

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.7
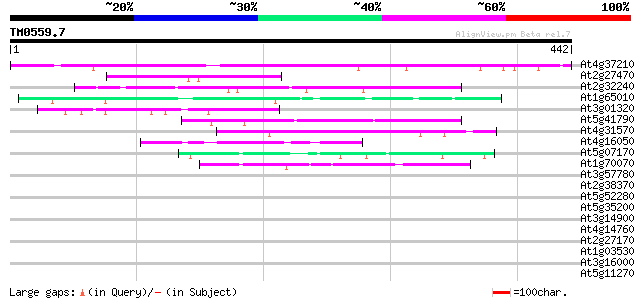
(442 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g37210 unknown protein 353 1e-97
At2g27470 putative CCAAT-binding transcription factor subunit 59 5e-09
At2g32240 putative myosin heavy chain 47 2e-05
At1g65010 hypothetical protein 46 4e-05
At3g01320 unknown protein 45 8e-05
At5g41790 myosin heavy chain-like protein 43 3e-04
At4g31570 putative protein 43 3e-04
At4g16050 hypothetical protein 43 4e-04
At5g07170 putative protein 42 7e-04
At1g70070 Similar to Synechocystis antiviral protein (At1g70070) 42 9e-04
At3g57780 putative protein 41 0.001
At2g38370 hypothetical protein 41 0.001
At5g52280 hyaluronan mediated motility receptor-like protein 41 0.002
At5g35200 unknown protein 41 0.002
At3g14900 hypothetical protein 41 0.002
At4g14760 centromere protein homolog 40 0.002
At2g27170 putative chromosome associated protein 40 0.002
At1g03530 unknown protein 40 0.002
At3g16000 myosin heavy chain like protein 40 0.003
At5g11270 unknown protein 40 0.003
>At4g37210 unknown protein
Length = 492
Score = 353 bits (906), Expect = 1e-97
Identities = 223/508 (43%), Positives = 303/508 (58%), Gaps = 82/508 (16%)
Query: 1 MVEE-APASETSVSMAPSEEAVAVDGTLNPSEHGKSEITNGATVDSAAIGGAESASNAET 59
MVEE A ASE SV +E A + TL P+ ATV+S GG ES N +
Sbjct: 1 MVEESASASEASVIQTLTEPATEIAQTLEPNLASIE-----ATVESVVQGGTESTCNNDA 55
Query: 60 SGKNS----------------LELAVELMDKGTKAMKEDDFGEAAENFSRALEIRVANYG 103
+ N+ LE A EL +KG+ +KE+DF EA + FSRALEIRVA+YG
Sbjct: 56 NNNNAADSAATEVCDEEREKTLEFAEELTEKGSVFLKENDFAEAVDCFSRALEIRVAHYG 115
Query: 104 ELAPECVNTYYKYGCALLYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTA 163
EL EC+N YY+YG ALL KAQ EADPLG++PKK+ Q SS ES+ ++
Sbjct: 116 ELDAECINAYYRYGLALLAKAQAEADPLGNMPKKEGEVQQESSNGESLAPSV-------- 167
Query: 164 ASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIA 223
S + E+ +S+ QE + + ++ ++ D+D D +ADEDESDLD+AWKMLDIA
Sbjct: 168 --VSGDPERQGSSSGQEGSGGKDQGEDGEDCQDDDLSDADGDADEDESDLDMAWKMLDIA 225
Query: 224 RAIVEKQCVNTIEHVDILSTLAEISLEREEFENSLSDYQKALSILEQLVD---------- 273
R I +KQ T+E VDIL +LAE+SLERE+ E+SLSDY+ ALSILE+LV+
Sbjct: 226 RVITDKQSTETMEKVDILCSLAEVSLEREDIESSLSDYKNALSILERLVEPDSRRTAELN 285
Query: 274 ----------SKPEEAIAYCEKAASVCKARLDRLTNEVKSITPASEAK------------ 311
+P+EAI YC+KA +CKAR++RL+NE+K + ++ +
Sbjct: 286 FRICICLETGCQPKEAIPYCQKALLICKARMERLSNEIKGASGSATSSTVSEIDEGIQQS 345
Query: 312 ------NKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKVAAKAGGGK 365
+KS DK+ EI L GL+ DLE KLEDL+ NPK L+EL+ V+AK
Sbjct: 346 SNVPYIDKSASDKEVEIGDLAGLAEDLEKKLEDLKQQAENPKQVLAELMGMVSAKPNASD 405
Query: 366 ESIP--AKVSSSQLATANSSGGFD--SPTISTAHS----NGSAAGVTHLGVVGRGVKR-- 415
+ +P A++SSS++ T N++ G D SPT+STAH+ G+A+GVTHLGVVGRGVKR
Sbjct: 406 KVVPAAAEMSSSRMGTVNTNFGKDLESPTVSTAHTGAAGGGAASGVTHLGVVGRGVKRVL 465
Query: 416 -TSLAPEASAPKKPALESTEGKGDSGSA 442
+ + E+SA KKPALE ++ K D S+
Sbjct: 466 MNTTSIESSASKKPALEFSD-KADGNSS 492
>At2g27470 putative CCAAT-binding transcription factor subunit
Length = 275
Score = 58.9 bits (141), Expect = 5e-09
Identities = 44/145 (30%), Positives = 68/145 (46%), Gaps = 7/145 (4%)
Query: 77 KAMKEDDFGEAAENFSRALE-IRVANYGELAPECVNTYYKYGCALLYKAQEEADPLGDVP 135
KA++E DF E E +LE + N G+ A +Y G AL + + P
Sbjct: 77 KALEEMDFSEFLEPLKSSLEDFKKKNAGKKAGAAAASYPAGGAALKSSSGTASKPKETKK 136
Query: 136 KKQE--GSQHGSSK---DESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDEST-K 189
+KQE +Q G+ K DE K E+ + N E+D ND+E DDE+T +
Sbjct: 137 RKQEEPSTQKGARKSKIDEETKRNDEETENDNTEEENGNDEEDENGNDEEDENDDENTEE 196
Query: 190 NYQEEDDEDSDAEDTEADEDESDLD 214
N +E+++D + E+ DE+ D
Sbjct: 197 NGNDEENDDENTEENGNDEENEKED 221
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 47.4 bits (111), Expect = 2e-05
Identities = 74/324 (22%), Positives = 134/324 (40%), Gaps = 28/324 (8%)
Query: 52 ESASNAETSGKNSLELAVELMDKGTKAMKEDDFGEAAENFSRALEIRVANYGELAPECVN 111
+SAS+A++ + +LE + EL+ ++ KE + A S EI+ N E V
Sbjct: 223 QSASHADSESQKALEFS-ELLKSTKESAKEMEEKMA----SLQQEIKELNEKMSENEKVE 277
Query: 112 TYYKYGCALLYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAE 171
K L QEE + E Q SS + + E A+ E
Sbjct: 278 AALKSSAGELAAVQEELAL--SKSRLLETEQKVSSTEALIDELTQELEQKKASESRFKEE 335
Query: 172 ----QDIASND---QESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIAR 224
QD+ + Q + E + E+ ++ + ++ + + E L A + L A
Sbjct: 336 LSVLQDLDAQTKGLQAKLSEQEGINSKLAEELKEKELLESLSKDQEEKLRTANEKL--AE 393
