

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0587.6
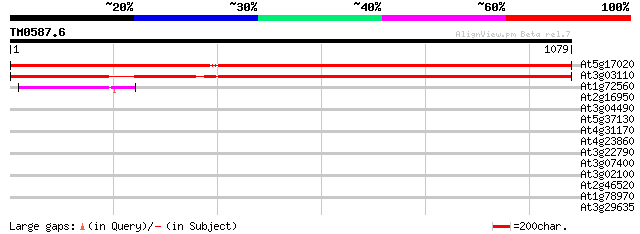
(1079 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g17020 Exportin1 (XPO1) protein 1837 0.0
At3g03110 exportin1 (XPO1) like protein 1611 0.0
At1g72560 PAUSED 46 1e-04
At2g16950 transportin like protein 42 0.002
At3g04490 hypothetical protein 38 0.027
At5g37130 putative protein 32 2.5
At4g31170 protein kinase - like protein 31 3.3
At4g23860 unknown protein 31 3.3
At3g22790 unknown protein 31 4.3
At3g07400 hypothetical protein 31 4.3
At3g02100 putative UDP-glucosyl transferase 31 4.3
At2g46520 putative cellular apoptosis susceptibility protein 31 4.3
At1g78970 unknown protein 31 4.3
At3g29635 hypothetical protein, 5' partial 30 9.7
>At5g17020 Exportin1 (XPO1) protein
Length = 1075
Score = 1837 bits (4759), Expect = 0.0
Identities = 919/1079 (85%), Positives = 996/1079 (92%), Gaps = 4/1079 (0%)
Query: 1 MAAEKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQN 60
MAAEKLRDLSQPIDV +LDATVAAF+ TGSKE+R AADQILRDLQ NPDMWLQV+HILQN
Sbjct: 1 MAAEKLRDLSQPIDVGVLDATVAAFFVTGSKEERAAADQILRDLQANPDMWLQVVHILQN 60
Query: 61 TQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNK 120
T +L+TKFFALQVLEGVIKYRWNALP+EQRDGMKN+IS++IVQLS NEASFR ERLYVNK
Sbjct: 61 TNSLDTKFFALQVLEGVIKYRWNALPVEQRDGMKNYISEVIVQLSSNEASFRSERLYVNK 120
Query: 121 LNIILVQILKHEWPLRWRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQ 180
LN+ILVQI+KH+WP +W SFIPDLV+AAKTSETICENCMAILKLLSEEVFDFSRGEMTQQ
Sbjct: 121 LNVILVQIVKHDWPAKWTSFIPDLVAAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQ 180
Query: 181 KIKELKQSLNSEFQLIHELCLYVLSASQRAELIRATLSTLHAFLSWIPLGYIFESPLLET 240
KIKELKQSLNSEF+LIHELCLYVLSASQR +LIRATLS LHA+LSWIPLGYIFES LLET
Sbjct: 181 KIKELKQSLNSEFKLIHELCLYVLSASQRQDLIRATLSALHAYLSWIPLGYIFESTLLET 240
Query: 241 LLKFFPLPVYRNLTLKCLIEVAALQFGDYYDTQYTKMYNIFMVQLQTILLPTTNIPEAYA 300
LLKFFP+P YRNLT++CL EVAAL FGD+Y+ QY KMY IF+ QL+ IL P+T IPEAY+
Sbjct: 241 LLKFFPVPAYRNLTIQCLTEVAALNFGDFYNVQYVKMYTIFIGQLRIILPPSTKIPEAYS 300
Query: 301 QGSSEEQTFIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFKVC 360
GS EEQ FIQNLALFFTSF+K HIR+LEST E ++ LL+GLEYLINISYVDDTEVFKVC
Sbjct: 301 SGSGEEQAFIQNLALFFTSFFKFHIRVLESTPEVVSLLLAGLEYLINISYVDDTEVFKVC 360
Query: 361 LDYWNILVSELFEPHRSLENPAAAATNMLGNQQAPLMPPGMVDGLGSQLLQRRQLYAGPM 420
LDYWN LV ELF+ H + +NPA +A+ ++G Q P +P GMVDGLGSQ++QRRQLY+ PM
