL---AVLKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLYN 319
+ L C+C F +W ++ H +YN
Sbjct: 286 LIDYKGKLGACIC------DHTLFELWVLENAEEHKWSKNIYN 322
>At1g14430 hypothetical protein
Length = 849
Score = 45.4 bits (106), Expect = 4e-05
Identities = 40/131 (30%), Positives = 63/131 (47%), Gaps = 10/131 (7%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNI-QSFPVSLSHISQLGVY 233
GFG D +N TYK V + S R + +V+D + W+ I S P + VY
Sbjct: 636 GFGKDIINGTYKPVCLYNSLERGLRNATTCEVFDFRTNVWKYITPSSPYRIIAFHD-PVY 694
Query: 234 ISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLS-LPYSPLYGEDFEFPFLAVLKDCLCILE 292
+ G+L+W T+ ++SLDL TE + +S +P+ ++ + L + LCI E
Sbjct: 695 VDGSLHW--FTECKEIKVLSLDLHTETFQVISKVPFDIANCDNIA---MCNLDNRLCISE 749
Query: 293 NNEGDNRFLVW 303
N D +VW
Sbjct: 750 NKWPDQ--VVW 758
>At5g10340 putative protein
Length = 445
Score = 45.1 bits (105), Expect = 5e-05
Identities = 42/150 (28%), Positives = 74/150 (49%), Gaps = 12/150 (8%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVV-KVYDMGGSFWRNIQSFPVSLSHI--SQLG 231
GFG D ++ TYK V + S + ++ +V+D + WR + FP S I +Q
Sbjct: 231 GFGKDKISGTYKPVWLYNSAELDLNDKPTTCEVFDFATNAWRYV--FPASPHLILHTQDP 288
Query: 232 VYISGTLNW-EVKTDNYTFIIVSLDLGTEEYSQLS-LPYSPLYGEDFEFPFLAVLKDCLC 289
VY+ G+L+W + +++SLDL +E + +S P+ L D + + L D LC
Sbjct: 289 VYVDGSLHWFTALSHEGETMVLSLDLHSETFQVISKAPF--LNVSDEYYIVMCNLGDRLC 346
Query: 290 ILENNEGDNRFLVWQMKKFGVHNSWIQLYN 319
+ E + ++W + H +W Q+Y+
Sbjct: 347 VSEQKWPNQ--VIWSLDD-SDHKTWKQIYS 373
>At1g12870 hypothetical protein
Length = 416
Score = 45.1 bits (105), Expect = 5e-05
Identities = 29/116 (25%), Positives = 57/116 (49%), Gaps = 6/116 (5%)
Query: 205 KVYDMGGSFWRNIQSFPVSLSHISQLGVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQL 264
+V+D + WR + P + Q +GTL W +T N +++LD+ TE + L
Sbjct: 214 EVFDFKANAWRYLTCIPSYRIYHDQKPASANGTLYWFTETYNAEIKVIALDIHTEIFRLL 273
Query: 265 SLPYSPLYGEDFEFPFLAVLKDCLCILENNEGDNRFL--VWQMKKFGVHNSWIQLY 318
P S + + + ++ + LC+ E EGD + + +W++K ++W ++Y
Sbjct: 274 PKP-SLIASSEPSHIDMCIIDNSLCMYE-TEGDKKIIQEIWRLK--SSEDAWEKIY 325
>At3g16210 hypothetical protein
Length = 360
Score = 44.7 bits (104), Expect = 7e-05
Identities = 37/144 (25%), Positives = 62/144 (42%), Gaps = 12/144 (8%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDM-GGSFWRNIQSFPVSLSHISQLGVY 233
GFGYD ++D YKVV F + + V++ GS+ +++ + + G +
Sbjct: 134 GFGYDPVHDDYKVVT-----FIDRLDVSTAHVFEFRTGSWGESLRISYPDWHYRDRRGTF 188
Query: 234 ISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCILEN 293
+ L W + I+ +L T EY +L L P+Y + +L V LCI E
Sbjct: 189 LDQYLYWIAYRSSADRFILCFNLSTHEYRKLPL---PVYNQGVTSSWLGVTSQKLCITEY 245
Query: 294 NEGDNRFLVWQMKKFGVHNSWIQL 317
+ M+K G SW ++
Sbjct: 246 EMCKKEIRISVMEKTG---SWSKI 266
>At1g32600 hypothetical protein
Length = 293
Score = 43.1 bits (100), Expect = 2e-04