Query: 225 AIVEKQCV--NTIEHVDILSTLAEISLEREE-FENSLSDYQKALSILEQLVDSKPE---- 277
+ EK+ + N E ++T+ E+ E EE + S ++ K ++L Q + + E
Sbjct: 394 VLKEKEALEANVAEVTSNVATVTEVCNELEEKLKTSDENFSKTDALLSQALSNNSELEQK 453
Query: 278 -----EAIAYCEKAASVCKARLDRLTNEVKSITPASEAKNKSIEDKQAEIETLTGLSSDL 332
E + AA+ + L + V+S + A+E I++ + + +++L
Sbjct: 454 LKSLEELHSEAGSAAAAATQKNLELEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAEL 513
Query: 333 ENKLEDLQLLVSNPKSTLSELLEK 356
E +L LQL S+ + L EL EK
Sbjct: 514 EQQLNLLQLKSSDAERELKELSEK 537
Score = 34.7 bits (78), Expect = 0.11
Identities = 86/383 (22%), Positives = 163/383 (42%), Gaps = 66/383 (17%)
Query: 1 MVEEAPASETSVSMAPSEEAVAVDGTLNPSEHGKSEITNGATVDSAAIGGAESASNAETS 60
+ EE T+ ++A ++ +L S SE+ + A GAE A T+
Sbjct: 547 VAEEEKKQATTQMQEYKQKASELELSLTQSSARNSELEEDLRI--ALQKGAEHEDRANTT 604
Query: 61 GKNSLELAVELMDKGTKAMKEDDFGEAAENFSRALEIRVANYGELAPECVNTYYKYGCAL 120
+ S+EL E + + +++ ED AE + LE+ +
Sbjct: 605 HQRSIEL--EGLCQSSQSKHED-----AEGRLKDLELLLQTEK----------------- 640
Query: 121 LYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAE-SSTAASFSSNAEQDIASNDQ 179
Y+ QE + + + KK HG ++ +S AE ST +F + S+
Sbjct: 641 -YRIQELEEQVSSLEKK-----HGETEADSKGYLGQVAELQSTLEAFQ------VKSSSL 688
Query: 180 ESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVD 239
E+A++ +T+N ++E E+ +A +E + E A V++ V E +
Sbjct: 689 EAALNI-ATEN-EKELTENLNAVTSEKKKLE--------------ATVDEYSVKISESEN 732
Query: 240 ILSTLA-EISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCE-KAASVCKARLDRL 297
+L ++ E+++ + + E+ +D KA + E V K + A E K + +A R+
Sbjct: 733 LLESIRNELNVTQGKLESIENDL-KAAGLQESEVMEKLKSAEESLEQKGREIDEATTKRM 791
Query: 298 TNEV--KSITPASEAK-NKSIED---KQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLS 351
E +S++ SE + K++E+ + +E +LT DLE K++ + ++ S
Sbjct: 792 ELEALHQSLSIDSEHRLQKAMEEFTSRDSEASSLTEKLRDLEGKIKSYEEQLAEASGKSS 851
Query: 352 ELLEKVAAKAG--GGKESIPAKV 372
L EK+ G ES+ K+
Sbjct: 852 SLKEKLEQTLGRLAAAESVNEKL 874
Score = 32.7 bits (73), Expect = 0.41
Identities = 76/411 (18%), Positives = 154/411 (36%), Gaps = 38/411 (9%)
Query: 1 MVEEAPASETSVSMAPSEEAVAVDGTLNPSEHGKSEITNGATVDSAAIGGAESASNAETS 60
+++E A E +V+ S A + E K+ N + D+ + S E
Sbjct: 394 VLKEKEALEANVAEVTSNVATVTEVCNELEEKLKTSDENFSKTDALLSQALSNNSELEQK 453
Query: 61 GKNSLELAVELMDKGTKAMKED-DFGEAAENFSRALEIRVANYGEL-----APECVNTYY 114
K+ EL E A +++ + + + S+A E + EL A E N
Sbjct: 454 LKSLEELHSEAGSAAAAATQKNLELEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAEL 513
Query: 115 KYGCALLYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDI 174
+ LL +A+ + S K +++ I AE + + E
Sbjct: 514 EQQLNLLQLKSSDAE---------RELKELSEKSSELQTAIEVAEEEKKQATTQMQEYKQ 564
Query: 175 ASNDQESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAI-VEKQCVN 233
+++ E ++ S +N + E+D + ED ++ R+I +E C +
Sbjct: 565 KASELELSLTQSSARNSELEEDLRIALQKGAEHEDRAN-------TTHQRSIELEGLCQS 617
Query: 234 T-IEHVDILSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAA----- 287
+ +H D L ++ L + + + + ++ +S LE+ ++ Y + A
Sbjct: 618 SQSKHEDAEGRLKDLELLLQTEKYRIQELEEQVSSLEKKHGETEADSKGYLGQVAELQST 677
Query: 288 --------SVCKARLDRLTNEVKSITPASEAKNKSIEDKQAEIETLTGLSSDLENKLEDL 339
S +A L+ T K +T A + +A ++ + S+ EN LE +
Sbjct: 678 LEAFQVKSSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLLESI 737
Query: 340 QLLVSNPKSTLSELLEKVAAKAGGGKESIPAKVSSSQLATANSSGGFDSPT 390
+ ++ + L E +E AG + + K+ S++ + D T
Sbjct: 738 RNELNVTQGKL-ESIENDLKAAGLQESEVMEKLKSAEESLEQKGREIDEAT 787
>At1g65010 hypothetical protein
Length = 1318
Score = 46.2 bits (108), Expect = 4e-05
Identities = 88/398 (22%), Positives = 157/398 (39%), Gaps = 45/398 (11%)
Query: 8 SETSVSMAPSEEAVAVDGTLNPSEH---GKSEITNGATVDSAAIGGAESASN-AETSGKN 63
S+ + S+ A+ + L+ +E K E++ A + A+ AE A+ AE +
Sbjct: 141 SKNELESIRSQHALDISALLSTTEELQRVKHELSMTADAKNKALSHAEEATKIAEIHAEK 200
Query: 64 SLELAVELMDK----GTKAMKEDDFG-EAAENFSRALEIRVANYGELAPECVNTYYKYGC 118
+ LA EL G+K KE G E +E+ +++ + + G
Sbjct: 201 AEILASELGRLKALLGSKEEKEAIEGNEIVSKLKSEIELLRGELEKVSILESSLKEQEGL 260
Query: 119 ALLYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASND 178
K EA + + S + SV+ N SN + AS
Sbjct: 261 VEQLKVDLEAAKMAE-----------SCTNSSVEEWKNKVHELEKEVEESNRSKSSASES 309
Query: 179 QESAVDDESTKNYQEEDDEDSDAEDTEADE--------DESDLDLAWKMLDIARAIVEKQ 230
ES + + N+ + + +A E E +DL+ + + IA+ K
Sbjct: 310 MESVMKQLAELNHVLHETKSDNAAQKEKIELLEKTIEAQRTDLEEYGRQVCIAKEEASK- 368
Query: 231 CVNTIEHVDILSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVC 290
