

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0550.4
(888 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
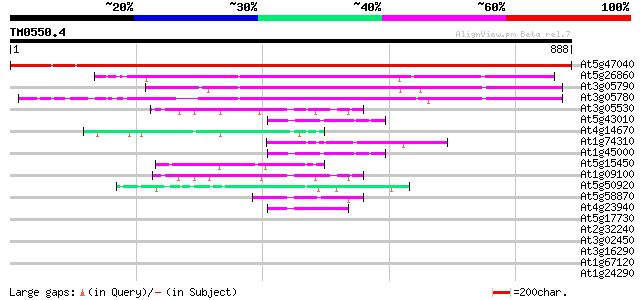
Score E
Sequences producing significant alignments: (bits) Value
At5g47040 Lon protease homolog 1 precursor 1425 0.0
At5g26860 LON protease homolog (LON_ARA) 493 e-139
At3g05790 putative mitochondrial LON ATP-dependent protease 464 e-131
At3g05780 putative mitochondrial LON ATP-dependent protease 442 e-124
At3g05530 26S proteasome AAA-ATPase subunit RPT5a 49 1e-05
At5g43010 26S proteasome AAA-ATPase subunit RPT4a (gb|AAF22524.1) 45 2e-04
At4g14670 heat shock protein like 45 2e-04
At1g74310 heat shock protein 101 45 2e-04
At1g45000 26S proteasome AAA-ATPase subunit RPT4a like protein 45 2e-04
At5g15450 clpB heat shock protein-like 44 3e-04
At1g09100 putative 26S protease regulatory subunit 6A 44 3e-04
At5g50920 ATP-dependent Clp protease, ATP-binding subunit 44 4e-04
At5g58870 cell division protein - like 43 7e-04
At4g23940 cell division protein - like 43 9e-04
At5g17730 BCS1 - like protein 42 0.001
At2g32240 putative myosin heavy chain 42 0.002
At3g02450 cell division protein FtsH-like protein 41 0.003
At3g16290 putative FtsH-like metalloprotease 41 0.003
At1g67120 hypothetical protein 40 0.004
At1g24290 unknown protein 40 0.008
>At5g47040 Lon protease homolog 1 precursor
Length = 888
Score = 1425 bits (3689), Expect = 0.0
Identities = 729/891 (81%), Positives = 807/891 (89%), Gaps = 6/891 (0%)
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MAE+VELPSRLAILPFRNKVLLPGAIIRIRCTS +SV LVEQELWQ+E+KGLIGILPVRD
Sbjct: 1 MAETVELPSRLAILPFRNKVLLPGAIIRIRCTSHSSVTLVEQELWQKEEKGLIGILPVRD 60
Query: 61 AAETEVKPVGPTISQDG--DTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALH 118
AE +G I+ D+ ++S K ++D+ K DAK Q D+ WH RGVAARALH
Sbjct: 61 DAEGS--SIGTMINPGAGSDSGERSLKFLVGTTDAQKSDAKDQQDL-QWHTRGVAARALH 117
Query: 119 LSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFI 178
LSRGVEKPSGRVTY+VVLEGL RF+VQEL RG Y ARI+SLEM K E+EQV+ DPDF+
Sbjct: 118 LSRGVEKPSGRVTYVVVLEGLSRFNVQELGKRGPYSVARITSLEMTKAELEQVKQDPDFV 177
Query: 179 MLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDS 238
LSRQFK TAMEL+SVLEQKQKTGGRTKVLLETVP+HKLADIFVASFE+SFEEQLSMLDS
Sbjct: 178 ALSRQFKTTAMELVSVLEQKQKTGGRTKVLLETVPIHKLADIFVASFEMSFEEQLSMLDS 237
Query: 239 VDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD 298
VD K+RLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKE+LLRQQMRAIKEELGDNDD
Sbjct: 238 VDLKVRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEYLLRQQMRAIKEELGDNDD 297
Query: 299 DEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKA 358
DEDD+AALERKMQ+AGMP +IWKHA RELRRLKKMQPQQPGY+SSR YL+LLADLPW KA
Sbjct: 298 DEDDVAALERKMQAAGMPSNIWKHAQRELRRLKKMQPQQPGYNSSRVYLELLADLPWDKA 357
Query: 359 SEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 418
SEEHE+DLK+A+ERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL
Sbjct: 358 SEEHELDLKAAKERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 417
Query: 419 ASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEI 478
ASSIAAAL RKFVR+SLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV VCNPVMLLDEI
Sbjct: 418 ASSIAAALGRKFVRLSLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVGVCNPVMLLDEI 477
Query: 479 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDR 538
DKTGSDVRGDPASALLEVLDPEQNK+FNDHYLNVP+DLSKV+FVATANR Q IPPPLLDR
Sbjct: 478 DKTGSDVRGDPASALLEVLDPEQNKSFNDHYLNVPYDLSKVVFVATANRVQPIPPPLLDR 537
Query: 539 MEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNL 598
ME+IELPGYT EEKLKIAM+HLIPRVLDQHGLSSE+L+IPE MV+ +IQRYTREAGVR+L
Sbjct: 538 MELIELPGYTQEEKLKIAMRHLIPRVLDQHGLSSEFLKIPEAMVKNIIQRYTREAGVRSL 597
Query: 599 ERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDI 658
ERNLA+LARAAAV VAE EQ +PL+K+VQ LT+PLL R+ +G EVEMEVIPM VN +I
Sbjct: 598 ERNLAALARAAAVMVAEHEQSLPLSKDVQKLTSPLLNGRMAEGGEVEMEVIPMGVNDHEI 657
Query: 659 SNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATE 718
TF+ S LVV+E +LEK+LGPP+FD EAA+RVA+ G+ VGLVWTTFGGEVQFVEAT
Sbjct: 658 GGTFQSPSALVVDETMLEKILGPPRFDDSEAADRVASAGVSVGLVWTTFGGEVQFVEATS 717
Query: 719 MVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAI 778
MVGKG++HLTGQLGDVIKESAQ+ALTWVRARA+D +LA A +N+L+GRDIHIHFPAGA+
Sbjct: 718 MVGKGEMHLTGQLGDVIKESAQLALTWVRARASDFKLALAGDMNVLDGRDIHIHFPAGAV 777
