

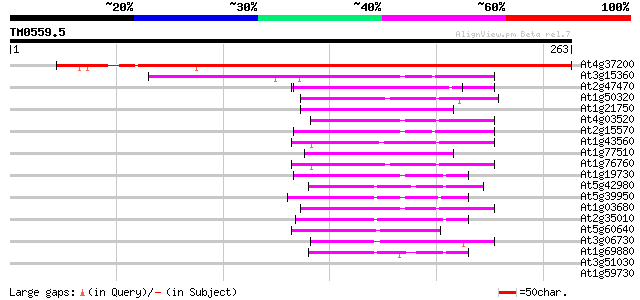
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.5
(263 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g37200 thiol-disulfide interchange like protein 327 4e-90
At3g15360 thioredoxin m4 55 4e-08
At2g47470 putative protein disulfide-isomerase 52 2e-07
At1g50320 thioredoxin like protein 52 4e-07
At1g21750 putative disulfide isomerase 52 4e-07
At4g03520 putative M-type thioredoxin 50 9e-07
At2g15570 putative thioredoxin M 50 9e-07
At1g43560 unknown protein 50 1e-06
At1g77510 putative thioredoxin 49 3e-06
At1g76760 thioredoxin-like protein 49 4e-06
At1g19730 thioredoxin 47 1e-05
At5g42980 thioredoxin (clone GIF1) (pir||S58118) 46 2e-05
At5g39950 thioredoxin 46 2e-05
At1g03680 putative thioredoxin-m 45 4e-05
At2g35010 thioredoxin like protein 44 7e-05
At5g60640 protein disulfide isomerase precursor - like 44 9e-05
At3g06730 thioredoxin, putative 42 3e-04
At1g69880 thioredoxin like protein 42 3e-04
At3g51030 thioredoxin h 40 0.001
At1g59730 unknown protein 40 0.001
>At4g37200 thiol-disulfide interchange like protein
Length = 261
Score = 327 bits (838), Expect = 4e-90
Identities = 166/252 (65%), Positives = 202/252 (79%), Gaps = 17/252 (6%)
Query: 23 FTVNPRHLH--FNTR----KFHTLACQTNPNLDEKDASATSQEIKIVVEPSTVNGESETC 76
F ++PR+L F T+ +F + C+ NP ++S T QE K+V++ + S+
Sbjct: 16 FNLSPRNLQSFFVTQTGAPRFRAVRCKPNP-----ESSETKQE-KLVIDNGETSSASKEV 69
Query: 77 KPTSSTADAS-----GLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVSLKDLTANAL 131
+ +SS AD+S G P+ P KDINR+VA + +AAL LF++TRLDFG+SLKDLTA+AL
Sbjct: 70 ESSSSVADSSSSSSSGFPESPNKDINRRVAAVTVIAALSLFVSTRLDFGISLKDLTASAL 129
Query: 132 PYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDE 191
PYE+ALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKD+VNFVMLNVDNT WEQELDE
Sbjct: 130 PYEEALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDKVNFVMLNVDNTKWEQELDE 189
Query: 192 FGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEAR 251
FGVEGIPHFAFLD+EGNEEGNVVGRLPRQ L+ENV+ALA G+ S+P+AR VG+YSS+E+R
Sbjct: 190 FGVEGIPHFAFLDREGNEEGNVVGRLPRQYLVENVNALAAGKQSIPYARAVGQYSSSESR 249
Query: 252 KVHQVVDPRSHG 263
KVHQV DP SHG
Sbjct: 250 KVHQVTDPLSHG 261
>At3g15360 thioredoxin m4
Length = 193
Score = 55.1 bits (131), Expect = 4e-08
Identities = 39/175 (22%), Positives = 79/175 (44%), Gaps = 16/175 (9%)
Query: 66 PSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVSL-- 123
P + S + P+ S S L + + ++ + A+LG TR+ G +
Sbjct: 17 PIAASVSSSSAAPSVSRRRISPARFLEFRGLKSSRSLVTQSASLGANRRTRIARGGRIAC 76
Query: 124 --KDLTANALPYE---------QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDR 172
+D TA A+ + L + P +VEF+A WC CR + P V ++ + + +
Sbjct: 77 EAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCGPCRMIHPIVDQLAKDFAGK 136
Query: 173 VNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
F +N D + + +G+ +P + K G ++ +++G +PR+ L + ++
Sbjct: 137 FKFYKINTDES--PNTANRYGIRSVP-TVIIFKGGEKKDSIIGAVPRETLEKTIE 188
>At2g47470 putative protein disulfide-isomerase
Length = 361
Score = 52.4 bits (124), Expect = 2e-07
Identities = 27/79 (34%), Positives = 40/79 (50%), Gaps = 1/79 (1%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L K +VEFYA WC C+ LAP K+ +K V+ N+D + +++G
Sbjct: 153 EIVLDQNKDVLVEFYAPWCGHCKSLAPTYEKVATVFKQEEGVVIANLDADAHKALGEKYG 212