N +E + +E+ + +EE +L + + A S ++ L+D + E +I C
Sbjct: 369 LENLVESIK-----SELEISQEEKTRALDNEKAATSNIQNLLDQRTELSI-----ELERC 418
Query: 291 KARLDRLTNEVKSITPA-SEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKST 349
K ++ +++S+T A EA +S E K TL +L+N + L K T
Sbjct: 419 KVEEEKSKKDMESLTLALQEASTESSEAK----ATLLVCQEELKNCESQVDSLKLASKET 474
Query: 350 LSELLEKVAAKAGGGKESIPAKVSSSQLATANSSGGFD 387
+E EK+ A +S+ + V S Q NS G++
Sbjct: 475 -NEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKAGWE 511
>At3g01320 unknown protein
Length = 1324
Score = 45.1 bits (105), Expect = 8e-05
Identities = 55/229 (24%), Positives = 95/229 (41%), Gaps = 45/229 (19%)
Query: 23 VDGTLNPSEHGKSEITNGAT--------VDSAAIGGAESAS----------NAETSGKNS 64
+D + S +G++ +++G + SAA G S+S N +++GK +
Sbjct: 767 LDVNHSTSPNGEAAVSSGGDTARLASRKLKSAANGDENSSSGTFKHGIGLLNKDSTGKEN 826
Query: 65 LELAVELMDK------GTKAMKEDDFG-EAAENFSRALEIRVANYGELAPECV-----NT 112
LE VE+ ++ K KE + G EA + F + + + ++ ++ + N
Sbjct: 827 LE-DVEIANRDGVACSAVKPQKEQETGNEAEKRFGKPIPMDISERAAISSISIPSGAENN 885
Query: 113 YYKYGCALL------YKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTI---NAAESSTA 163
+ G +L K + E P GD G KD VKST N+AE+
Sbjct: 886 HCVVGKEVLPGPSRNEKEEGELSPNGDFE-----DNFGVYKDHGVKSTSKPENSAEAEVE 940
Query: 164 ASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEADEDESD 212
A E D D E+A + T++ + +D D E+ + DE D
Sbjct: 941 ADAEVENEDDADDVDSENASEASGTESGGDVCSQDEDREEENGEHDEID 989
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 43.1 bits (100), Expect = 3e-04
Identities = 45/229 (19%), Positives = 104/229 (44%), Gaps = 11/229 (4%)
Query: 136 KKQEGSQHGSSKDESVKSTINA--AESSTAASFSSNAEQDIASNDQESAV-----DDEST 188
K ++ + SS E++ + I+ AE + + E+ + +E++V DDE
Sbjct: 816 KLEDNDKQSSSSIETLTAEIDGLRAELDSMSVQKEEVEKQMVCKSEEASVKIKRLDDEVN 875
Query: 189 KNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIE-HVDILSTLAEI 247
Q+ DS + E ++ +++ + I ++++ +N ++ H IL + +
Sbjct: 876 GLRQQVASLDSQRAELEIQLEKKSEEISEYLSQITN--LKEEIINKVKVHESILEEINGL 933
Query: 248 SLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLTNEVKSITPA 307
S + + E L K S L++ + +K EE + +K +V + + LT + ++
Sbjct: 934 SEKIKGRELELETLGKQRSELDEELRTKKEENVQMHDKI-NVASSEIMALTELINNLKNE 992
Query: 308 SEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEK 356
++ + +AE+E S+L N++ D+Q + ++ + L E+
Sbjct: 993 LDSLQVQKSETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEE 1041
Score = 32.0 bits (71), Expect = 0.70
Identities = 37/215 (17%), Positives = 93/215 (43%), Gaps = 22/215 (10%)
Query: 145 SSKDESVKSTINAAESSTAASFSSNAEQDIASN-----DQESAVDDE--STKNYQEEDDE 197
++++E + AE S + N Q++ S + S + E S ++ E
Sbjct: 91 NAEEEKKLLSQKIAELSNEIQEAQNTMQELMSESGQLKESHSVKERELFSLRDIHEIHQR 150
Query: 198 DSDAEDTEAD-------EDESDLDLAWKMLDIARAIVEKQCVNTIEHVD-ILSTLAEISL 249
DS +E + + SDL + K + + + V T+ ++ +T+ E+
Sbjct: 151 DSSTRASELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQNTIQELMA 210
Query: 250 EREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLTNEVKSITPASE 309
E + ++S + + LS L ++ ++ ++ + ++ +++ + K + ++
Sbjct: 211 ELGKLKDSHREKESELSSLVEVHETHQRDSSIHVKELE-------EQVESSKKLVAELNQ 263
Query: 310 AKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVS 344
N + E+K+ + + LS++++ +Q LVS
Sbjct: 264 TLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVS 298
Score = 30.4 bits (67), Expect = 2.0
Identities = 83/430 (19%), Positives = 152/430 (35%), Gaps = 84/430 (19%)
Query: 2 VEEAPASETSVSMAPSEEAVA--------VDGTLNPSEHGKSEITNGATVDSAAIGGAES 53
V E ++S+ + EE V ++ TLN +E K ++ S I A++
Sbjct: 232 VHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQN 291
Query: 54 ASNAETSGKNSLELAVELMDKGTKAMKEDDFGEAAENFSRALEIRVANYGELAPECVNTY 113
S L+ + + D+ ++++ E+ +R E+
Sbjct: 292 TIQELVSESGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQ------------- 338
Query: 114 YKYGCALLYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQD 173
L Q +D D+ +E ++ SSK+ + + A+++ E
Sbjct: 339 ------LESSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELMDELGELK 392
Query: 174 IASNDQESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVN 233
++ES E + + D + +D + + + +E L+ ++LDI+ I E Q
Sbjct: 393 DRHKEKES----ELSSLVKSADQQVADMKQSLDNAEEEKKMLSQRILDISNEIQEAQ--K 446
Query: 234 TI-EHVDILSTLAEIS--LERE-------------EFENSLSDYQKALSILEQLV----- 272
TI EH+ L E ERE E LS+ + L +LEQ V
Sbjct: 447 TIQEHMSESEQLKESHGVKERELTGLRDIHETHQRESSTRLSELETQLKLLEQRVVDLSA 506
Query: 273 --------------------------DSKPEEAIAYCEKAASVCKARLDRLTNEVKSITP 306
