

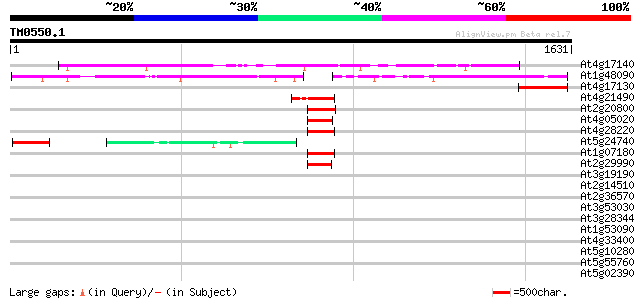
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0550.1
(1631 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g17140 hypothetical protein 1101 0.0
At1g48090 unknown protein 487 e-137
At4g17130 hypothetical protein 142 2e-33
At4g21490 NADH dehydrogenase like protein 125 2e-28
At2g20800 putative NADH-ubiquinone oxireductase 117 7e-26
At4g05020 unknown protein 114 4e-25
At4g28220 putative NADH dehydrogenase 110 9e-24
At5g24740 VPS13 - like protein 89 2e-17
At1g07180 unknown protein 62 2e-09
At2g29990 putative NADH dehydrogenase (ubiquinone oxidoreductase) 59 2e-08
At3g19190 hypothetical protein 42 0.003
At2g14510 putative receptor-like protein kinase 34 0.61
At2g36570 putative receptor-like protein kinase 33 1.0
At3g53030 serine protein kinase - like 33 1.8
At3g28344 P-glycoprotein, 5' partial 33 1.8
At1g53090 phytochrome A supressor spa1, putative 33 1.8
At4g33400 Dem -like protein 32 3.0
At5g10280 putative transcription factor MYB92 32 3.9
At5g55760 transcription regulator Sir2-like protein 31 5.2
At5g02390 putative protein 31 5.2
>At4g17140 hypothetical protein
Length = 1322
Score = 1101 bits (2848), Expect = 0.0
Identities = 659/1456 (45%), Positives = 856/1456 (58%), Gaps = 263/1456 (18%)
Query: 143 LFSTIIGNLKLSISNIHIRYEDGES------NPGHPFAAGVMLDKLSAVTVDDTGKETFI 196
+ +TI+GNLKLSISNIHIRYED ES NPGHPF+AGV L+KLSAVT+D++GKETFI
Sbjct: 4 VINTIVGNLKLSISNIHIRYEDLESLCYKCSNPGHPFSAGVTLEKLSAVTIDESGKETFI 63
Query: 197 TGGALDLIQKL-----SSMDLFAYQRITKNINSFL---NCKMRIVKQPLNPKSLSCWSVE 248
TGG LD IQK+ + ++L N L N M + + + SVE
Sbjct: 64 TGGTLDSIQKVPIVRSTEIELGCSLEGLVFCNRSLYETNIWMTALGKEYPCQETKAGSVE 123
Query: 249 LDRLAVYLDSDIIPWHASKEWEDLLPSEWFQIFKFGTKDGKPADRLLQKHSYVLEPVTGK 308
LDRLA YLDSD+ PWH K WE L P EW QIF++GTKDGKPAD L +KH Y+L+PV+G
Sbjct: 124 LDRLAFYLDSDMSPWHIDKPWEVLTPFEWDQIFRYGTKDGKPADCLTRKHFYILQPVSGN 183
Query: 309 GNYSKLLLNEVADSKQPLQKAVVNLDDVTISLSKDGYRDIMKLADNFAAFNQRLKYAHFR 368
YSK NE +++ QPLQKA VNLDDVT+ LSK GYRD+MKLADNFAAFNQRLKYAH+R
Sbjct: 184 AKYSKSQANESSNAVQPLQKAYVNLDDVTLCLSKGGYRDVMKLADNFAAFNQRLKYAHYR 243
Query: 369 PPVPVKADPRSWWKYAYRAVSDQMKKA--------------------------------- 395
P VPVK D +SWWKYAYR VS+Q+K A
Sbjct: 244 PSVPVKIDAKSWWKYAYRVVSEQIKIARYLNVNLYLFFLTDAGFLRNLHRLWKHLPLQFL 303
Query: 396 SGKMSWEQVLRYTSLRKRYIYLYASLLKSDPSQVTISGNKEIEDLDHELDIELILQWRML 455
+G+MSWE VL+YTSLRKRYI YASLLKSD S++ + ++EIE LD ELD ++ILQWRML
Sbjct: 304 TGRMSWEHVLKYTSLRKRYITQYASLLKSDISRIVVDDDEEIEALDRELDTDVILQWRML 363