Query: 194 VEGIPHFAFLDKEGNEEGN 212
V G P F K+ N+ G+
Sbjct: 213 VSGFPTLKFFPKD-NKAGH 230
Score = 51.2 bits (121), Expect = 6e-07
Identities = 27/95 (28%), Positives = 45/95 (46%)
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+E+ + K +VEFYA WC C++LAP+ K+ +K + ++ VD + ++
Sbjct: 33 FEKEVGKDKGALVEFYAPWCGHCKKLAPEYEKLGASFKKAKSVLIAKVDCDEQKSVCTKY 92
Query: 193 GVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
GV G P + K E G + L E V+
Sbjct: 93 GVSGYPTIQWFPKGSLEPQKYEGPRNAEALAEYVN 127
>At1g50320 thioredoxin like protein
Length = 182
Score = 51.6 bits (122), Expect = 4e-07
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 5/95 (5%)
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + +P +VEF A WC C+ + P + + Q+Y D++ V +D+ + + EF V G
Sbjct: 84 LESAQPVLVEFVATWCGPCKLIYPAMEALSQEYGDKLTIV--KIDHDANPKLIAEFKVYG 141
Query: 197 IPHFAFLDKEGNE--EGNVVGRLPRQLLLENVDAL 229
+PHF L K+G E G + + L E +D L
Sbjct: 142 LPHF-ILFKDGKEVPGSRREGAITKAKLKEYIDGL 175
>At1g21750 putative disulfide isomerase
Length = 501
Score = 51.6 bits (122), Expect = 4e-07
Identities = 25/72 (34%), Positives = 40/72 (54%)
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L++GK ++EFYA WC C++LAP + ++ Y+ + V+ +D T + D F V+G
Sbjct: 389 LNSGKNVLLEFYAPWCGHCQKLAPILDEVAVSYQSDSSVVIAKLDATANDFPKDTFDVKG 448
Query: 197 IPHFAFLDKEGN 208
P F GN
Sbjct: 449 FPTIYFKSASGN 460
Score = 37.0 bits (84), Expect = 0.011
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 3/76 (3%)
Query: 126 LTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNW 185
LT + + ++ VVEFYA WC C++LAP+ K V V+L + +
Sbjct: 33 LTLDHTNFTDTINKHDFIVVEFYAPWCGHCKQLAPEYEKAASALSSNVPPVVLAKIDASE 92
Query: 186 EQELD---EFGVEGIP 198
E + ++ V+G P
Sbjct: 93 ETNREFATQYEVQGFP 108
>At4g03520 putative M-type thioredoxin
Length = 186
Score = 50.4 bits (119), Expect = 9e-07
Identities = 25/86 (29%), Positives = 44/86 (51%), Gaps = 3/86 (3%)
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFA 201
P VV+F+A WC C+ + P V + Q Y ++ F LN D + ++GV IP
Sbjct: 100 PVVVDFWAPWCGPCKMIDPLVNDLAQHYTGKIKFYKLNTDES--PNTPGQYGVRSIPTI- 156
Query: 202 FLDKEGNEEGNVVGRLPRQLLLENVD 227
+ G ++ ++G +P+ L ++D
Sbjct: 157 MIFVGGEKKDTIIGAVPKTTLTSSLD 182
>At2g15570 putative thioredoxin M
Length = 173
Score = 50.4 bits (119), Expect = 9e-07
Identities = 24/94 (25%), Positives = 48/94 (50%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
+ L + P +VEFY WC CR + + +I Y ++N +LN DN +E+
Sbjct: 78 DSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYLLNADND--LPVAEEYE 135
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
++ +P L K G + +++G +P++ + ++
Sbjct: 136 IKAVP-VVLLFKNGEKRESIMGTMPKEFYISAIE 168
>At1g43560 unknown protein
Length = 167
Score = 50.1 bits (118), Expect = 1e-06
Identities = 26/96 (27%), Positives = 50/96 (52%), Gaps = 4/96 (4%)
Query: 133 YEQALSNG-KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDE 191
++ L N KP +V+FYA WC C+ + P + ++ + KD + ++ +D + ++
Sbjct: 68 FDDLLQNSDKPVLVDFYATWCGPCQLMVPILNEVSETLKDII--AVVKIDTEKYPSLANK 125
Query: 192 FGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ +E +P F L K+G G LP L+E ++
Sbjct: 126 YQIEALPTF-ILFKDGKLWDRFEGALPANQLVERIE 160
>At1g77510 putative thioredoxin
Length = 508
Score = 48.9 bits (115), Expect = 3e-06
Identities = 22/70 (31%), Positives = 39/70 (55%)