Sbjct: 361 LDYWNSLVLELFDAHHNSDNPAVSAS-LMGLQ--PFLP-GMVDGLGSQVMQRRQLYSHPM 416
Query: 421 SKLRTLMICRMAKPEEVLIIEDENGNIVRETLKDNDVLVQYKIMRETLIYLSHLDHDDTE 480
SKLR LMI RMAKPEEVLI+EDENGNIVRET+KDNDVLVQYKIMRETLIYLSHLDHDDTE
Sbjct: 417 SKLRGLMINRMAKPEEVLIVEDENGNIVRETMKDNDVLVQYKIMRETLIYLSHLDHDDTE 476
Query: 481 RQMLRKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKG 540
+QMLRKL+KQL GE+W WNNLNTLCWAIGSISGSM EDQENRFLVMVIRDLLNLCE TKG
Sbjct: 477 KQMLRKLNKQLSGEEWAWNNLNTLCWAIGSISGSMAEDQENRFLVMVIRDLLNLCEITKG 536
Query: 541 KDNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQ 600
KDNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHE+HPGVQDMACDTFLKIVQ
Sbjct: 537 KDNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHETHPGVQDMACDTFLKIVQ 596
Query: 601 KCKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEY 660
KCKRKFVI Q+GENEPFVSELL+GL+TT+ DLEPHQIH+FYESVG+M+QAESD QKRDEY
Sbjct: 597 KCKRKFVIVQVGENEPFVSELLTGLATTVQDLEPHQIHSFYESVGNMIQAESDPQKRDEY 656
Query: 661 LQRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQISLI 720
LQRLM LPNQKW EIIGQAR +V+FLKD VIRTVLNILQTNTS A+SLGTYFL QISLI
Sbjct: 657 LQRLMALPNQKWAEIIGQARHSVEFLKDQVVIRTVLNILQTNTSAATSLGTYFLSQISLI 716
Query: 721 FLDMLNVYRMYSELISKSIAEGGPYASRTSFVKLLRSVKRETLKLIETFLDKAEDQPQIG 780
FLDMLNVYRMYSEL+S +I EGGPYAS+TSFVKLLRSVKRETLKLIETFLDKAEDQP IG
Sbjct: 717 FLDMLNVYRMYSELVSTNITEGGPYASKTSFVKLLRSVKRETLKLIETFLDKAEDQPHIG 776
Query: 781 KQFVPPMMDPVLGDYARNVPDARESEVLSLFATIVNKYKAAMVEDVPRIFEAVFQCTLEM 840
KQFVPPMM+ VLGDYARNVPDARESEVLSLFATI+NKYKA M++DVP IFEAVFQCTLEM
Sbjct: 777 KQFVPPMMESVLGDYARNVPDARESEVLSLFATIINKYKATMLDDVPHIFEAVFQCTLEM 836
Query: 841 ITKNFEDYPEHRLKFFSLLCAIATHCFHALICLSSQQLKFVMDSIIWAFRHTERNIAETG 900
ITKNFEDYPEHRLKFFSLL AIAT CF ALI LSS QLK VMDSIIWAFRHTERNIAETG
Sbjct: 837 ITKNFEDYPEHRLKFFSLLRAIATFCFPALIKLSSPQLKLVMDSIIWAFRHTERNIAETG 896
Query: 901 LNLLLEMLKKFQATEFCNQFYRTYFLTIEQEIFAVLTDTFHKPGFKLHVLVLQHLFCLAE 960
LNLLLEMLK FQ +EFCNQFYR+YF+ IEQEIFAVLTDTFHKPGFKLHVLVLQ LFCL E
Sbjct: 897 LNLLLEMLKNFQQSEFCNQFYRSYFMQIEQEIFAVLTDTFHKPGFKLHVLVLQQLFCLPE 956
Query: 961 TGALTEPLWDAATNPYPYPSNAAFVLEYTIKLLSTSFPNMTAAEVTQFVNGLFESTNDLS 1020
+GALTEPLWDA T PYPYP N AFV EYTIKLLS+SFPNMTAAEVTQFVNGL+ES ND S
Sbjct: 957 SGALTEPLWDATTVPYPYPDNVAFVREYTIKLLSSSFPNMTAAEVTQFVNGLYESRNDPS 1016
Query: 1021 TFKNHIRDFLVQSKEFSAQDNKDLYAEEAAAQRERERQRMLSVPGLIAPSELQDEMLDS 1079