SK +E + E A S K L + NE+ S
Sbjct: 507 SLNAAEEEKKSLSSMILEITDELKQAQSKVQELVT--ELAES--KDTLTQKENELSSFVE 562
Query: 307 ASEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKVAAKAGGGKE 366
EA + + E+E + + +L K LS+ + +++ K +
Sbjct: 563 VHEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKILSQQISEMSIKIKRAES 622
Query: 367 SIPAKVSSSQ 376
+I S S+
Sbjct: 623 TIQELSSESE 632
>At4g31570 putative protein
Length = 2712
Score = 43.1 bits (100), Expect = 3e-04
Identities = 51/237 (21%), Positives = 104/237 (43%), Gaps = 21/237 (8%)
Query: 164 ASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAED-----TEADEDESDLDLAWK 218
AS S E+ + +D +A D E Q D+E+S ED TE +++ +L +
Sbjct: 2359 ASHSELQEKVTSLSDLLAAKDLEIEALMQALDEEESQMEDLKLRVTELEQEVQQKNLDLQ 2418
Query: 219 MLDIARAIVEKQCVNTIEHVDILSTLAE-ISLEREEFENSLSDYQKALSILEQLVDSKPE 277
+ +R + K+ T++ D L L+E + E E+ + + D +S L Q V
Sbjct: 2419 KAEASRGKISKKLSITVDKFDELHHLSENLLAEIEKLQQQVQDRDTEVSFLRQEVTRCTN 2478
Query: 278 EAIAYCEKAASVCKARLDRLTNEVKSITPASEAKNKSIEDKQAEI----ETLTGLSSDLE 333
EA+A + + + + +I ++ D + I ET + +
Sbjct: 2479 EALAASQMGTKRDSEEIQTVLSWFDTIASLLGIEDSLSTDADSHINHYMETFEKRIASML 2538
Query: 334 NKLEDLQL-------LVSNPKSTLSELLEKVAAKAGGGKESIPAKVSSSQLATANSS 383
+++++L+L L+ +S ++EL +K A ++ + K S ++T+++S
Sbjct: 2539 SEIDELRLVGQSKDVLLEGERSRVAELRQKEATL----EKFLLEKESQQDISTSSTS 2591
Score = 31.6 bits (70), Expect = 0.92
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 9/136 (6%)
Query: 227 VEKQCVNTIEHVDILSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKA 286
V KQ + +E+ + L E+ EF + + +K +++ L EEAI
Sbjct: 812 VSKQSASFLENTQY-TNLEEVREYTSEFSALMKNLEKGEKMVQNL-----EEAIKQILTD 865
Query: 287 ASVCKARLDRLTNEVKSITPASEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNP 346
+SV K+ T V + A E+K K E+ ++E LT S+ + + + + + N
Sbjct: 866 SSVSKSSDKGATPAVSKLIQAFESKRKP-EEPESENAQLTDDLSEAD-QFVSVNVQIRNL 923
Query: 347 KSTLSELLEKVAAKAG 362
+ L +LL A KAG
Sbjct: 924 RGLLDQLLLN-ARKAG 938
Score = 30.4 bits (67), Expect = 2.0
Identities = 45/247 (18%), Positives = 103/247 (41%), Gaps = 35/247 (14%)
Query: 143 HGSSKDESVKST--INAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSD 200
+G+ + + ++ +N + T+ F +N + +++ +ES+V + +++ E S
Sbjct: 301 NGNFRSQQIREAAELNEEKPETSIDFPNNRDHVLSAEPEESSVAEMASQLQLPESVSISG 360
Query: 201 AEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVDILSTLAE----ISLEREEFEN 256
E L+L+ ++ + + E + V+ ++ +DI+ L + I E +
Sbjct: 361 VLSHEETRKIDTLNLSAELT--SAHVHEGRSVSFLQLMDIVKGLGQDEYQILCNAREAAS 418
Query: 257 SLSDYQKALSIL-EQLVDSKPEEAIAYCEKAA------------SVCKARLDRLTNEVKS 303
S +L L E+L S E I + + + A + +L +
Sbjct: 419 STEPGTSSLERLREELFVSSTMEDILHVQLTEQSHLQIEFDHQHNQFVAEISQLRASYSA 478
Query: 304 ITPASEAKNKSIEDKQAEIETLTGLSSDLEN--------------KLEDLQLLVSNPKST 349
+T +++ + + + Q+++ T +++LEN K+ +LQL +
Sbjct: 479 VTERNDSLAEELSECQSKLYAATSSNTNLENQLLATEAQVEDFTAKMNELQLSLEKSLLD 538
Query: 350 LSELLEK 356
LSE EK
Sbjct: 539 LSETKEK 545
>At4g16050 hypothetical protein
Length = 900
Score = 42.7 bits (99), Expect = 4e-04
Identities = 45/175 (25%), Positives = 74/175 (41%), Gaps = 26/175 (14%)
Query: 104 ELAPECVNTYYKYGCALLYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTA 163
E+ E + T+ Y ++ PL VP SQ D+ +K
Sbjct: 682 EMRKESMETFNVSSTVDHYDDSDDDIPLKIVPL----SQVYQKLDDEMKK---------- 727
Query: 164 ASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIA 223
A S+N + A D ESA + E ++ EDDE +D ED E+ E E D ++ IA
Sbjct: 728 AKHSTNKRRKRAREDDESAAETEDDESADTEDDESADTEDDESAETEDDDNMT-----IA 782
Query: 224 RAIVEKQCVNTIEHVDILSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEE 278
+ I ++ + IE+ E R +N++S + L+ ++ V + EE
Sbjct: 783 QRINSRKKSDDIEN-------TEGERSRLVADNNVSGLPQKLAYGDETVATTQEE 830
>At5g07170 putative protein
Length = 542
Score = 42.0 bits (97), Expect = 7e-04
Identities = 62/284 (21%), Positives = 115/284 (39%), Gaps = 53/284 (18%)
Query: 134 VPKKQEGS--QHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNY 191
+P Q+ SS +E + + + A + + E D +D + DD+ +
Sbjct: 78 IPSSQDDPLIDDSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDD--DDDDDDDD 135
Query: 192 QEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVDILSTLAEISLER 251
++DD+D +++D+E +E+E D DL + +D E +DI + + SLE+
Sbjct: 136 DDDDDDDDESKDSEVEEEEGDDDLRMRKID-------------PETMDIFA--EDFSLEK 180
Query: 252 EEFENSLS-----DYQKALSILEQLVDSKPEEA------------------IAYCEKAAS 288
+ + D + + +DS+ EE + Y EK
Sbjct: 181 FDIDFEFDIDFEFDAPRFYDFSKPELDSETEETELWFQSAGNYPPSPFSIHLRYEEKHLE 240