Query: 139 NGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIP 198
+GK ++EFYA WC C++LAP + ++ +++ + ++ +D T + D F V+G P
Sbjct: 389 SGKNVLIEFYAPWCGHCQKLAPILDEVALSFQNDPSVIIAKLDATANDIPSDTFDVKGFP 448
Query: 199 HFAFLDKEGN 208
F GN
Sbjct: 449 TIYFRSASGN 458
Score = 38.9 bits (89), Expect = 0.003
Identities = 27/110 (24%), Positives = 48/110 (43%), Gaps = 3/110 (2%)
Query: 101 AIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAP 160
A S L L LF+++ LT + + + +S VVEFYA WC C++LAP
Sbjct: 7 ACFSILLLLSLFVSSIRSEETKEFVLTLDHSNFTETISKHDFIVVEFYAPWCGHCQKLAP 66
Query: 161 DVYKIEQQ---YKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEG 207
+ K + + + ++ ++ +E+ ++G P L G
Sbjct: 67 EYEKAASELSSHNPPLALAKIDASEEANKEFANEYKIQGFPTLKILRNGG 116
>At1g76760 thioredoxin-like protein
Length = 172
Score = 48.5 bits (114), Expect = 4e-06
Identities = 25/96 (26%), Positives = 50/96 (52%), Gaps = 4/96 (4%)
Query: 133 YEQALSNG-KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDE 191
+E L N KP +V++YA WC C+ + P + ++ + KD++ V +D + ++
Sbjct: 73 FEDLLVNSDKPVLVDYYATWCGPCQFMVPILNEVSETLKDKIQVV--KIDTEKYPSIANK 130
Query: 192 FGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ +E +P F L K+G G L + L++ ++
Sbjct: 131 YKIEALPTF-ILFKDGEPCDRFEGALTAKQLIQRIE 165
>At1g19730 thioredoxin
Length = 119
Score = 47.0 bits (110), Expect = 1e-05
Identities = 26/82 (31%), Positives = 42/82 (50%), Gaps = 3/82 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
++A + K V++F A WC CR +AP + +++ F ++VD + EFG
Sbjct: 22 DKAKESNKLIVIDFTASWCPPCRMIAPIFNDLAKKFMSSAIFFKVDVDEL--QSVAKEFG 79
Query: 194 VEGIPHFAFLDKEGNEEGNVVG 215
VE +P F F+ K G +VG
Sbjct: 80 VEAMPTFVFI-KAGEVVDKLVG 100
>At5g42980 thioredoxin (clone GIF1) (pir||S58118)
Length = 118
Score = 46.2 bits (108), Expect = 2e-05
Identities = 29/82 (35%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Query: 141 KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHF 200
K V++F A WC CR +AP + +++ D V F ++VD N E EF V+ +P F
Sbjct: 28 KLIVIDFTATWCPPCRFIAPVFADLAKKHLD-VVFFKVDVDELNTVAE--EFKVQAMPTF 84
Query: 201 AFLDKEGNEEGNVVGRLPRQLL 222
F+ KEG + VVG +++
Sbjct: 85 IFM-KEGEIKETVVGAAKEEII 105
>At5g39950 thioredoxin
Length = 133
Score = 46.2 bits (108), Expect = 2e-05
Identities = 28/85 (32%), Positives = 43/85 (49%), Gaps = 4/85 (4%)
Query: 131 LPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELD 190
L + + + K VV+F A WC CR + P ++ + ++ D V+FV L+VD
Sbjct: 38 LHFNEIKESNKLLVVDFSASWCGPCRMIEPAIHAMADKFND-VDFVKLDVDEL--PDVAK 94
Query: 191 EFGVEGIPHFAFLDKEGNEEGNVVG 215
EF V +P F L K G E ++G
Sbjct: 95 EFNVTAMPTFV-LVKRGKEIERIIG 118
>At1g03680 putative thioredoxin-m
Length = 179
Score = 45.1 bits (105), Expect = 4e-05
Identities = 23/91 (25%), Positives = 45/91 (49%), Gaps = 3/91 (3%)
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L +P V+F+A WC C+ + P V ++ Q+Y + F LN D + ++GV
Sbjct: 89 LKADEPVFVDFWAPWCGPCKMIDPIVNELAQKYAGQFKFYKLNTDES--PATPGQYGVRS 146
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP + G ++ ++G + + L +++
Sbjct: 147 IPTI-MIFVNGEKKDTIIGAVSKDTLATSIN 176
>At2g35010 thioredoxin like protein
Length = 194
Score = 44.3 bits (103), Expect = 7e-05
Identities = 23/81 (28%), Positives = 41/81 (50%), Gaps = 2/81 (2%)
Query: 135 QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGV 194