FKN+IRDFLVQSKEFSAQDNKDLYAEEAAAQRERERQRMLS+PGLIAP+E+QDEM+DS
Sbjct: 1017 GFKNNIRDFLVQSKEFSAQDNKDLYAEEAAAQRERERQRMLSIPGLIAPNEIQDEMVDS 1075
>At3g03110 exportin1 (XPO1) like protein
Length = 1022
Score = 1611 bits (4172), Expect = 0.0
Identities = 827/1081 (76%), Positives = 913/1081 (83%), Gaps = 61/1081 (5%)
Query: 1 MAAEKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQN 60
MAAEKLRDLSQPIDV LLDATV AFY TGSKE+R +AD ILRDL+ NPD WLQV+HILQN
Sbjct: 1 MAAEKLRDLSQPIDVVLLDATVEAFYSTGSKEERASADNILRDLKANPDTWLQVVHILQN 60
Query: 61 TQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNK 120
T + +TKFFALQVLEGVIKYRWNALP+EQRDGMKN+ISD+IVQLS +EASFR ERLYVNK
Sbjct: 61 TSSTHTKFFALQVLEGVIKYRWNALPVEQRDGMKNYISDVIVQLSRDEASFRTERLYVNK 120
Query: 121 LNIILVQILKHEWPLRWRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQ 180
LNIILVQI+K EWP +W+SFIPDLV AAKTSETICENCMAILKLLSEEVFDFS+GEMTQQ
Sbjct: 121 LNIILVQIVKQEWPAKWKSFIPDLVIAAKTSETICENCMAILKLLSEEVFDFSKGEMTQQ 180
Query: 181 KIKELKQSLNSEFQLIHELCLYVLSASQRAELIRATLSTLHAFLSWIPLGYIFESPLLET 240
KIKELKQSLN + LE
Sbjct: 181 KIKELKQSLNRQ---------------------------------------------LEI 195
Query: 241 LLKFFPLPVYRNLTLKCLIEVAALQFGDYYDTQYTKMYNIFMVQLQTILLPTTNIPEAYA 300
LLKFFP+P YRNLTL+CL EVA+L FGD+YD QY KMY+IFM QLQ IL NIPEAY+
Sbjct: 196 LLKFFPVPAYRNLTLQCLSEVASLNFGDFYDMQYVKMYSIFMNQLQAILPLNLNIPEAYS 255
Query: 301 QGSSEEQT--FIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFK 358
GSSEEQ FIQNLALFFTSF+K+HI+ILES ENI+ LL+GL YLI+ISYVDDTEVFK
Sbjct: 256 TGSSEEQASAFIQNLALFFTSFFKLHIKILESAPENISLLLAGLGYLISISYVDDTEVFK 315
Query: 359 VCLDYWNILVSELFEPHRSLENPAAAATNMLGNQQAPLMPPGMVDGLGSQLLQRRQLYAG 418
P S++ + +L ++Q +P VDG+ S++ +R++LY+
Sbjct: 316 N-------------GPMLSVDALETDSFCLLTSEQMAFLP-STVDGVKSEVTERQKLYSD 361
Query: 419 PMSKLRTLMICRMAKPEEVLIIEDENGNIVRETLKDNDVLVQYKIMRETLIYLSHLDHDD 478
PMSKLR LMI R AKPEEVLI+EDENGNIVRET+KDNDVLVQYKIMRETLIYLSHLDH+D
Sbjct: 362 PMSKLRGLMISRTAKPEEVLIVEDENGNIVRETMKDNDVLVQYKIMRETLIYLSHLDHED 421
Query: 479 TERQMLRKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFT 538
TE+QML KLSKQL GE+W WNNLNTLCWAIGSISGSM+ +QENRFLVMVIRDLL+LCE
Sbjct: 422 TEKQMLSKLSKQLSGEEWAWNNLNTLCWAIGSISGSMVVEQENRFLVMVIRDLLSLCEVV 481
Query: 539 KGKDNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKI 598
KGKDNKAVIASNIMYVVGQY RFLRAHWKFLKTVV+KLFEFMHE+HPGVQDMACDTFLKI
Sbjct: 482 KGKDNKAVIASNIMYVVGQYSRFLRAHWKFLKTVVHKLFEFMHETHPGVQDMACDTFLKI 541