Query: 779 PKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKR 838
PKDGPSAGVTLVTALVSLFS+KRVR+DTAMTGEMTLRGLVLPVGGIKDKILAAHR GIKR
Sbjct: 778 PKDGPSAGVTLVTALVSLFSQKRVRADTAMTGEMTLRGLVLPVGGIKDKILAAHRYGIKR 837
Query: 839 VIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQ-SKL 888
VI+P+RN KDLVEVP++VL +LE++LAKRMEDVLE A EGGCPWR+ SKL
Sbjct: 838 VILPQRNSKDLVEVPAAVLSSLEVILAKRMEDVLENAFEGGCPWRNNYSKL 888
>At5g26860 LON protease homolog (LON_ARA)
Length = 985
Score = 493 bits (1268), Expect = e-139
Identities = 289/748 (38%), Positives = 435/748 (57%), Gaps = 39/748 (5%)
Query: 134 VVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSRQFKATAMELIS 193
V+L G R ++E+ + T ++ L+ N +M D D + KAT+ E+IS
Sbjct: 237 VILVGHRRLRIKEMVSEEPL-TVKVDHLKDNPFDM-----DDDVV------KATSFEVIS 284
Query: 194 VLEQKQKTGGRTKVLLETVPVH-------KLADIFVASFEISFEEQLSMLDSVDAKLRLS 246
L KT + ++T H +LAD A + + +L+ +D RL
Sbjct: 285 TLRDVLKTSSLWRDHVQTYTQHIGDFTYPRLADFGAAICGANRHQAQEVLEELDVHKRLR 344
Query: 247 KATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDLAAL 306
EL+ + ++ ++ E I + +E ++S Q+ +LL +Q++AIK+ELG DD+ L+A
Sbjct: 345 LTLELMKKEMEISKIQETIAKAIEEKISGEQRRYLLNEQLKAIKKELGVETDDKSALSAK 404
Query: 307 ERKM---QSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHE 363
++ +P + + EL +L+ ++ ++ +R YLD L LPW S E+
Sbjct: 405 FKERIEPNKEKIPAHVLQVIEEELTKLQLLEASSSEFNVTRNYLDWLTILPWGNYSNEN- 463
Query: 364 MDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIA 423
D+ AQ LD DHYGL+ VK+RI+E++AV +L+ ++G ++C GPPGVGKTS+ SIA
Sbjct: 464 FDVARAQTILDEDHYGLSDVKERILEFIAVGRLRGTSQGKIICLSGPPGVGKTSIGRSIA 523
Query: 424 AALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGS 483
AL+RKF R S+GG+ D A+I+GHRRTY+G+MPG+++ LK V NP++L+DEIDK G
Sbjct: 524 RALNRKFFRFSVGGLADVAEIKGHRRTYVGAMPGKMVQCLKSVGTANPLVLIDEIDKLGR 583
Query: 484 DVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIE 543
GDPASALLE+LDPEQN F DHYL+V DLSKV+FV TAN IP PLLDRMEVI
Sbjct: 584 GHAGDPASALLELLDPEQNANFLDHYLDVTIDLSKVLFVCTANVIDMIPNPLLDRMEVIS 643
Query: 544 LPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLA 603
+ GY +EK+ IA +L G+ E +++ + + +I+ Y REAGVRNL++ +
Sbjct: 644 IAGYITDEKVHIARDYLEKTARGDCGVKPEQVEVSDAALLSLIENYCREAGVRNLQKQIE 703
Query: 604 SLARAAAVRVAE-----QEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDI 658
+ R A+++ +E V + E + + E+ + VE + ++
Sbjct: 704 KIYRKIALKLVREGAVPEEPAVASDPEEAEIVADVGES--IENHTVEENTVSSAEEPKEE 761
Query: 659 SNTFRIT-SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEAT 717
+ T +I ++++E+ L +G P F + E+ G+ +GL WT+ GG ++E T
Sbjct: 762 AQTEKIAIETVMIDESNLADYVGKPVFHAEKLYEQTPV-GVVMGLAWTSMGGSTLYIETT 820
Query: 718 ---EMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFP 774
E GKG L++TGQLGDV+KESAQIA T R + L +H+H P
Sbjct: 821 VVEEGEGKGGLNITGQLGDVMKESAQIAHTVAR----KIMLEKEPENQFFANSKLHLHVP 876
Query: 775 AGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRC 834
AGA PKDGPSAG T++T+L+SL +KK VR D AMTGE+TL G +LP+GG+K+K +AA R
Sbjct: 877 AGATPKDGPSAGCTMITSLLSLATKKPVRKDLAMTGEVTLTGRILPIGGVKEKTIAARRS 936
Query: 835 GIKRVIIPERNLKDLVEVPSSVLVNLEI 862
IK +I PE N +D E+ +V L +
Sbjct: 937 QIKTIIFPEANRRDFDELAENVKEGLNV 964
>At3g05790 putative mitochondrial LON ATP-dependent protease
Length = 942
Score = 464 bits (1195), Expect = e-131
Identities = 270/686 (39%), Positives = 396/686 (57%), Gaps = 35/686 (5%)
Query: 216 KLADIFVASFEISFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSK 275
KLAD + + +L+ +D RL ELV + ++ ++ E I + VE + S
Sbjct: 261 KLADFGAGISGANKHQNQGVLEELDVHKRLELTLELVKKEVEINKIQESIAKAVEEKFSG 320
Query: 276 SQKEFLLRQQMRAIKEELGDNDDDEDDLAALERKMQSA------GMPQSIWKHAHRELRR 329
++ +L++Q+ AIK+ELG D + +AL K + +P + K EL++
Sbjct: 321 DRRRIILKEQINAIKKELGGETDSK---SALSEKFRGRIDPIKDKIPGHVLKVIEEELKK 377
Query: 330 LKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIE 389
L+ ++ + + YLD L LPW S+E+ ++ A++ LD DHYGL+ VK+RI+E
Sbjct: 378 LQLLETSSSEFDVTCNYLDWLTVLPWGNFSDEN-FNVLRAEKILDEDHYGLSDVKERILE 436
Query: 390 YLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRR 449
++AV L+ ++G ++C GP GVGKTS+ SIA ALDRKF R S+GG+ D A+I+GHRR
Sbjct: 437 FIAVGGLRGTSQGKIICLSGPTGVGKTSIGRSIARALDRKFFRFSVGGLSDVAEIKGHRR 496
Query: 450 TYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTG-SDVRGDPASALLEVLDPEQNKTFNDH 508
TYIG+MPG+++ LK V NP++L+DEIDK G GDPASA+LE+LDPEQN F DH