Query: 289 VCKARLDRLTNEVKSITPAS-EAKNKSIEDKQAEIETLTGLSSDLENKLEDL---QLLVS 344
+ K D+ N K + A+ ++K K+ K + + T +NK D+ +LL S
Sbjct: 241 IPKPVSDQY-NAPKDVPKATHKSKTKTYLRKNSTLTRPTASLLARQNKPLDIYSVRLLTS 299
Query: 345 NPKSTLSELLEKVAAKAGGGKESIPAKV------SSSQLATANS 382
S+ ++ + K GK S PA +SS+L N+
Sbjct: 300 VMTSSHLTYIQCLKPKITKGKNSKPASCARCCSQTSSKLTALNT 343
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/69 (30%), Positives = 39/69 (56%), Gaps = 2/69 (2%)
Query: 148 DESVK--STINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTE 205
DE++K + I +++ SS+ E+D + + A DD++ +EDD+D D +D +
Sbjct: 69 DENMKIAAAIPSSQDDPLIDDSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDDD 128
Query: 206 ADEDESDLD 214
D+D+ D D
Sbjct: 129 DDDDDDDDD 137
Score = 32.7 bits (73), Expect = 0.41
Identities = 26/90 (28%), Positives = 39/90 (42%), Gaps = 25/90 (27%)
Query: 150 SVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTK-----------------NYQ 192
S+K + ++ ST + S +I+S+D DDE+ K +
Sbjct: 34 SLKPGLKDSQDSTFVTSVSYDMGNISSDDHYCGDDDENMKIAAAIPSSQDDPLIDDSSSD 93
Query: 193 EEDDE--------DSDAEDTEADEDESDLD 214
EEDD D DA+DT+ DED+ D D
Sbjct: 94 EEDDSESTHCYAADDDADDTDDDEDDDDDD 123
>At1g70070 Similar to Synechocystis antiviral protein (At1g70070)
Length = 1171
Score = 41.6 bits (96), Expect = 9e-04
Identities = 44/226 (19%), Positives = 99/226 (43%), Gaps = 21/226 (9%)
Query: 150 SVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEADED 209
S KST+N+ S + + +++ D++ DDE+ Y DE +++D + DE+
Sbjct: 47 SFKSTLNSLSPSQSQLYEEEDDEEEEEEDEDD--DDEAADEYDNISDEIRNSDDDDDDEE 104
Query: 210 -ESDLDLA-----------WKMLDIARAIVEKQCVNTIEHVDILSTLAEISLEREEFENS 257
E +DL W+ ++ R++V V I+ +D L ++ + +++ + +
Sbjct: 105 TEFSVDLPTESARERVEFRWQRVEKLRSLVRDFGVEMID-IDELISIYDFRIDKFQ-RLA 162
Query: 258 LSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLTNEVKSITPASEAKNKSIED 317
+ + + S++ S + IA ++V K R T +K++ + K E
Sbjct: 163 IEAFLRGSSVVVSAPTSSGKTLIAEAAAVSTVAKGRRLFYTTPLKAL-----SNQKFREF 217
Query: 318 KQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKVAAKAGG 363
++ + GL + +D Q+++ + + L + V + G
Sbjct: 218 RETFGDDNVGLLTGDSAINKDAQIVIMTTEILRNMLYQSVGMASSG 263
>At3g57780 putative protein
Length = 670
Score = 41.2 bits (95), Expect = 0.001
Identities = 46/189 (24%), Positives = 79/189 (41%), Gaps = 19/189 (10%)
Query: 135 PKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEE 194
P+K G++ SS+ E +K+T ES+ + Q S+D + AV+D S K +EE
Sbjct: 25 PQKTNGAKR-SSEQERLKAT--KEESNVSVVVDDTTTQSKLSDDDDHAVNDSSEKTEKEE 81
Query: 195 --DDEDSDAEDTEADEDESDLD-LAWKMLDIARAIVEKQCVNTIEHVDIL---------- 241
+ D ED E E+ +LD +A + D + + VN + V++
Sbjct: 82 TINGLACDDEDEEEKEESKELDAIAHEKTDSVSSPETCEGVNVDKVVEVWDDASNGGLSG 141
Query: 242 ---STLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLT 298
+ ++ + E FE ++ + LE V+ EE S+ D +
Sbjct: 142 GSENEAGDVKEKNENFEEDEEMLKQMVETLETRVEKLEEELREVAALEISLYSVVPDHSS 201
Query: 299 NEVKSITPA 307
+ K TPA
Sbjct: 202 SAHKLHTPA 210
>At2g38370 hypothetical protein
Length = 539
Score = 41.2 bits (95), Expect = 0.001
Identities = 83/378 (21%), Positives = 148/378 (38%), Gaps = 41/378 (10%)
Query: 8 SETSVSMA--PSEEAVAVDGTLNPSEHGKSEITNGATVDSAAIGGAESASNAETSG--KN 63
S +S SMA P V D L+ K EI A +S E+A+ G +
Sbjct: 12 SSSSASMAEFPEPGTVNPDSDLSNGRAEKPEIDTSAPFESVR----EAATRFGGFGFWRP 67
Query: 64 SLELAVELMDKGTKAMKEDDF-GEAAENFS--RALEIRVANYGELAPECVNTYYKYGCAL 120
SL +L D + ++E D G A+ F R L ++ E+ E T A
Sbjct: 68 SLN---KLPDASQENIQEPDIMGLKAQAFELQRELIVKERETLEVLKELEATK-----AT 119
Query: 121 LYKAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAES-STAASFSSNAEQ-DIASND 178
+ K Q+ + + ++E H +K A + AS + EQ N+
Sbjct: 120 VLKLQQRNEAYEEDTLREEVDSHIKPAGVVLKDLSQAKMNLCKIASIRESVEQLKNKLNE 179
Query: 179 QESAVD-------DESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQC 231
+ +A++ ++S K + E++E E E DL + ++ ++R E +
Sbjct: 180 ERAALEKTRERLMEKSLKVFSLEEEEVRVRFAKEGQTGEKDLGMLNEVQRLSRQAQEVKK 239
Query: 232 VNTIEHVDILSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAA---- 287
++++ +AE R++ + A + E ++ EA+A E A
Sbjct: 240 TGENAELEVVKAMAETESTRDKIRTAKIRLVAARKMKEA---AREAEAVAIAEIEAVTGS 296
Query: 288 -SVCKARLDRLTNE-----VKSITPASEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQL 341
+V KA ++ E +S A E K +ED + +E D+ K+++
Sbjct: 297 MNVGKAEAVTISAEEYSVLARSARDAEEEARKRVEDAMSRVEEANVSKKDVLKKVDEAAQ 356
Query: 342 LVSNPKSTLSELLEKVAA 359
+ K L E +E+V A
Sbjct: 357 EIETSKRVLEEAVERVDA 374
>At5g52280 hyaluronan mediated motility receptor-like protein
Length = 853