Query: 599 VQKCKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRD 658
VQKCKRKFVI Q+GE+EPFVSELLSGL+T + DL+PHQIHTFYESVGSM+QAESD QKR
Sbjct: 542 VQKCKRKFVIVQVGESEPFVSELLSGLATIVGDLQPHQIHTFYESVGSMIQAESDPQKRG 601
Query: 659 EYLQRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQIS 718
EYLQRLM LPNQKW EIIGQARQ+ D LK+ DVIRTVLNILQTNT VA+SLGT+FL QIS
Sbjct: 602 EYLQRLMALPNQKWAEIIGQARQSADILKEPDVIRTVLNILQTNTRVATSLGTFFLSQIS 661
Query: 719 LIFLDMLNVYRMYSELISKSIAEGGPYASRTSFVKLLRSVKRETLKLIETFLDKAEDQPQ 778
LIFLDMLNVYRMYSEL+S SIA GGPYASRTS VKLLRSVKRE LKLIETFLDKAE+QP
Sbjct: 662 LIFLDMLNVYRMYSELVSSSIANGGPYASRTSLVKLLRSVKREILKLIETFLDKAENQPH 721
Query: 779 IGKQFVPPMMDPVLGDYARNVPDARESEVLSLFATIVNKYKAAMVEDVPRIFEAVFQCTL 838
IGKQFVPPMMD VLGDYARNVPDARESEVLSLFATI+NKYK M ++VP IFEAVFQCTL
Sbjct: 722 IGKQFVPPMMDQVLGDYARNVPDARESEVLSLFATIINKYKVVMRDEVPLIFEAVFQCTL 781
Query: 839 EMITKNFEDYPEHRLKFFSLLCAIATHCFHALICLSSQQLKFVMDSIIWAFRHTERNIAE 898
EMITKNFEDYPEHRLKFFSLL AIAT CF ALI LSS+QLK VMDS+IWAFRHTERNIAE
Sbjct: 782 EMITKNFEDYPEHRLKFFSLLRAIATFCFRALIQLSSEQLKLVMDSVIWAFRHTERNIAE 841
Query: 899 TGLNLLLEMLKKFQATEFCNQFYRTYFLTIEQEIFAVLTDTFHKPGFKLHVLVLQHLFCL 958
TGLNLLLEMLK FQ ++FCN+FY+TYFL IEQE+FAVLTDTFHKPGFKLHVLVLQHLF L
Sbjct: 842 TGLNLLLEMLKNFQKSDFCNKFYQTYFLQIEQEVFAVLTDTFHKPGFKLHVLVLQHLFSL 901
Query: 959 AETGALTEPLWDAATNPYPYPSNAAFVLEYTIKLLSTSFPNMTAAEVTQFVNGLFESTND 1018
E+G+L EPLWDAAT P+PY +N AFVLEYT KLLS+SFPNMT EVTQFVNGL+ES ND
Sbjct: 902 VESGSLAEPLWDAATVPHPYSNNVAFVLEYTTKLLSSSFPNMTTTEVTQFVNGLYESRND 961
Query: 1019 LSTFKNHIRDFLVQSKEFSAQDNKDLYAEEAAAQRERERQRMLSVPGLIAPSELQDEMLD 1078
+ FK++IRDFL+QSKEFSAQDNKDLYAEEAAAQ ERERQRMLS+PGLIAPSE+QD+M D
Sbjct: 962 VGRFKDNIRDFLIQSKEFSAQDNKDLYAEEAAAQMERERQRMLSIPGLIAPSEIQDDMAD 1021
Query: 1079 S 1079
S
Sbjct: 1022 S 1022
>At1g72560 PAUSED
Length = 988
Score = 46.2 bits (108), Expect = 1e-04
Identities = 45/241 (18%), Positives = 106/241 (43%), Gaps = 18/241 (7%)
Query: 18 LDATVAAFYGTGSKEQ--RTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLE 75
L+ + + TG+ + ++ A + ++ P + + L ++ + +F+ LQ L+
Sbjct: 4 LEQAIVISFETGAVDSALKSQAVTYCQQIKETPSICSICIEKLWFSKLVQVQFWCLQTLQ 63
Query: 76 GVIKYRWNALPLEQRDGM-KNFISDIIVQLSGNEASFRMER---LYVNKLNIILVQILKH 131
V++ ++ ++ L+++ + K+ S +++ NE + R+ NKL +L ++ +
Sbjct: 64 DVLRVKYGSMSLDEQSYVRKSVFSMACLEVIDNENAGRVVEGPPFVKNKLAQVLATLIYY 123
Query: 132 EWPLRWRSFIPDLVSAAKTSETICENCMAILKLLSEEVF--DFSRGEMTQQKIKELKQSL 189