Sbjct: 497 TYIGAMPGKMVQCLKNVGTENPLVLIDEIDKLGVRGHHGDPASAMLELLDPEQNANFLDH 556
Query: 509 YLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQH 568
YL+VP DLSKV+FV TAN IP PLLDRMEVI L GY +EK+ IA +L
Sbjct: 557 YLDVPIDLSKVLFVCTANVTDTIPGPLLDRMEVITLSGYITDEKMHIARDYLEKTARRDC 616
Query: 569 GLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQ----------EQ 618
G+ E + + + +I+ Y REAGVRNL++ + + R A+++ + +
Sbjct: 617 GIKPEQVDVSDAAFLSLIEHYCREAGVRNLQKQIEKIFRKIALKLVRKAASTEVPRISDD 676
Query: 619 VVPLNKEVQGLTTPLLENRLT--DGAEVEMEVI----PMDVNSRDISNTFRITSPLVVNE 672
V +E + L LE+ T +G+ V + + P + + +++E
Sbjct: 677 VTTDTEETKSLAKTDLESPETSAEGSTVLTDELATGDPTESTTEQSGEVAETVEKYMIDE 736
Query: 673 AILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEAT---EMVGKGDLHLTG 729
+ L +G P F + E+ G+ +GL WT+ GG ++E T E GKG LH+TG
Sbjct: 737 SNLSDYVGKPVFQEEKIYEQTPV-GVVMGLAWTSMGGSTLYIETTFVEEGEGKGGLHITG 795
Query: 730 QLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAIPKDGPSAGVTL 789
+LGDV+KESA+IA T R + L L +H+H PAGA PKDGPSAG T+
Sbjct: 796 RLGDVMKESAEIAHTVARR----IMLEKEPENKLFANSKLHLHVPAGATPKDGPSAGCTM 851
Query: 790 VTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVIIPERNLKDL 849
+T+L+SL KK VR D AMTGE+TL G +L +GG+K+K +AA R +K +I PE N +D
Sbjct: 852 ITSLLSLALKKPVRKDLAMTGEVTLTGRILAIGGVKEKTIAARRSQVKVIIFPEANRRDF 911
Query: 850 VEVPSSVLVNLEILLAKRMEDVLEQA 875
E+ +V LE+ E + E A
Sbjct: 912 DELARNVKEGLEVHFVDEYEQIFELA 937
>At3g05780 putative mitochondrial LON ATP-dependent protease
Length = 924
Score = 442 bits (1138), Expect = e-124
Identities = 298/880 (33%), Positives = 458/880 (51%), Gaps = 93/880 (10%)
Query: 14 LPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRDAAETEVKPVGPTI 73
LP +K L+PG + I P + +++ R+ +G ++D A T+ +
Sbjct: 115 LPLPHKPLIPGFYMPIHVKDPKVLAALQEST--RQQSPYVGAFLLKDCASTD----SSSR 168
Query: 74 SQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHW-HNRGVAARALHLSRGVEKPSGRVTY 132
S+ D + + KV G K K + ++++ H G A+ + +G +
Sbjct: 169 SETEDNVVEKFKVKG------KPKKKRRKELLNRIHQVGTLAQISSI-QGEQ-------- 213
Query: 133 IVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSRQFKATAMELI 192
V+L G R ++E+ + T R+ L+ +D D ++ KA+ +E+I
Sbjct: 214 -VILVGRRRLIIEEMVSEDPL-TVRVDHLK-------DKPYDKDNAVI----KASYVEVI 260
Query: 193 SVLEQKQKTGGRTKVL-LETVPVHKLADIFVASFEISFEEQLSMLDSVDAKLRLSKATEL 251
S L + KT + + LAD + + +L +D RL EL
Sbjct: 261 STLREVLKTNSLWRDQDIGDFSYQHLADFGAGISGANKHKNQGVLTELDVHKRLELTLEL 320
Query: 252 VDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDLAALER--- 308
V + ++ ++ E DD L+A R
Sbjct: 321 VKKQVEINKIKET---------------------------------DDGSSLSAKIRVRI 347
Query: 309 KMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKS 368
+ +P+ + K E +L+ ++ + + YL L LPW S E+ D+
Sbjct: 348 DTKRDKIPKHVIKVMEEEFTKLEMLEENYSDFDLTYNYLHWLTVLPWGNFSYEN-FDVLR 406
Query: 369 AQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAALDR 428
A++ LD DHYGL+ VK+RI+E++AV +L+ ++G ++C GPPGVGKTS+ SIA ALDR
Sbjct: 407 AKKILDEDHYGLSDVKERILEFIAVGRLRGTSQGKIICLSGPPGVGKTSIGRSIARALDR 466
Query: 429 KFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGD 488
KF R S+GG+ D A+I+GH +TY+G+MPG+++ LK V NP++L DEIDK G GD
Sbjct: 467 KFFRFSVGGLSDVAEIKGHCQTYVGAMPGKMVQCLKSVGTANPLILFDEIDKLGRCHTGD 526
Query: 489 PASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIELPGYT 548
PASALLEV+DPEQN F DH+LNV DLSKV+FV TAN + IP PLLDRMEVI+L GY
Sbjct: 527 PASALLEVMDPEQNAKFLDHFLNVTIDLSKVLFVCTANVIEMIPGPLLDRMEVIDLSGYV 586
Query: 549 PEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARA 608
+EK+ IA +L+ + G+ E++ + + + +I+ Y REAGVRNL++ + + R
Sbjct: 587 TDEKMHIARDYLVKKTCRDCGIKPEHVDLSDAALLSLIENYCREAGVRNLQKQIEKIYRK 646
Query: 609 AAVRVAEQEQV---VPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNT---- 661
A+ + Q V V K+ + L E + A++ M+++ S T
Sbjct: 647 VALELVRQGAVSFDVTDTKDTKSLAKTDSEVKRMKVADI-MKILESATGDSTESKTKQSG 705
Query: 662 --FRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEAT-- 717
+ ++++E+ L +G P F + E+ G+ +GL WT+ GG ++E T
Sbjct: 706 LVAKTFEKVMIDESNLADYVGKPVFQEEKIYEQTPV-GVVMGLAWTSMGGSTLYIETTFV 764
Query: 718 -EMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINL-LEGRDIHIHFPA 775
E +GKG LH+TGQLGDV+KESAQIA T R + E NL +H+H P
Sbjct: 765 EEGLGKGGLHITGQLGDVMKESAQIAHTVARR-----IMFEKEPENLFFANSKLHLHVPE 819
Query: 776 GAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCG 835