+A P+V F A WC CR ++P + ++ +QY D V +++D + + +
Sbjct: 101 KAQDGSLPSVFYFTAAWCGPCRFISPVIVELSKQYPD-VTTYKVDIDEGGISNTISKLNI 159
Query: 195 EGIPHFAFLDKEGNEEGNVVG 215
+P F K G+++G VVG
Sbjct: 160 TAVPTLHFF-KGGSKKGEVVG 179
>At5g60640 protein disulfide isomerase precursor - like
Length = 597
Score = 43.9 bits (102), Expect = 9e-05
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 1/70 (1%)
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+ + N + +VEFYA WC C+ LAP+ + K+ V+ +D T + E+
Sbjct: 113 FTDVIENNQYVLVEFYAPWCGHCQSLAPEYAAAATELKED-GVVLAKIDATEENELAQEY 171
Query: 193 GVEGIPHFAF 202
V+G P F
Sbjct: 172 RVQGFPTLLF 181
Score = 35.4 bits (80), Expect = 0.031
Identities = 20/69 (28%), Positives = 31/69 (43%), Gaps = 2/69 (2%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L + K ++E YA WC C+ L P K+ + + + V+ +D T E +
Sbjct: 453 EIVLDDSKDVLLEVYAPWCGHCQALEPMYNKLAKHLRSIDSLVITKMDGTT--NEHPKAK 510
Query: 194 VEGIPHFAF 202
EG P F
Sbjct: 511 AEGFPTILF 519
>At3g06730 thioredoxin, putative
Length = 183
Score = 42.4 bits (98), Expect = 3e-04
Identities = 21/87 (24%), Positives = 46/87 (52%), Gaps = 3/87 (3%)
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFA 201
P +V+FYA WC C +A ++ + +Y+ N +++ VD + + + V G+P
Sbjct: 96 PLIVDFYATWCGPCILMAQELEMLAVEYES--NAIIVKVDTDDEYEFARDMQVRGLPTLF 153
Query: 202 FLDKEGNEEG-NVVGRLPRQLLLENVD 227
F+ + +++ G +P Q++ + +D
Sbjct: 154 FISPDPSKDAIRTEGLIPLQMMHDIID 180
>At1g69880 thioredoxin like protein
Length = 148
Score = 42.0 bits (97), Expect = 3e-04
Identities = 28/77 (36%), Positives = 40/77 (51%), Gaps = 8/77 (10%)
Query: 141 KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVD--NTNWEQELDEFGVEGIP 198
K V+EF A WC C+ L P + ++ +Y D V FV ++VD + W EF + +P
Sbjct: 60 KLLVIEFTAKWCGPCKTLEPKLEELAAKYTD-VEFVKIDVDVLMSVWM----EFNLSTLP 114
Query: 199 HFAFLDKEGNEEGNVVG 215
F+ K G E VVG
Sbjct: 115 AIVFM-KRGREVDMVVG 130
>At3g51030 thioredoxin h
Length = 114
Score = 40.4 bits (93), Expect = 0.001
Identities = 25/72 (34%), Positives = 35/72 (47%), Gaps = 4/72 (5%)
Query: 144 VVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFL 203
VV+F A WC CR +AP + K N + L VD + ++ ++ +P F FL
Sbjct: 32 VVDFTASWCGPCRFIAPFFADLA---KKLPNVLFLKVDTDELKSVASDWAIQAMPTFMFL 88
Query: 204 DKEGNEEGNVVG 215
KEG VVG
Sbjct: 89 -KEGKILDKVVG 99
>At1g59730 unknown protein
Length = 129
Score = 40.0 bits (92), Expect = 0.001
Identities = 26/90 (28%), Positives = 43/90 (46%), Gaps = 4/90 (4%)
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
++ + K V++F A WC C+ + P V +I +Y + V F ++VD +
Sbjct: 36 FDSMKGSNKLLVIDFTAVWCGPCKAMEPRVREIASKYSEAV-FARVDVDRL--MDVAGTY 92
Query: 193 GVEGIPHFAFLDKEGNEEGNVVGRLPRQLL 222
+P F F+ K G E VVG P +L+
Sbjct: 93 RAITLPAFVFV-KRGEEIDRVVGAKPDELV 121
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,929,188
Number of Sequences: 26719
Number of extensions: 252333
Number of successful extensions: 738
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 686
Number of HSP's gapped (non-prelim): 62
length of query: 263
length of database: 11,318,596
effective HSP length: 97
effective length of query: 166
effective length of database: 8,726,853
effective search space: 1448657598
effective search space used: 1448657598
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0559.5