E+PL W S D + + + +L L +E+ D+ R +K ++
Sbjct: 124 EYPLIWSSVFLDFMLHLCKGAVVIDMFCRVLNALDDELISLDYPRTPEEISVAARVKDAM 183
Query: 190 NSEFQLIHELC-----LYVLSASQRAELIRATLSTLHAFLSWIPLGYIFES---PLLETL 241
Q + ++ + + + +L L + F+SWI +G + PLL L
Sbjct: 184 RQ--QCVPQIARAWYDIVSMYKNSDPDLSATVLDCMRRFVSWIDIGLVANDAFVPLLFEL 241
Query: 242 L 242
+
Sbjct: 242 I 242
>At2g16950 transportin like protein
Length = 827
Score = 41.6 bits (96), Expect = 0.002
Identities = 55/295 (18%), Positives = 115/295 (38%), Gaps = 44/295 (14%)
Query: 471 LSHLDHDDTERQMLRKLSKQLGGE-DWTWNNLNTLCWAIGSISGSMMED-----QENRFL 524
LS++ D+ ++ + K L D W A+G+I+ M E +
Sbjct: 379 LSNVFGDEILPALMPLIQKNLSASGDEAWKQREAAVLALGAIAEGCMNGLYPHLSEASLI 438
Query: 525 VMVIRDLLNLCEFTKGKDNKAVIASNIMYVVGQYPRFL-------RAHWKFLKTVVNKLF 577
V + LL+ D +I S + + ++ ++L + + +F K ++ L
Sbjct: 439 VAFLLPLLD--------DKFPLIRSISCWTLSRFGKYLIQESGNPKGYEQFEKVLMGLLR 490
Query: 578 EFMHESHPGVQDMACDTFLKIVQKCKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQI 637
+ +++ VQ+ AC F + + + V P + +L L + +
Sbjct: 491 RLL-DTNKRVQEAACSAFATVEEDAAEELV--------PHLGVILQHLMCAFGKYQRRNL 541
Query: 638 HTFYESVGSMVQAESDSQKRDEYLQRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLN 697
Y+++G++ + + + YL+ LM KW Q N D + +
Sbjct: 542 RIVYDAIGTLADSVREELNKPAYLEILMPPLVAKW-----QQLSNSD--------KDLFP 588
Query: 698 ILQTNTSVASSLGTYFLPQISLIFLDMLNVYRMYSELISKSIAEGGPYASRTSFV 752
+L+ TS++ +LG F P +F +++ ++ +L + A G R V
Sbjct: 589 LLECFTSISQALGVGFAPFAQPVFQRCMDIIQL-QQLAKVNPASAGAQYDREFIV 642
>At3g04490 hypothetical protein
Length = 1248
Score = 38.1 bits (87), Expect = 0.027
Identities = 21/98 (21%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query: 36 AADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKN 95
AA+ + L +P + +IL+N+Q N +F A + W+ L + + G+ +
Sbjct: 256 AAEATILSLHQSPQPYKACRYILENSQVANARFQAAAAIRESAIREWSFLATDDKGGLIS 315
Query: 96 FISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEW 133
F ++Q + + E ++K++ + Q++K W
Sbjct: 316 FCLGYVMQHANSS-----EGYVLSKVSSVAAQLMKRGW 348
>At5g37130 putative protein
Length = 887
Score = 31.6 bits (70), Expect = 2.5
Identities = 48/199 (24%), Positives = 80/199 (40%), Gaps = 26/199 (13%)
Query: 30 SKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLEGVIKYRWNALPLEQ 89
SKE A ++L+ +++ +W H+ + N + F A+Q + + + + ++ L
Sbjct: 629 SKESFIAFKEVLKLNRDSWQIWENFSHVAMDVGNTDQAFEAIQQIMRLTQNKSISVVLLD 688