GA PKDGPSAG T++T+ +SL KK VR D AMTGE+TL G +LP+GG+K+K +AA R
Sbjct: 820 GATPKDGPSAGCTMITSFLSLAMKKLVRKDLAMTGEVTLTGRILPIGGVKEKTIAARRSQ 879
Query: 836 IKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQA 875
IK +I PE N +D E+ ++ L++ E + + A
Sbjct: 880 IKTIIFPEANRRDFEELAENMKEGLDVHFVDEYEKIFDLA 919
>At3g05530 26S proteasome AAA-ATPase subunit RPT5a
Length = 424
Score = 49.3 bits (116), Expect = 1e-05
Identities = 89/377 (23%), Positives = 161/377 (42%), Gaps = 72/377 (19%)
Query: 223 ASFEISFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKITQ----KVEGQLSKSQK 278
+SFE E+QL+ + + D +++AT L+D ++ ++ + T + ++ ++Q+
Sbjct: 10 SSFE---EDQLASMSTED----ITRATRLLDNEIRILKEDAQRTNLECDSYKEKIKENQE 62
Query: 279 EFLLRQQMRAIKE------ELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLK- 331
+ L +Q+ + E+ DD E+D A ++ Q G + K + R+ L
Sbjct: 63 KIKLNKQLPYLVGNIVEILEMNPEDDAEEDGANIDLDSQRKGKCV-VLKTSTRQTIFLPV 121
Query: 332 -------KMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVK 384
++P + +YL +L LP + S M++ +D GL K
Sbjct: 122 VGLVDPDSLKPGDLVGVNKDSYL-ILDTLPSEYDSRVKAMEVDEKPTEDYNDIGGLEKQI 180
Query: 385 QRIIEYLAV------RKLKPDARGPV-LCFVGPPGVGKTSLASSIAAALDRKFVRISLGG 437
Q ++E + + R K R P + GPPG GKT +A + AA + F++++
Sbjct: 181 QELVEAIVLPMTHKERFEKLGVRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAGPQ 240
Query: 438 VKDEADIRGHRRTYIGSMPGRLIDGL---KRVAVCNPVMLLDEIDKTG-----SDVRGD- 488
+ + +IG + D K A C ++ +DEID G S+V GD
Sbjct: 241 LV---------QMFIGDGAKLVRDAFQLAKEKAPC--IIFIDEIDAIGTKRFDSEVSGDR 289
Query: 489 -PASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPL-----LDRMEVI 542
+LE+L+ + ++ ++ +A NRA + P L LDR I
Sbjct: 290 EVQRTMLELLNQLDGFSSDE----------RIKVIAATNRADILDPALMRSGRLDRK--I 337
Query: 543 ELPGYTPEEKLKIAMKH 559
E P T E + +I H
Sbjct: 338 EFPHPTEEARARILQIH 354
>At5g43010 26S proteasome AAA-ATPase subunit RPT4a (gb|AAF22524.1)
Length = 399
Score = 45.1 bits (105), Expect = 2e-04
Identities = 51/196 (26%), Positives = 89/196 (45%), Gaps = 26/196 (13%)
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA +IA+ +D F+++ + D+ YIG + +
Sbjct: 180 GPPGTGKTLLARAIASNIDANFLKVVSSAIIDK---------YIGESARLIREMFNYARE 230
Query: 469 CNP-VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFND--HYLNVPFDLSKVIFVATA 525
P ++ +DEID G + SA D E +T + + L+ +L KV +
Sbjct: 231 HQPCIIFMDEIDAIGGRRFSEGTSA-----DREIQRTLMELLNQLDGFDNLGKVKMIMAT 285
Query: 526 NRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEY---LQIPE 579
NR + P LL R++ IE+P P E+ ++ + + + +HG +Y +++ E
Sbjct: 286 NRPDVLDPALLRPGRLDRKIEIP--LPNEQSRMDILKIHAAGIAKHG-EIDYEAIVKLAE 342
Query: 580 GMVQLVIQRYTREAGV 595
G ++ EAG+
Sbjct: 343 GFNGADLRNICTEAGM 358
>At4g14670 heat shock protein like
Length = 668
Score = 44.7 bits (104), Expect = 2e-04
Identities = 91/430 (21%), Positives = 168/430 (38%), Gaps = 61/430 (14%)
Query: 117 LHLSRGVEKPSGRVTYIVVLE-----GLCRFSVQELSTRGTYHTARISSLEMNKTEMEQV 171
+H++ G K SG +L+ G RF H + ++ E ++
Sbjct: 246 IHMALGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERRFQQVFVA 305
Query: 172 EHD-PDFIMLSRQFKAT------------AMELISVLEQKQKTGGRTKV----LLETVPV 214
E PD I + R K A+ L + L ++ TG R L++
Sbjct: 306 EPSVPDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKAIDLVDESCA 365
Query: 215 HKLADIFVASFEI-SFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVA-EKITQKVEGQ 272
H A + + EI S E ++ L+ L K + + L +R + + K+E
Sbjct: 366 HVKAQLDIQPEEIDSLERKVMQLEIEIHALEKEKDDKASEARLSEVRKELDDLRDKLEPL 425
Query: 273 LSKSQKEFLLRQQMRAIKEELGDNDDDEDDLAALERKM---QSAGMPQSIWKHAHRELRR 329
K +KE + + R +K+ + DD L ER+ ++A + + + +
Sbjct: 426 TIKYKKEKKIINETRRLKQ---NRDDLMIALQEAERQHDVPKAAVLKYGAIQEVESAIAK 482
Query: 330 LKK------MQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKV 383
L+K M + G + + +P + + + L S ++L G +
Sbjct: 483 LEKSAKDNVMLTETVGPENIAEVVSRWTGIPVTRLDQNEKKRLISLADKLHERVVGQDEA 542
Query: 384 KQRIIEYLAVRKL---KPDARGPVLCFVGPPGVGKTSLASSIAAAL---DRKFVRISLGG 437
+ + + ++ +P F+GP GVGKT LA ++A L + VR+ +
Sbjct: 543 VKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMSE 602
Query: 438 VKDEADIRGHRRTYIGSMP---------GRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGD 488
D+ + IG+ P G+L + ++R C V+L DE++KT V
Sbjct: 603 YNDKFSV----NKLIGAPPGYYIGHEEGGQLTEPVRRRPYC--VVLFDEVEKTHVTV--- 653
Query: 489 PASALLEVLD 498
+ LL+VL+
Sbjct: 654 -FNTLLQVLE 662