Query: 456 AHKFVEQSAEPNLSVRKQKAGNSWWSFGWTGKSPKEDSEELSFSEEDWNRLNKIIGYKEG 515
AHKFVE+S + +KQ+A +SWW FG + + E + F++EDW RLNK+IGYKEG
Sbjct: 364 AHKFVERSVQAENYSKKQQAKSSWWPFGGKSEVSGGEGESIQFTDEDWERLNKVIGYKEG 423
Query: 516 DDGQSPVNSKADVMHTFLVVHMNHNASKLIGEAQDLVAELSCEDLSCSVKLYPETKVFDI 575
D+ N+K D +HTFL V M +ASKL ++ +AELSCE L+CSVKL+PETK+ DI
Sbjct: 424 DEQSIINNAKPDALHTFLEVQMKRSASKLYDGEKECLAELSCEGLNCSVKLFPETKIADI 483
Query: 576 KLGSYQLSSPKGLLAESAASFDSLVGVFKYKPFDDKVDWSMVAKASPCYMTYMKDSINQI 635
KLG Y+LSSP GLLAE Y+KDSI+ I
Sbjct: 484 KLGRYRLSSPSGLLAE-----------------------------------YLKDSIDGI 508
Query: 636 VKFFETNATVSQTIALETAAAVQLKIDEVKRTAQQQMNRALKDHARFSLDL-DIAAPKIT 694
V FFE++ VSQTIALETAAAVQ D+ K ++M+ L +F L L D++A
Sbjct: 509 VNFFESSTAVSQTIALETAAAVQ---DDYKHELTEEMDMYL----QFDLVLSDVSA---- 557
Query: 695 IPTDFYPDNTHATKLLLDLGNLMIRTQDDSRQESAEDNMYLRFDLVLSDVSAFLFDGDYH 754
LL+D G+ + R S+ + F
Sbjct: 558 --------------LLVD-GDYSWKQLSSKRASSSGRESSVTF----------------- 585
Query: 755 WSEISVNKLTHSTNTSFFPIIDRCGVILQLQQILLETPYYPSTRLAVRLPSLAFHFSPAR 814
P+ID+CGV+L+LQQI P YPSTRLAVRLPSL FHFSPAR
Sbjct: 586 -----------------LPVIDKCGVLLKLQQIRRPNPAYPSTRLAVRLPSLGFHFSPAR 628
Query: 815 YHRLMHVIKIFEEGDDGSSEFLRPWNQADLEGWLSLLTWK------------VMVSFICA 862
YHRLM V +IF+ DD SS+ LRPW +AD EGWLS+L+WK ++ FI
Sbjct: 629 YHRLMQVAQIFQTKDDESSQILRPWEEADFEGWLSILSWKGREASWQRRYLCLVGPFIYV 688
Query: 863 SLTIGGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKI-TLLEAGDHILSMFDKRITAFAD 921
+ G + ++ + + + V L G++ ++ I + ++ + I F
Sbjct: 689 LESPGSKSYKKYTSLRGKHIYKVPVELAGGVEHVLSIRNASRISEKVMEDVNALILMFDS 748
Query: 922 DKFQRDGIDVKTGSVV-VKSTDPISGLSETSSDHDDIESELDNQGVIDVAISERLFVTGV 980
+ ++ G+V + PI+GLS+TSSD + ESE + + D++ E ++VTGV
Sbjct: 749 EDSRKTWHSRLQGAVYRASGSAPIAGLSDTSSDSE--ESETEQKDGFDLSNLESVYVTGV 806
Query: 981 LDELKVCFSYSYQPDQSLMKVLLNQEKRLFEFRAIGG----------------------- 1017
LDELK+CFSY +Q D S M VLL +E +LFEFRA+GG
Sbjct: 807 LDELKICFSYGHQDDASFMAVLLARESKLFEFRALGGKVRWWYASKLSFCHFLFFLLKVC 866
Query: 1018 -----QVEVSMKDNDIFIGTILKSLEVEDLVCYSQPSQPRYLARSFIGAADEKSLFYDTM 1072
QVEVSM+ +D+FIGT+LKSLE+EDL PS F D
Sbjct: 867 LPLTNQVEVSMRGSDMFIGTVLKSLEIEDLSSEMLPS------------------FEDAE 908
Query: 1073 RENVESSGLIPTESDDKFYEAPETLADSDVYMQSPGGTSEYPSSSSNEIKFNYSSLEPPK 1132
+ E +E ++KFYEAPE L DS +Y+SL P
Sbjct: 909 SRSPERLDPTSSEGEEKFYEAPEILVDS----------------------IDYTSLRTPS 946
Query: 1133 FSRIIGLLPTDAPSTSTKEHELNDTLESFVKAQIIIYDQNSTRYNNIDKQVIVTLATLTF 1192
FSRI GLLP D + + +E ++L+SFVKAQI+IY Q S +Y NID QV+VTLATL+F
Sbjct: 947 FSRIDGLLPVDNKNITKPSNETTESLDSFVKAQIVIYHQTSPQYKNIDNQVMVTLATLSF 1006