Score = 40.8 bits (94), Expect = 0.002
Identities = 57/249 (22%), Positives = 107/249 (42%), Gaps = 26/249 (10%)
Query: 126 EEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDD 185
EEA L ++ +G G+++ +++K I + + N EQ+I ++ +
Sbjct: 408 EEAKKL----EEHKGMDSGNNEIDTLKQQIEDLDWELDSYKKKNEEQEILLDELTQEYES 463
Query: 186 ESTKNYQE-----EDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVDI 240
+NY+ E E S+AED D + +D ++I +++Q ++E+ +
Sbjct: 464 LKEENYKNVSSKLEQQECSNAEDEYLDSKDI-IDELKSQIEILEGKLKQQ---SLEYSEC 519
Query: 241 LSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCK-ARLDRLTN 299
L T+ E+ + +E + L D +A ++ +D+ E ++A + R R N
Sbjct: 520 LITVNELESQVKELKKELEDQAQA---YDEDIDTMMREKTEQEQRAIKAEENLRKTRWNN 576
Query: 300 EVKSITPASEAKNKSI--EDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKV 357
+ + + K S+ E K +E E LT + N L LQ TL E+ EK
Sbjct: 577 AITAERLQEKCKRLSLEMESKLSEHENLTKKTLAEANNLR-LQ------NKTLEEMQEKT 629
Query: 358 AAKAGGGKE 366
+ KE
Sbjct: 630 HTEITQEKE 638
Score = 30.8 bits (68), Expect = 1.6
Identities = 52/255 (20%), Positives = 103/255 (40%), Gaps = 24/255 (9%)
Query: 145 SSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDT 204
S+ DES + N+ E+S FSS E + + ++ E+ + E + + +
Sbjct: 235 STSDESYIESRNSPENSFQRGFSSVTES--SDPIERLKMELEALRRQSELSELEKQSLRK 292
Query: 205 EADEDESDLDLAWKML-------DIARAIVEK-QCVNTIEHVDILSTLAEISLEREEFEN 256
+A ++ + K + D A EK + N+ + D S L IS E+ N
Sbjct: 293 QAIKESKRIQELSKEVSCLKGERDGAMEECEKLRLQNSRDEADAESRLRCIS---EDSSN 349
Query: 257 SLSDYQKALSILEQLVDS------KPEEAIAYCEKAASVCKARLDRLTNEVKSITPASEA 310
+ + + LS + L + + +E+ + A L++ NE+ S+ E
Sbjct: 350 MIEEIRDELSCEKDLTSNLKLQLQRTQESNSNLILAVRDLNEMLEQKNNEISSLNSLLEE 409
Query: 311 KNKSIEDK-----QAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKVAAKAGGGK 365
K E K EI+TL DL+ +L+ + + L EL ++ +
Sbjct: 410 AKKLEEHKGMDSGNNEIDTLKQQIEDLDWELDSYKKKNEEQEILLDELTQEYESLKEENY 469
Query: 366 ESIPAKVSSSQLATA 380
+++ +K+ + + A
Sbjct: 470 KNVSSKLEQQECSNA 484
>At5g35200 unknown protein
Length = 544
Score = 40.8 bits (94), Expect = 0.002
Identities = 61/227 (26%), Positives = 93/227 (40%), Gaps = 17/227 (7%)
Query: 181 SAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVDI 240
S V ESTK YQ D + D D +D A K LD+ R V KQ E ++
Sbjct: 215 SMVISESTKIYQALTDGIDNLVDKFFDMQRND---AVKALDMYRRAV-KQAGRLSEFFEV 270
Query: 241 LSTLAEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARL-DRLTN 299
S+ E + Q S L+ + EE + AA V K ++ ++LT
Sbjct: 271 CK-----SVNVGRGERFIKIEQPPTSFLQAM-----EEYVKEAPLAAGVKKEQVVEKLTA 320
Query: 300 EVKSITPASEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKVAA 359
+ + E K +E+K A E + + K DL L + +P +SEL EK A
Sbjct: 321 PKEILAIEYEIPPKVVEEKPASPEPVKAEAEKPVEKQPDL-LSMDDPAPMVSELEEKNAL 379
Query: 360 KAGGGKESIPAKVSSSQLATANSSGGFDSPTISTAHSNGSAAGVTHL 406
S+ S++ NS+ G++ ++ SN AA + L
Sbjct: 380 ALAIVPVSVEQPHSTTDFTNGNST-GWELALVTAPSSNEGAAADSKL 425
>At3g14900 hypothetical protein
Length = 611
Score = 40.8 bits (94), Expect = 0.002
Identities = 54/194 (27%), Positives = 81/194 (40%), Gaps = 29/194 (14%)
Query: 82 DDFGEAAENFSRALEIRVANY--GELAPECVNTYYKYGCALLYKAQEEADPLG-DVPKKQ 138
DD A E S + RV + G E ++ Y K +KA+ E LG D P +
Sbjct: 362 DDVVNAYEKLSDEKKSRVLKFLLGNHPNELLHPYTKE-----WKAKLEEMELGCDAPDED 416
Query: 139 EGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDED 198
E DE I+ + SS A FS E D A ND + DD+ +E DD+D
Sbjct: 417 E--------DE-----ISISGSSEKAEFSEWIE-DEADNDDDDDDDDDDDGEVEEVDDDD 462
Query: 199 SDAEDTEADEDESDLDLAWKMLDIA--RAIVEKQCVNTIEHVDILSTLAEISLEREEFEN 256
+ D E + +E L+ + D E+Q + + + LAE+S+ +
Sbjct: 463 NMVVDVEGNVEEDSLEDEIEESDPEEDERYWEEQFNKATNNAERMEKLAEMSMVVSD--- 519
Query: 257 SLSDYQKALSILEQ 270
Y+K L LE+
Sbjct: 520 --KFYEKQLKALEE 531
>At4g14760 centromere protein homolog
Length = 1676
Score = 40.4 bits (93), Expect = 0.002
Identities = 55/251 (21%), Positives = 97/251 (37%), Gaps = 32/251 (12%)
Query: 132 GDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASN-DQESAVD------ 184
GD+ K+E D S+ I E S E+++A +Q SA+
Sbjct: 454 GDISSKEENRNLSEINDTSISLEIQKNEISCLKKMKEKLEEEVAKQMNQSSALQVEIHCV 513
Query: 185 ----DESTKNYQEEDDEDS----DAEDTE------ADEDESDLDLAWKMLDIARAIVEKQ 230
D + YQ+ D+ S D E DE+ ++L D A+ K
Sbjct: 514 KGNIDSMNRRYQKLIDQVSLTGFDPESLSYSVKKLQDENSKLVELCTNQRDENNAVTGKL 573
Query: 231 CVNTIEHVDILSTLAEISLEREEFENSL-SDYQKALSILEQLVDSKPEEAIAYCEKAASV 289
C E IL A++ E L +KA ++E+ + E++ E+A V
Sbjct: 574 C----EMDSILKRNADLEKLLLESNTKLDGSREKAKDLIERCESLRGEKSELAAERANLV 629