Query: 90 RDGMKNFISDIIVQLSGNE----ASFRMERLYVNKLNIILVQILKHE-----WPL--RWR 138
R +I + S NE ERLY+ I+ QI+K E W L RW
Sbjct: 689 RLMTDLENRNISYESSSNELIKTKPTTTERLYIELFGKIIQQIVKTESTFENWGLYARWS 748
Query: 139 SFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKIKELKQSLNSEFQLIHE 198
DL TIC A+LK +V + EM + K + K+ + +L
Sbjct: 749 RINGDL--------TICSE--ALLK----QVRSYLGVEMWKDK-ERFKKFARASLELCRV 793
Query: 199 LCLYVLSASQRAELIRATL 217
S + EL A +
Sbjct: 794 YIEISASVESKRELFSAEM 812
>At4g31170 protein kinase - like protein
Length = 412
Score = 31.2 bits (69), Expect = 3.3
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query: 784 VPPMMDPVLGDYARNVPDARESEVLSLFATIVNKYKAAMVEDVPRIFEAVFQCTL 838
VP PVLG+ DA + EV FA IVN +AA E + + +A F+C +
Sbjct: 352 VPADCLPVLGEIMTRCWDA-DPEVRPCFAEIVNLLEAAETEIMTNVRKARFRCCM 405
>At4g23860 unknown protein
Length = 452
Score = 31.2 bits (69), Expect = 3.3
Identities = 15/46 (32%), Positives = 24/46 (51%)
Query: 570 KTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVITQLGENE 615
K+VV K E + +S PG + + +VQKC K ++ G+ E
Sbjct: 246 KSVVGKCSETISDSEPGQPENGTEAEKSVVQKCSEKIDESEAGQPE 291
>At3g22790 unknown protein
Length = 1694
Score = 30.8 bits (68), Expect = 4.3
Identities = 17/60 (28%), Positives = 31/60 (51%)
Query: 403 DGLGSQLLQRRQLYAGPMSKLRTLMICRMAKPEEVLIIEDENGNIVRETLKDNDVLVQYK 462
+ L + Q Y+ + K ++L + E+ I+E+ENG I+ E + N+V V Y+
Sbjct: 1023 ENLHESYMALHQDYSDALGKNKSLHLKFSELKGEICILEEENGAILEEAIALNNVSVVYQ 1082
>At3g07400 hypothetical protein
Length = 1003
Score = 30.8 bits (68), Expect = 4.3
Identities = 22/75 (29%), Positives = 33/75 (43%), Gaps = 3/75 (4%)
Query: 176 EMTQQKIKELKQSLNSEFQLIHELCLYV---LSASQRAELIRATLSTLHAFLSWIPLGYI 232
E+ ++ +S S FQ IH+LC+ V Q+ + A I LG+I
Sbjct: 490 ELRERLQSHSMKSYRSRFQRIHDLCMDVDGFFGVDQQKQFPHLQQWLGLAVGGSIELGHI 549
Query: 233 FESPLLETLLKFFPL 247
ESP++ T PL
Sbjct: 550 VESPVIRTATSIAPL 564
>At3g02100 putative UDP-glucosyl transferase
Length = 464
Score = 30.8 bits (68), Expect = 4.3
Identities = 25/93 (26%), Positives = 43/93 (45%), Gaps = 5/93 (5%)
Query: 364 WNILVSELFEPHRSLENPAAAATNMLGNQQAPLMPPGMVDGLGSQLLQRRQLYAGPMSKL 423
W I V+ F R+ PAAAA+ +LG L+ G++D G+ + + + M K+
Sbjct: 133 WAIEVAAKFGIRRTAFCPAAAASMVLGFSIQKLIDDGLIDSDGTVRVNKTIQLSPGMPKM 192
Query: 424 RT---LMICRMAKPEEVLIIE--DENGNIVRET 451
T + +C K + I + +N N + T
Sbjct: 193 ETDKFVWVCLKNKESQKNIFQLMLQNNNSIEST 225