Score = 34.3 bits (77), Expect = 0.32
Identities = 43/148 (29%), Positives = 65/148 (43%), Gaps = 12/148 (8%)
Query: 357 KASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKT 416
+A + + DL +LD G + +R+IE L+ R PVL +G PGVGKT
Sbjct: 127 RALKTYGTDLVEQAGKLDPV-IGRHREIRRVIEVLSRRT----KNNPVL--IGEPGVGKT 179
Query: 417 SLASSIAAALDRKFVRISLGGVK---DEADIRGHRRTYIGSMPGRLIDGLKRV--AVCNP 471
++ +A + + V I+L GVK E T G RL LK V A
Sbjct: 180 AVVEGLAQRILKGDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQGKV 239
Query: 472 VMLLDEIDKTGSDVRGDPASALLEVLDP 499
V+ +DEI + ++ ++L P
Sbjct: 240 VLFIDEIHMALGACKASGSTDAAKLLKP 267
>At1g74310 heat shock protein 101
Length = 911
Score = 44.7 bits (104), Expect = 2e-04
Identities = 68/307 (22%), Positives = 127/307 (41%), Gaps = 35/307 (11%)
Query: 407 FVGPPGVGKTSLASSIAAAL---DRKFVRISLGGVKDEADIR---GHRRTYIG-SMPGRL 459
F+GP GVGKT LA ++A L + VRI + ++ + G Y+G G+L
Sbjct: 604 FLGPTGVGKTELAKALAEQLFDDENLLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEGGQL 663
Query: 460 IDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKV 519
+ ++R C V+L DE++K V + LL+VLD + + V F S +
Sbjct: 664 TEAVRRRPYC--VILFDEVEKAHVAV----FNTLLQVLD--DGRLTDGQGRTVDFRNSVI 715
Query: 520 IFVATANRAQQIPPPLLDR--MEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQI 577
I + A+ + L + MEV + ++ KH P +L++ + +
Sbjct: 716 IMTSNLG-AEHLLAGLTGKVTMEVAR------DCVMREVRKHFRPELLNRLDEIVVFDPL 768
Query: 578 PEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQVVPL-----------NKEV 626
++ V + ++ VR ER +A AA+ E P+ K V
Sbjct: 769 SHDQLRKVARLQMKDVAVRLAERGVALAVTDAALDYILAESYDPVYGARPIRRWMEKKVV 828
Query: 627 QGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNTFRITSPLVVNEAILEKVLGPPKFDG 686
L+ ++ + + + V ++ D+ R S S ++ ++ GP + D
Sbjct: 829 TELSKMVVREEIDENSTVYIDAGAGDLVYRVESGGLVDASTGKKSDVLIHIANGPKRSDA 888
Query: 687 REAAERV 693
+A +++
Sbjct: 889 AQAVKKM 895
>At1g45000 26S proteasome AAA-ATPase subunit RPT4a like protein
Length = 399
Score = 44.7 bits (104), Expect = 2e-04
Identities = 51/196 (26%), Positives = 88/196 (44%), Gaps = 26/196 (13%)
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA +IA+ +D F+++ + D+ YIG + +
Sbjct: 180 GPPGTGKTLLARAIASNIDANFLKVVSSAIIDK---------YIGESARLIREMFNYARE 230
Query: 469 CNP-VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFND--HYLNVPFDLSKVIFVATA 525
P ++ +DEID G + SA D E +T + + L+ L KV +
Sbjct: 231 HQPCIIFMDEIDAIGGRRFSEGTSA-----DREIQRTLMELLNQLDGFDQLGKVKMIMAT 285
Query: 526 NRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEY---LQIPE 579
NR + P LL R++ IE+P P E+ ++ + + + +HG +Y +++ E
Sbjct: 286 NRPDVLDPALLRPGRLDRKIEIP--LPNEQSRMEILKIHASGIAKHG-EIDYEAIVKLGE 342
Query: 580 GMVQLVIQRYTREAGV 595
G ++ EAG+
Sbjct: 343 GFNGADLRNICTEAGM 358
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 44.3 bits (103), Expect = 3e-04
Identities = 69/287 (24%), Positives = 124/287 (43%), Gaps = 31/287 (10%)
Query: 231 EQLSMLDSVD--AKLRLSKA-TELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMR 287
E+LS+ + D ++ RL++ TELV L + AE +T++ E + S + +++++
Sbjct: 501 ERLSLTNDTDKASRERLNRIETELV---LLKEKQAE-LTEQWEHERSVMSRLQSIKEEID 556
Query: 288 AIKEELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRL----KKMQPQQPGYSSS 343
+ E+ + + D A E K S Q A +EL K M ++ S
Sbjct: 557 RVNLEIQQAEREYDLNRAAELKYGSLNSLQRQLNEAEKELNEYLSSGKSMFREEVLGSDI 616
Query: 344 RAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGP 403
+ +P K + L +E L G + E A+++ + P
Sbjct: 617 AEIVSKWTGIPVSKLQQSERDKLLHLEEELHKRVVGQNPAVTAVAE--AIQRSRAGLSDP 674
Query: 404 -----VLCFVGPPGVGKTSLASSIAAAL---DRKFVRISLGGVKDE---ADIRGHRRTYI 452
F+GP GVGKT LA ++A+ + + VRI + ++ + + G Y+
Sbjct: 675 GRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMSEYMEKHAVSRLIGAPPGYV 734
Query: 453 G-SMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLD 498
G G+L + ++R V+L DEI+K GD + L++LD
Sbjct: 735 GYEEGGQLTETVRRRPY--SVILFDEIEK----AHGDVFNVFLQILD 775
Score = 31.2 bits (69), Expect = 2.7
Identities = 22/86 (25%), Positives = 42/86 (48%), Gaps = 17/86 (19%)
Query: 403 PVLCFVGPPGVGKTSLASSIA----------AALDRKFVRISLGGVKDEADIRGHRRTYI 452
PVL +G PGVGKT+++ +A A ++RK + + +G + A RG +
Sbjct: 278 PVL--IGEPGVGKTAISEGLAQRIVQGDVPQALMNRKLISLDMGALIAGAKYRGEFEDRL 335
Query: 453 GSMPGRLIDGLKRVAVCNPVMLLDEI 478
++ + D ++ ++ +DEI
Sbjct: 336 KAVLKEVTDSEGQI-----ILFIDEI 356