Query: 290 CKARLDRLTNEVKSITPASEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKST 349
++L +T ++++ + KS+ E+E+L D +D + N KS
Sbjct: 630 --SQLQIMTANMQTLLEKNSVLEKSLSCANIELESL----RDKSKCFDDFFQFLKNDKSE 683
Query: 350 LSELLEKVAAK 360
L + E + ++
Sbjct: 684 LMKERESLVSQ 694
Score = 35.4 bits (80), Expect = 0.063
Identities = 59/329 (17%), Positives = 130/329 (38%), Gaps = 36/329 (10%)
Query: 63 NSLELAVELMDKGTKAMKEDDFGEAAENFSRALEIRVANYGELAPECVNTYYKYGCALLY 122
N+ L+ E++ K ++ E+F++ +++ N +
Sbjct: 336 NAKRLSSEVLAGAAKIKTVEEQCALLESFNQTMKVEAENLAH--------------KMSA 381
Query: 123 KAQEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESA 182
K QE + ++ K Q Q + + +++ ES + S EQ + +++ S
Sbjct: 382 KDQELSQKQNEIEKLQAVMQEEQLRFSELGASLRNLESLHS---QSQEEQKVLTSELHSR 438
Query: 183 VD---DESTKNYQEEDDEDSDAED---TEADEDESDLDLAWKMLDIARAIVEKQCVNTIE 236
+ + +N + E D S E+ +E ++ L++ + + + EK +
Sbjct: 439 IQMLRELEMRNSKLEGDISSKEENRNLSEINDTSISLEIQKNEISCLKKMKEKLEEEVAK 498
Query: 237 HVDILSTL-AEISLEREEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARLD 295
++ S L EI + ++ YQK + + L PE +++Y K ++L
Sbjct: 499 QMNQSSALQVEIHCVKGNIDSMNRRYQKLIDQVS-LTGFDPE-SLSYSVKKLQDENSKLV 556
Query: 296 RLTNEVKSITPASEAKNKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLE 355
L + +N ++ K E++++ ++DLE L + + + +L+E
Sbjct: 557 ELCTNQRD-------ENNAVTGKLCEMDSILKRNADLEKLLLESNTKLDGSREKAKDLIE 609
Query: 356 KVAAKAGGGKESIPAK---VSSSQLATAN 381
+ + G E + VS Q+ TAN
Sbjct: 610 RCESLRGEKSELAAERANLVSQLQIMTAN 638
>At2g27170 putative chromosome associated protein
Length = 1163
Score = 40.4 bits (93), Expect = 0.002
Identities = 37/179 (20%), Positives = 76/179 (41%), Gaps = 12/179 (6%)
Query: 184 DDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVDILST 243
+ E + YQ+ D + E T D++ D + +++AR ++ + V+
Sbjct: 206 EKEELRKYQQLDKQRKSLEYTIYDKELHDAREKLEQVEVARTKASEESTKMYDRVE---- 261
Query: 244 LAEISLEREEFENSLSDYQKALSIL---EQLVDSKPEEAIAYCEKAASVCKARLDRLTNE 300
+ + + + SL + K L L ++ V+++ +A+ K K DR+T
Sbjct: 262 --KAQDDSKSLDESLKELTKELQTLYKEKETVEAQQTKALKKKTKLELDVKDFQDRITGN 319
Query: 301 VKSITPASEAKN---KSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEK 356
++S A E N + ++D E+E + L +K ++ + TLS L +K
Sbjct: 320 IQSKNDALEQLNTVEREMQDSLRELEAIKPLYESQVDKENQTSKRINELEKTLSILYQK 378
Score = 32.3 bits (72), Expect = 0.54
Identities = 40/209 (19%), Positives = 82/209 (39%), Gaps = 7/209 (3%)
Query: 149 ESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEADE 208
E +K I A A + ++ D + +D + +E + ++ D E
Sbjct: 682 EQLKQEIANANKQKHAIHKAIEYKEKLLGDIRTRIDQVRSSMSMKEAEMGTELVDHLTPE 741
Query: 209 DESDLDLAWKMLDIARAIVEKQCVNTIEHVDILSTLAEISLE-REEFENSLSDYQKALSI 267
+ L K+ + + EK+ + ++ + AE+ + +++ Q ++
Sbjct: 742 EREQLS---KLNPEIKDLKEKKFAYQADRIERETRKAELEANIATNLKRRITELQATIAS 798
Query: 268 LEQLVDSKPEEAIAYCEKAASVCKARLDRLTNEVKSITPASEAKNKSIEDKQAEIETLTG 327
++ DS P A E+ K ++ E+KS+ + + K K I+ + E L
Sbjct: 799 IDD--DSLPSSA-GTKEQELDDAKLSVNEAAKELKSVCDSIDEKTKQIKKIKDEKAKLKT 855
Query: 328 LSSDLENKLEDLQLLVSNPKSTLSELLEK 356
L D + L+DL + S + LL K
Sbjct: 856 LEDDCKGTLQDLDKKLEELFSLRNTLLAK 884
>At1g03530 unknown protein
Length = 801
Score = 40.4 bits (93), Expect = 0.002
Identities = 27/115 (23%), Positives = 48/115 (41%), Gaps = 7/115 (6%)
Query: 148 DESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEAD 207
DE + +I + + + + D A + +SA + T + + S +E+ E+D
Sbjct: 211 DEGLACSIEVGLEKVSLAVDDDEKSDEAKGEMDSAESESETSSSSASSSDSSSSEEEESD 270
Query: 208 EDESDLDLAWKMLDIARAIVEKQCVNTIEHVDILSTLAEISLEREEFENSLSDYQ 262
EDESD + K +V K+ D+ L E +E + EN D +
Sbjct: 271 EDESDKEENKKEEKFEHMVVGKE-------DDLAGELEEGEIENLDEENGDDDIE 318
Score = 35.8 bits (81), Expect = 0.049
Identities = 25/95 (26%), Positives = 44/95 (46%), Gaps = 27/95 (28%)
Query: 147 KDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVDDESTK--NYQEE---------- 194
K + K +++AES + S SS + D +S+++E + +DES K N +EE
Sbjct: 234 KSDEAKGEMDSAESESETSSSSASSSDSSSSEEEESDEDESDKEENKKEEKFEHMVVGKE 293
Query: 195 ---------------DDEDSDAEDTEADEDESDLD 214
D+E+ D + + D+D+ D D
Sbjct: 294 DDLAGELEEGEIENLDEENGDDDIEDEDDDDDDDD 328
Score = 32.0 bits (71), Expect = 0.70
Identities = 25/111 (22%), Positives = 47/111 (41%), Gaps = 26/111 (23%)
Query: 133 DVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQE------------ 180
D +K + ++ ES T +++ SS+ +S S E D +D+E
Sbjct: 230 DDDEKSDEAKGEMDSAESESETSSSSASSSDSSSSEEEESDEDESDKEENKKEEKFEHMV 289
Query: 181 --------SAVDDESTKNYQEE------DDEDSDAEDTEADEDESDLDLAW 217
+++ +N EE +DED D +D + D+D+ + +AW