Identities = 32/120 (26%), Positives = 53/120 (43%), Gaps = 12/120 (10%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQ-LGVY 233
GFGYD + D YK+V + ++ ++ S WR + P S + G
Sbjct: 88 GFGYDRVLDDYKIVTII---------DKKTYIFTFKESSWRESKLIPSSDCFFKERTGTV 138
Query: 234 ISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCILEN 293
+ + W N I+ D EEYS+L++P L G +F +L V + LC++ +
Sbjct: 139 VDNCMYWIANRFNKEKFILCFDFVNEEYSKLNVPMM-LSGLEFN-SWLDVSRGELCVINH 196
>At5g62660 putative protein
Length = 379
Score = 42.0 bits (97), Expect = 4e-04
Identities = 31/95 (32%), Positives = 48/95 (49%), Gaps = 6/95 (6%)
Query: 176 FGYDCMNDTYKVVAVAVSCFREHKE---ERVVKVYDMGGSFWRNIQSFPVSLSHISQLGV 232
FGYD + D YKVV + K E V V + GGS W+ I+ L ++LG+
Sbjct: 190 FGYDPVLDQYKVVCTVALFSKRLKRITSEHWVFVLEPGGS-WKRIEFDQPHLP--TRLGL 246
Query: 233 YISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLP 267
++G + + T I+VS D+ +EE+S + P
Sbjct: 247 CVNGVIYYLASTWKRRDIVVSFDVRSEEFSMIQGP 281
>At1g33530 hypothetical protein
Length = 441
Score = 41.2 bits (95), Expect = 8e-04
Identities = 42/152 (27%), Positives = 78/152 (50%), Gaps = 23/152 (15%)
Query: 173 SFGFGYDCMNDTYKVVAVAVSCFREHKEERV-VKVYDMGGSFWRN--IQSFPVSLSHI-- 227
S GFG D + TYKV+ + + +RV V+D+ + WR + P+ LS I
Sbjct: 226 SVGFGIDVVTGTYKVMVL-------YGFDRVGTVVFDLDTNKWRQRYKTAGPMPLSCIPT 278
Query: 228 -SQLGVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFE----FPFLA 282
+ V+++G+L W + +D ++ I+V +DL TE++ LS P +D + + ++
Sbjct: 279 PERNPVFVNGSLFWLLASD-FSEILV-MDLHTEKFRTLSQPNDM---DDVDVSSGYIYMW 333
Query: 283 VLKDCLCILENNEGDNRFLVWQMKKFGVHNSW 314
L+D LC+ +G + + VW + + + W
Sbjct: 334 SLEDRLCVSNVRQGLHSY-VWVLVQDELSEKW 364
>At3g62380 putative protein
Length = 325
Score = 40.0 bits (92), Expect = 0.002
Identities = 39/156 (25%), Positives = 70/156 (44%), Gaps = 23/156 (14%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMG-GSFWRNIQSFPVSLSHI--SQLG 231
GFG D + TYKV+ + + + VK + + R FP+S + +
Sbjct: 126 GFGRDMVTGTYKVILMYLF------DRNFVKTEALNLKNGERRYVYFPISYGQLGNDKRS 179
Query: 232 VYISGTLNWEVKTD-----NYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKD 286
++ +G+L W K D N + ++DL TE + + LP Y + + +L LKD
Sbjct: 180 IFANGSLYWLPKPDVDFYTNQLIKLAAIDLHTETFRYVPLP--SWYTKYCKSVYLWSLKD 237
Query: 287 CLC---ILENNEGDNRFLVWQMKKFGVHNSWIQLYN 319
LC +L+N D VW +++ W ++++
Sbjct: 238 SLCVSDVLQNPSVD----VWSLQQEEPSVKWEKMFS 269
>At2g40910 unknown protein
Length = 449
Score = 40.0 bits (92), Expect = 0.002
Identities = 29/93 (31%), Positives = 51/93 (54%), Gaps = 6/93 (6%)
Query: 177 GYDCMNDTYKVV---AVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHI-SQLGV 232
G+D +ND YK+V ++ + F K E V V + GGS WR +++ H S +G
Sbjct: 213 GHDPVNDQYKLVCTIGISSAFFTNLKSEHWVFVLEAGGS-WRKVRTLESYHPHAPSTVGQ 271
Query: 233 YISGTLNWEVK-TDNYTFIIVSLDLGTEEYSQL 264