>At1g09100 putative 26S protease regulatory subunit 6A
Length = 423
Score = 44.3 bits (103), Expect = 3e-04
Identities = 85/372 (22%), Positives = 157/372 (41%), Gaps = 64/372 (17%)
Query: 226 EISFE-EQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKIT----QKVEGQLSKSQKEF 280
+ SFE +QL+ + + D + +A+ L+ ++ ++ + T + V+ ++ ++Q++
Sbjct: 8 DTSFEGDQLASMTTDD----IGRASRLLANEIRILKEESQRTNLDLESVKEKIKENQEKI 63
Query: 281 LLRQQMRAIKE------ELGDNDDDEDDLAALERKMQSAG----MPQSIWKHAHRELRRL 330
L +Q+ + E+ DD E+D A ++ Q G + S + + L
Sbjct: 64 KLNKQLPYLVGNIVEILEMSPEDDAEEDGANIDLDSQRKGKCVVLKTSTRQTIFLPVVGL 123
Query: 331 KKMQPQQPGYS---SSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRI 387
+PG + +YL +L LP + S M++ +D GL K Q +
Sbjct: 124 VDPDTLKPGDLVGVNKDSYL-ILDTLPSEYDSRVKAMEVDEKPTEDYNDIGGLEKQIQEL 182
Query: 388 IEYLAVRKL------KPDARGPV-LCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKD 440
+E + + K R P + GPPG GKT +A + AA + F++++ +
Sbjct: 183 VEAIVLPMTHKEQFEKLGIRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAGPQLV- 241
Query: 441 EADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VMLLDEIDKTG-----SDVRGD--PASA 492
+ +IG + D +P ++ +DEID G S+V GD
Sbjct: 242 --------QMFIGDGAKLVRDAFLLAKEKSPCIIFIDEIDAIGTKRFDSEVSGDREVQRT 293
Query: 493 LLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPL-----LDRMEVIELPGY 547
+LE+L+ + +D ++ +A NRA + P L LDR IE P
Sbjct: 294 MLELLNQLDGFSSDD----------RIKVIAATNRADILDPALMRSGRLDRK--IEFPHP 341
Query: 548 TPEEKLKIAMKH 559
T E + +I H
Sbjct: 342 TEEARGRILQIH 353
>At5g50920 ATP-dependent Clp protease, ATP-binding subunit
Length = 929
Score = 43.9 bits (102), Expect = 4e-04
Identities = 110/522 (21%), Positives = 200/522 (38%), Gaps = 86/522 (16%)
Query: 169 EQVEHDPDFIMLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVH-KLA---DIFVAS 224
+ +E DP L R+F+ + +V E Q G L E +H KL + VA+
Sbjct: 416 KHIEKDP---ALERRFQPVKVPEPTVDETIQILKG----LRERYEIHHKLRYTDESLVAA 468
Query: 225 FEISFE---------EQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSK 275
++S++ + + ++D +++RL RH Q A ++ +++ Q++K
Sbjct: 469 AQLSYQYISDRFLPDKAIDLIDEAGSRVRL--------RHAQVPEEARELEKELR-QITK 519
Query: 276 SQKEFLLRQQMRAIKEELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQP 335
+ E + Q E+ G D E +L A +Q+ G S A E M
Sbjct: 520 EKNEAVRGQDF----EKAGTLRDREIELRAEVSAIQAKGKEMS---KAESETGEEGPMVT 572
Query: 336 QQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYG---LAKVKQRIIEYLA 392
+ S + + +P +K S + L +E L G K R I
Sbjct: 573 E----SDIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKRIIGQDEAVKAISRAIRRAR 628
Query: 393 VRKLKPDARGPVLCFVGPPGVGKTSLASSIAA---ALDRKFVRISLGGVKDE---ADIRG 446
V P+ F GP GVGK+ LA ++AA + +R+ + + + + G
Sbjct: 629 VGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIG 688
Query: 447 HRRTYIG-SMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRG------------------ 487
Y+G + G+L + ++R V+L DEI+K DV
Sbjct: 689 SPPGYVGYTEGGQLTEAVRRRPY--TVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRT 746
Query: 488 -DPASALLEVLDPEQNKTFNDHYLNVPFDL---------SKVIFVATANRAQQIPPPLLD 537
D + LL + + + FDL +++ + T Q P L+
Sbjct: 747 VDFKNTLLIMTSNVGSSVIEKGGRRIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLN 806
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVI-QRYTREAGVR 596
R++ + + + ++K L+ V ++ LQ+ E + V+ + Y G R
Sbjct: 807 RLDEMIVFRQLTKLEVKEIADILLKEVFERLKKKEIELQVTERFKERVVDEGYNPSYGAR 866
Query: 597 NLERNLA-----SLARAAAVRVAEQEQVVPLNKEVQGLTTPL 633
L R + S+A R ++ V ++ + +G T L
Sbjct: 867 PLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDVDAEGNVTVL 908
>At5g58870 cell division protein - like
Length = 806
Score = 43.1 bits (100), Expect = 7e-04
Identities = 53/190 (27%), Positives = 79/190 (40%), Gaps = 27/190 (14%)
Query: 385 QRIIEYLAV--RKLKPDARGPV-LCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDE 441
+ I+E+L R ++ AR P + VG PG GKT LA ++A D F+ S +
Sbjct: 342 EEIVEFLKNPDRYVRLGARPPRGVLLVGLPGTGKTLLAKAVAGESDVPFISCSASEFVE- 400
Query: 442 ADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VMLLDEIDKTGSDVRGDPASALLEVLDPE 500
Y+G R+ D R P ++ +DEID G V + E
Sbjct: 401 --------LYVGMGASRVRDLFARAKKEAPSIIFIDEIDAVAKSRDG----KFRMVSNDE 448
Query: 501 QNKTFNDHYLNVP-FDLSK-VIFVATANRAQQIPPPL-----LDRMEVIELPGYTPEE-- 551
+ +T N + FD S VI + NRA + P L DR+ +E P E
Sbjct: 449 REQTLNQLLTEMDGFDSSSAVIVLGATNRADVLDPALRRPGRFDRVVTVESPDKVGRESI 508