Sbjct: 290 VGKEDDLAGELEEGEIENLDEENGDDDIEDEDDDDDDDDDDDDDVNEMVAW 340
Score = 32.0 bits (71), Expect = 0.70
Identities = 22/116 (18%), Positives = 51/116 (43%), Gaps = 8/116 (6%)
Query: 125 QEEADPLGDVPKKQEGSQHGSSKDESVKSTINAAESSTAASFSSNAEQDIASNDQESAVD 184
+EE+D + E + + K+E + + E A +++ + + ++
Sbjct: 266 EEESD-------EDESDKEENKKEEKFEHMVVGKEDDLAGELEEGEIENLDEENGDDDIE 318
Query: 185 DESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTIEHVDI 240
DE + ++DD+D D + A ++ D DL + + R+ E + + + VD+
Sbjct: 319 DEDDDDDDDDDDDD-DVNEMVAWSNDEDDDLGLQTKEPIRSKNELKELPPVPPVDV 373
>At3g16000 myosin heavy chain like protein
Length = 727
Score = 40.0 bits (92), Expect = 0.003
Identities = 43/215 (20%), Positives = 88/215 (40%), Gaps = 14/215 (6%)
Query: 149 ESVKSTINAAES--STAASFSSNAEQDIASNDQESAVDDESTKNYQEEDDEDSDAEDTEA 206
E K T+ A+ S + + + + E A+ E K +E + + DAE +
Sbjct: 445 EGTKKTLQASRDRVSDLETMLDESRALCSKLESELAIVHEEWKEAKERYERNLDAEKQKN 504
Query: 207 DEDESDL----DLAWKMLDIARAIVEKQCVNTIEHVDILSTLAEISLEREEFENSLSDYQ 262
+ S+L DL ++ D + + +++++ + L EI + E L + +
Sbjct: 505 EISASELALEKDLRRRVKDELEGVTHELKESSVKNQSLQKELVEIYKKVETSNKELEEEK 564
Query: 263 KALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLTNEVKSITPASEAKNKSIEDKQAEI 322
K + L + V ++ + E S L +++ + + NK+ E+
Sbjct: 565 KTVLSLNKEVKGMEKQILMEREARKS--------LETDLEEAVKSLDEMNKNTSILSREL 616
Query: 323 ETLTGLSSDLENKLEDLQLLVSNPKSTLSELLEKV 357
E + +S+LE++ E LQ + K+ E E V
Sbjct: 617 EKVNTHASNLEDEKEVLQRSLGEAKNASKEAKENV 651
Score = 29.3 bits (64), Expect = 4.5
Identities = 30/112 (26%), Positives = 49/112 (42%), Gaps = 12/112 (10%)
Query: 252 EEFENSLSDYQKALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLTNEVKSITPASEAK 311
E ENSLS + LE +K E + E R++ L+ E+K ++
Sbjct: 238 ESLENSLSKAGEDKEALE----TKLREKLDLVEGLQD----RINLLSLELKDSEEKAQRF 289
Query: 312 NKSIEDKQAEIETLTGLSSDLENKLEDLQLLVSNPKS----TLSELLEKVAA 359
N S+ K+AE++ L + + L + +L + K T SEL K +A
Sbjct: 290 NASLAKKEAELKELNSIYTQTSRDLAEAKLEIKQQKEELIRTQSELDSKNSA 341
Score = 28.9 bits (63), Expect = 5.9
Identities = 40/201 (19%), Positives = 84/201 (40%), Gaps = 20/201 (9%)
Query: 176 SNDQESAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLAWKMLDIARAIVEKQCVNTI 235
S ++ + K E + +S T D E+ L++ + ++ R E N+
Sbjct: 282 SEEKAQRFNASLAKKEAELKELNSIYTQTSRDLAEAKLEIKQQKEELIRTQSELDSKNSA 341
Query: 236 EHVDILST-LAEISLEREEFENSLSDYQK---ALSILEQLVDSKPEEAIAYCEKAASVCK 291
++ L+T + + E+E + L K AL + + + E I+ E+
Sbjct: 342 --IEELNTRITTLVAEKESYIQKLDSISKDYSALKLTSETQAAADAELISRKEQEIQQLN 399
Query: 292 ARLDRLTNEV-KSITPASEAKNKSIEDKQ---AEIETLTGLSSDLE----------NKLE 337
LDR ++V KS ++ K + K+ E+ T+ L +LE +++
Sbjct: 400 ENLDRALDDVNKSKDKVADLTEKYEDSKRMLDIELTTVKNLRHELEGTKKTLQASRDRVS 459
Query: 338 DLQLLVSNPKSTLSELLEKVA 358
DL+ ++ ++ S+L ++A
Sbjct: 460 DLETMLDESRALCSKLESELA 480
>At5g11270 unknown protein
Length = 354
Score = 39.7 bits (91), Expect = 0.003
Identities = 50/218 (22%), Positives = 94/218 (42%), Gaps = 25/218 (11%)
Query: 125 QEEADPLGDVPKK-QEGSQHGSSKDESVKST---INAAESSTAASFSSNAEQDIASNDQE 180
+EE DP + +E ++ +S DE + A E + A + +
Sbjct: 92 EEEEDPFEALFNLLEEDLKNDNSDDEEISEEELEALADELARALGVGDDVDDIDLFGSVT 151
Query: 181 SAVDDESTKNYQEEDDEDSDAEDTEADEDESDLDLA-WKMLDIARAIVEKQCVNTIEHVD 239
VD + + + DD+D+D +D +++EDE L W++ +A A+ + +I+++
Sbjct: 152 GDVDVDVDNDDDDNDDDDNDDDDDDSEEDERPTKLKNWQLKRLAYALKAGRRKTSIKNL- 210
Query: 240 ILSTLAEISLEREEFENSLSD-YQKALSILEQLVDSKPEEAIAYCEKAASVCKARLDRLT 298
AE+ L+R L D K L + L D KP A + +S + ++ L+
Sbjct: 211 ----AAEVCLDRAYVLELLRDPPPKLLMLSATLPDEKPPVA---APENSSPDPSPVESLS 263
Query: 299 NEVKSITPASEAKNKSI-----------EDKQAEIETL 325
E + P + K++++ K+A IETL
Sbjct: 264 AEDVVVEPKEKVKDEAVHVMQQRWSAQKRVKKAHIETL 301
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.301 0.120 0.309
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,336,404
Number of Sequences: 26719
Number of extensions: 399466
Number of successful extensions: 5909
Number of sequences better than 10.0: 508
Number of HSP's better than 10.0 without gapping: 201
Number of HSP's successfully gapped in prelim test: 318
Number of HSP's that attempted gapping in prelim test: 3146
Number of HSP's gapped (non-prelim): 1530
length of query: 442
length of database: 11,318,596
effective HSP length: 102
effective length of query: 340
effective length of database: 8,593,258
effective search space: 2921707720
effective search space used: 2921707720
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0559.7