+I+G++ + + D T +VS D+ +EE + +
Sbjct: 272 FINGSVVYYMAWLDMDTCAVVSFDITSEELTTI 304
>At5g50220 putative protein
Length = 357
Score = 39.7 bits (91), Expect = 0.002
Identities = 32/144 (22%), Positives = 61/144 (42%), Gaps = 16/144 (11%)
Query: 177 GYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSF--WRNIQSFPVSLSHISQLGVYI 234
GYD + YKV+ + ++ + R V+ +G + WR IQ + L V I
Sbjct: 151 GYDPFKNQYKVICL------DNYKRRCCHVFTLGDAIRKWRKIQ-YNFGLYFPLLPPVCI 203
Query: 235 SGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCILENN 294
GT+ ++ K T++++ D+ +E++ Q+ P + + + L C
Sbjct: 204 KGTIYYQAKQYGSTYVLLCFDVISEKFDQVEAPKTMMDHRYTLINYQGKLGFMCC----- 258
Query: 295 EGDNRFLVWQMKKFGVHNSWIQLY 318
NR +W MK W +++
Sbjct: 259 --QNRVEIWVMKNDEKKQEWSKIF 280
>At4g17780 MYB transcription factor like protein
Length = 745
Score = 39.7 bits (91), Expect = 0.002
Identities = 34/134 (25%), Positives = 59/134 (43%), Gaps = 8/134 (5%)
Query: 176 FGYDCMNDTYKVVAVAVSCFRE---HKEERVVKVYDMGGSFWRNIQSFPVSLSHISQLGV 232
FG D + +YKVV + + F E E V D+ WR + P + + +
Sbjct: 498 FGVDKVTGSYKVVKMCLISFSEICARDPEVEYSVLDVETGEWRMLSPPPYKVFEVRK-SE 556
Query: 233 YISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCILE 292
+G++ W K + I++LDL EE +S+P + E F+ + L+D L I
Sbjct: 557 CANGSIYWLHKPTERAWTILALDLHKEELHNISVPDMSVTQETFQ---IVNLEDRLAIAN 613
Query: 293 N-NEGDNRFLVWQM 305
+ + + +W M
Sbjct: 614 TYTKTEWKLEIWSM 627
>At4g01640 hypothetical protein
Length = 300
Score = 39.7 bits (91), Expect = 0.002
Identities = 43/173 (24%), Positives = 73/173 (41%), Gaps = 38/173 (21%)
Query: 171 YSSFGFGYDCMNDTYKVV-------------------AVAVSCFREHKEERVVKVYDMGG 211
++ GFG D +N TYK V +++ C + +V+D
Sbjct: 68 WAKLGFGKDKINGTYKPVWLYNSAELGGLNDDDEDYDDISIICQGNNNTSTFCQVFDFTT 127
Query: 212 SFWRNIQSFPVSLSHI--SQLGVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLS-LPY 268
WR + P S I Q VY+ G+L+W T ++S DL TE + +S P+
Sbjct: 128 KAWRFV--VPASPYRILPYQDPVYVDGSLHW------LTTNVLSFDLHTETFQVVSKAPF 179
Query: 269 SPLYGEDFEFPFLAV--LKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLYN 319
L+ D+ L + L D LC+ E + ++W + H +W ++Y+
Sbjct: 180 --LHHHDYSSRELIMCNLDDRLCVSEKMWPNQ--VIWSLD--SDHKTWKEIYS 226
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.330 0.141 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,241,931
Number of Sequences: 26719
Number of extensions: 295725
Number of successful extensions: 886
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 857
Number of HSP's gapped (non-prelim): 71
length of query: 319
length of database: 11,318,596
effective HSP length: 99
effective length of query: 220
effective length of database: 8,673,415
effective search space: 1908151300
effective search space used: 1908151300
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0587.7