Query: 552 -KLKIAMKHL 560
K+ ++ K L
Sbjct: 509 LKVHVSKKEL 518
>At4g23940 cell division protein - like
Length = 946
Score = 42.7 bits (99), Expect = 9e-04
Identities = 39/134 (29%), Positives = 58/134 (43%), Gaps = 15/134 (11%)
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT +A +IA F +++ + +G R+ D KR V
Sbjct: 470 GPPGCGKTLVAKAIAGEAGVPFYQMAGSEFVE---------VLVGVGSARIRDLFKRAKV 520
Query: 469 CNP-VMLLDEIDKTGSDVRG---DPASALLEVLDPEQNKTFNDHYLNVP-FDLSK-VIFV 522
P V+ +DEID + +G + + L E+ T N + + FD K VIF+
Sbjct: 521 NKPSVIFIDEIDALATRRQGIFKENSDQLYNAATQERETTLNQLLIELDGFDTGKGVIFL 580
Query: 523 ATANRAQQIPPPLL 536
NR + P LL
Sbjct: 581 GATNRRDLLDPALL 594
>At5g17730 BCS1 - like protein
Length = 470
Score = 42.4 bits (98), Expect = 0.001
Identities = 35/96 (36%), Positives = 47/96 (48%), Gaps = 7/96 (7%)
Query: 353 LPWQKASEEHE--MDLKSAQERLDSDHYG-LAKVKQRIIEYLAVRKLKPDARGPVLCFVG 409
L WQ + EH D + E L G L + +R Y V K P RG +L G
Sbjct: 197 LKWQSVNLEHPSTFDTMAMNEELKRSVMGDLDRFIRRKDFYKRVGK--PWKRGYLL--YG 252
Query: 410 PPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIR 445
PPG GKTSL ++IA L + L V+++AD+R
Sbjct: 253 PPGTGKTSLVAAIANYLKFDIYDLQLASVREDADLR 288
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 42.0 bits (97), Expect = 0.002
Identities = 75/337 (22%), Positives = 143/337 (42%), Gaps = 49/337 (14%)
Query: 75 QDGDTLDQSSKVHGASSDSHKLD---AKAQNDVVHWHNRGVAARALHLSRGVEKPSGRVT 131
+DG+ + + A D+ K D + Q +V+ + G + R LH S+ EK
Sbjct: 44 EDGEFIKVEKEAFDAKDDAEKADHVPVEEQKEVIERSSSG-SQRELHESQ--EKAKELEL 100
Query: 132 YIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSRQFKATAMEL 191
+ + G ++ + T+ + S + E E+ D + + +Q K
Sbjct: 101 ELERVAG----ELKRYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQEK------ 150
Query: 192 ISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAKL-------- 243
++E +++ + K L + + H D + + +F+ L+S KL
Sbjct: 151 --IVEGEERHSSQLKSLEDALQSHDAKDKELTEVKEAFDALGIELESSRKKLIELEEGLK 208
Query: 244 -RLSKATELVDRHLQSIRVAEKITQKVE--GQLSKSQKEFL---------LRQQMRAIKE 291
+A + + H QS A+ +QK +L KS KE L+Q+++ + E
Sbjct: 209 RSAEEAQKFEELHKQSASHADSESQKALEFSELLKSTKESAKEMEEKMASLQQEIKELNE 268
Query: 292 ELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLD-LL 350
++ +N+ E AAL+ SAG ++ + RL + + + SS+ A +D L
Sbjct: 269 KMSENEKVE---AALK---SSAGELAAVQEELALSKSRLLETEQK---VSSTEALIDELT 319
Query: 351 ADLPWQKASEEHEMDLKSAQERLDSDHYGL-AKVKQR 386
+L +KASE + S + LD+ GL AK+ ++
Sbjct: 320 QELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQ 356
Score = 31.6 bits (70), Expect = 2.1
Identities = 62/280 (22%), Positives = 115/280 (40%), Gaps = 38/280 (13%)
Query: 38 KLVE-QELWQREDKGLIGILPVRDAAETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKL 96
K+VE +E + K L L DA + E+ V G L+ S K + K
Sbjct: 150 KIVEGEERHSSQLKSLEDALQSHDAKDKELTEVKEAFDALGIELESSRKKLIELEEGLKR 209
Query: 97 DAKAQNDVVHWHNRGVAARALHLSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTA 156
A+ H + + + +E + +L+ + S +E+ +
Sbjct: 210 SAEEAQKFEELHKQSASHADSESQKALE-------FSELLKST-KESAKEMEEKMASLQQ 261
Query: 157 RISSLEMNKTEMEQVEHDPDFIMLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHK 216
I L +E E+VE K++A EL +V E+ + R LLET
Sbjct: 262 EIKELNEKMSENEKVE---------AALKSSAGELAAVQEELALSKSR---LLETEQKVS 309
Query: 217 LADIFVASF----------EISFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKIT 266
+ + E F+E+LS+L +DA+ + +A +L ++ + ++AE++
Sbjct: 310 STEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQA-KLSEQEGINSKLAEELK 368
Query: 267 QK-VEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDLAA 305
+K + LSK Q+E ++R E+L + +++ L A
Sbjct: 369 EKELLESLSKDQEE-----KLRTANEKLAEVLKEKEALEA 403
>At3g02450 cell division protein FtsH-like protein
Length = 622
Score = 41.2 bits (95), Expect = 0.003
Identities = 62/276 (22%), Positives = 104/276 (37%), Gaps = 45/276 (16%)
Query: 376 DHYGLAKVKQRIIEYLA-----VRKLKPDARGPV-LCFVGPPGVGKTSLASSIAAALDRK 429
D G+ K ++E ++ + K AR P + VGPPG GKT LA ++A
Sbjct: 335 DVEGVDSAKDELVEIVSCLQGSINYKKLGARLPRGVLLVGPPGTGKTLLARAVAGEAGVP 394
Query: 430 FVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VMLLDEIDKTG------ 482
F +S + ++G R+ D +P ++ +DE+D G
Sbjct: 395 FFSVSASEFVE---------LFVGRGAARIRDLFNAARKNSPSIIFIDELDAVGGKRGRS 445
Query: 483 -SDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIP-----PPLL 536