>At2g46520 putative cellular apoptosis susceptibility protein
Length = 972
Score = 30.8 bits (68), Expect = 4.3
Identities = 42/171 (24%), Positives = 75/171 (43%), Gaps = 20/171 (11%)
Query: 32 EQRTAADQILRDLQNNPDMWLQVMHILQNTQ-NLNTKFFALQVLEGVIKYRWNAL----- 85
E R A++ L D + + L V+ ++ + T+ A + ++ RW+
Sbjct: 24 EPRRTAERALSDAADQANYGLAVLRLVAEPAIDEQTRHAAAVNFKNHLRSRWHPAGDSGI 83
Query: 86 -PLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEWPLRWRSFIPDL 144
P+ D K I +IV L + AS R++ ++L+ L I KH++P W + +P+L
Sbjct: 84 SPIV--DSEKEQIKTLIVSLMLS-ASPRIQ----SQLSEALTVIGKHDFPKAWPALLPEL 136
Query: 145 V----SAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKIKELKQSLNS 191
+ +AA + + N IL S FS T +LK L++
Sbjct: 137 IANLQNAALAGDYVSVN--GILGTASSIFKKFSYEYRTDALFVDLKYCLDN 185
>At1g78970 unknown protein
Length = 757
Score = 30.8 bits (68), Expect = 4.3
Identities = 28/122 (22%), Positives = 50/122 (40%), Gaps = 12/122 (9%)
Query: 565 HWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVITQLGENEPFVSELLSG 624
+WK + + K E M+ +HP VQD+ DT V+ ++ + +L V E
Sbjct: 297 NWKKSRRLYAK--EDMYYAHPLVQDLLSDTLQNFVEPLLTRWPLNKL------VREKALQ 348
Query: 625 LSTTIVDLEPHQIHTF----YESVGSMVQAESDSQKRDEYLQRLMHLPNQKWLEIIGQAR 680
L+ + E H E V M+ ++ D + + L +P+ W+ G
Sbjct: 349 LTMKHIHYEDENSHYITIGCVEKVLCMLACWVENPNGDYFKKHLARIPDYMWVAEDGMKM 408
Query: 681 QN 682
Q+
Sbjct: 409 QS 410
>At3g29635 hypothetical protein, 5' partial
Length = 458
Score = 29.6 bits (65), Expect = 9.7
Identities = 20/74 (27%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query: 780 GKQFVPPMMDPVLGDYARNVP---DARESEVLSLFATIVNKYKAAMVEDVPRIFEAVFQC 836
G +P + PVL NVP DA+ E+LS F+ + + +++ + I + +
Sbjct: 188 GAMDLPEDLTPVLDRTVINVPASLDAKIIELLSYFSEVKDSFRSLKLLPPKEISPDLVRI 247
Query: 837 TLEMITKNFEDYPE 850
+LE+ +N E E
Sbjct: 248 SLELTRENIEKLRE 261
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,663,524
Number of Sequences: 26719
Number of extensions: 941167
Number of successful extensions: 2459
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 2437
Number of HSP's gapped (non-prelim): 24
length of query: 1079
length of database: 11,318,596
effective HSP length: 110
effective length of query: 969
effective length of database: 8,379,506
effective search space: 8119741314
effective search space used: 8119741314
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0587.6