+D R + LL +D ++ T KVI +A NR + + P
Sbjct: 446 FNDERDQTLNQLLTEMDGFESDT-------------KVIVIAATNRPEALDSALCRPGRF 492
Query: 537 DRMEVIELPGYTPEEKLKIAMKHL--IPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAG 594
R ++ P E + KI HL +P D + + G V + EA
Sbjct: 493 SRKVLVAEP--DQEGRRKILAIHLRDVPLEEDAFLICDLVASLTPGFVGADLANIVNEAA 550
Query: 595 VRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGLT 630
+ R ++AR + E+ + +KE + T
Sbjct: 551 LLAARRGGEAVAREDIMEAIERAKFGINDKEARPRT 586
>At3g16290 putative FtsH-like metalloprotease
Length = 876
Score = 40.8 bits (94), Expect = 0.003
Identities = 90/387 (23%), Positives = 148/387 (37%), Gaps = 79/387 (20%)
Query: 270 EGQLSKSQKEFLLRQQMRAIKEEL-GDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELR 328
E +L + E R++MR ++ E+ G ++DE+ K M K R R
Sbjct: 332 EDRLKIEKAEADERKKMRELEREMEGIEEEDEEVEEGTGEKNPYLQMAMQFMKSGARVRR 391
Query: 329 RLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRII 388
K P+ YL+ D+ + +D GL K++ +
Sbjct: 392 ASNKRLPE---------YLERGVDVKF-------------------TDVAGLGKIRLELE 423
Query: 389 EYLAV--------RKLKPDARGPVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKD 440
E + R+ G +LC GPPGVGKT LA ++A F IS +
Sbjct: 424 EIVKFFTHGEMYRRRGVKIPGGILLC--GPPGVGKTLLAKAVAGEAGVNFFSISASQFVE 481
Query: 441 EADIRGHRRTYIGSMPGRLIDGLKRVAVCN--PVMLLDEIDKTGSD---VRGDPASALLE 495
Y+G R + L + A N V+ +DE+D G + ++G
Sbjct: 482 ---------IYVGVGASR-VRALYQEARENAPSVVFIDELDAVGRERGLIKGSGGQ---- 527
Query: 496 VLDPEQNKTFNDHYLNVPFD----LSKVIFVATANRAQQIPPPLL-----DRMEVIELPG 546
E++ T N L V D +VI +A+ NR + P L+ DR I PG
Sbjct: 528 ----ERDATLNQ--LLVSLDGFEGRGEVITIASTNRPDILDPALVRPGRFDRKIFIPKPG 581
Query: 547 YTPE-EKLKI-AMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLAS 604
E L++ A K + LD ++S + +GMV + A + +
Sbjct: 582 LIGRMEILQVHARKKPMAEDLDYMAVAS----MTDGMVGAELANIVEIAAINMMRDGRTE 637
Query: 605 LARAAAVRVAEQEQVVPLNKEVQGLTT 631
L ++ A+ E+ L+++ + L T
Sbjct: 638 LTTDDLLQAAQIEERGMLDRKDRSLET 664
>At1g67120 hypothetical protein
Length = 5138
Score = 40.4 bits (93), Expect = 0.004
Identities = 37/115 (32%), Positives = 58/115 (50%), Gaps = 12/115 (10%)
Query: 401 RGPVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLI 460
R PVL GP GKTSL +AA KFVRI+ ++ DI+ + +Y+ G+L+
Sbjct: 959 RYPVL-LQGPTSSGKTSLVKYLAAISGNKFVRIN---NHEQTDIQEYLGSYMTDSSGKLV 1014
Query: 461 ---DGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALL----EVLDPEQNKTFNDH 508
L + ++LDE++ SDV + + LL E+ PE ++T + H
Sbjct: 1015 FHEGALVKAVRGGHWIVLDELNLAPSDVL-EALNRLLDDNRELFVPELSETISAH 1068
Score = 32.3 bits (72), Expect = 1.2
Identities = 17/38 (44%), Positives = 21/38 (54%)
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRG 446
G PGVGKTSL ++ K VRI+L D D+ G
Sbjct: 1596 GSPGVGKTSLILALGKYSGHKVVRINLSEQTDMMDLLG 1633
>At1g24290 unknown protein
Length = 525
Score = 39.7 bits (91), Expect = 0.008
Identities = 57/239 (23%), Positives = 99/239 (40%), Gaps = 52/239 (21%)
Query: 401 RGPVLCFVGPPGVGKTSLASSIAAALD----RKFVRIS--LGGVKDEADIRGHRRTYIGS 454
R P + F GPPG GKTS+A S+ + +FV +S GVKD D + S
Sbjct: 136 RLPSIVFWGPPGTGKTSIAKSLINSSKDPSLYRFVSLSAVTSGVKDVRDA-------VES 188
Query: 455 MPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPF 514
++G KR V+ +DE+ + NK+ D +L V
Sbjct: 189 AKRLNLEGRKRT-----VLFMDEVHRF--------------------NKSQQDTFLPVIE 223
Query: 515 DLSKVIFV--ATANRAQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIPRVLD--QHGL 570
D ++F+ T N + + PLL R V+ L P ++ L+ R +D + GL
Sbjct: 224 D-GSILFIGATTENPSFHLITPLLSRCRVLTLNPLKPNH-----VETLLRRAVDDSERGL 277
Query: 571 SSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGL 629
+ +++ + +++ + +A V N ++ A A + VV ++ + L
Sbjct: 278 PNS-VEVDDSVIEFLANNCDGDARV---ALNALEISATMATTRAGTDAVVSIDDAKEAL 332
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,547,359
Number of Sequences: 26719
Number of extensions: 775813
Number of successful extensions: 2990
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 80
Number of HSP's that attempted gapping in prelim test: 2873
Number of HSP's gapped (non-prelim): 178
length of query: 888
length of database: 11,318,596
effective HSP length: 108
effective length of query: 780
effective length of database: 8,432,944
effective search space: 6577696320
effective search space used: 6577696320
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0550.4