Query: 1193 FCRRPTILAIMEFINSINIENGNLATSSDSSSTSMVKNDLSNDLDVLHVTTVEEHAVKGL 1252
FCRRPTILAI+EF+N+IN+E+ + + D+S + D + AVKGL
Sbjct: 1007 FCRRPTILAILEFVNAINVEDPSCESFEDNSPVAGEHTSPRRD----GFEDSRDAAVKGL 1062
Query: 1253 LGKGKSRVMFSLTLKMAQAQILLMKENETKLACLSQESLLAEIKVFPSSFSIKAALGN-L 1311
LGKGKSR++F+L L MA+AQI LM EN TK A LSQ++LL +IKV SF ++ L L
Sbjct: 1063 LGKGKSRIIFNLELNMARAQIFLMNENGTKFATLSQDNLLTDIKV---SFRTRSVLRQVL 1119
Query: 1312 KISDDSLSSSH---------------------------FYYWACDMRNPGGRSFVELEFT 1344
+I + +++S F++ C MR + L FT
Sbjct: 1120 EIYESVMTASQTTICTFGSVICETLEELLLLRYWLFVSFHFSLCGMRMDCVDE-IHLVFT 1178
Query: 1345 SFSCDDEDYEGYDFSLFGELSEVRIVYLNRFLQEIVGYFMGLVPNSPRSVVKVTDQVTNS 1404
SFS DEDYEG+D+ L G+ SEVRIVYLNRF+QE+ YFMGLVP+ + VVK+ DQ+T+S
Sbjct: 1179 SFSIIDEDYEGFDYCLSGQFSEVRIVYLNRFIQEVAEYFMGLVPSDSKGVVKMKDQITDS 1238
Query: 1405 EKWFSASDIEGSPAVKFDLSLRKPIILMPRNRDSLDFLRLDIVHITVRNTFQWIGGSKSE 1464
EKWF+ S+IEGSPA+K DLSL+KPII+MPR+ DS D+L+LDIVHITV NTFQW G K+E
Sbjct: 1239 EKWFTTSEIEGSPALKLDLSLKKPIIVMPRHTDSPDYLKLDIVHITVDNTFQWFAGDKNE 1298
Query: 1465 INAVHLETLMVQVEDI 1480
+NAVH+ET+ + V +
Sbjct: 1299 LNAVHVETMKIMVRTL 1314
>At1g48090 unknown protein
Length = 4099
Score = 487 bits (1254), Expect = e-137
Identities = 308/933 (33%), Positives = 490/933 (52%), Gaps = 142/933 (15%)
Query: 5 QVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGFLGSV 64
+V +LL+RYLG YV GL+ EAL+ISVWKGDV LK+++LK EALN+LKLPV VK+GF+G++
Sbjct: 7 EVLHLLRRYLGEYVHGLSTEALRISVWKGDVVLKDLKLKAEALNSLKLPVAVKSGFVGTI 66
Query: 65 KLKVPWSRLGQDPVLVYLDRIFLLAEPAT-------------------------QVEGCS 99
LKVPW LG++PV+V +DR+F+LA PA Q +
Sbjct: 67 TLKVPWKSLGKEPVIVLIDRVFVLAYPAPDDRTLKFFTLVGTEFAYTNYIPGGRQGKASR 126
Query: 100 EDAVQEAKKIRIEEMELKLWE-KSQQLKSEMKT-INPSSQGSA--KKLFSTIIGNLKLSI 155
A + MEL +E ++ L++ K+ + QG++ + +TIIGNLK+SI
Sbjct: 127 NQASADRGTSYFWLMELHGYEAETATLEARAKSKLGSPPQGNSWLGSIIATIIGNLKVSI 186
Query: 156 SNIHIRYEDG-------------------ESNPGHPFAAGVMLDKLSAVTVDDTGKETFI 196
SN+HIRYED SNPGHPFAAG+ L KL+AVT+D+ G ETF
Sbjct: 187 SNVHIRYEDSTRDSSEILASFFSYFNNICSSNPGHPFAAGITLAKLAAVTMDEEGNETFD 246
Query: 197 TGGALDLIQKLSSMDLFAYQRITKNINSFLNCKMRIVKQPLNPKSLSCWSVELDRLAVYL 256
T GALD ++K S++L+RLA+Y
Sbjct: 247 TSGALDKLRK---------------------------------------SLQLERLALYH 267
Query: 257 DSDIIPWHASKEWEDLLPSEWFQIFKFGTKDG---KPADRLLQKHSYVLEPVTGKGNYSK 313
DS+ PW K+W+++ P EW ++F+ G K+ K + Y+L P+ G Y +
Sbjct: 268 DSNSFPWEIEKQWDNITPEEWIEMFEDGIKEQTEHKIKSKWALNRHYLLSPINGSLKYHR 327
Query: 314 LLLNEVADSKQPLQKAVVNLDDVTISLSKDGYRDIMKLADNFAAFNQRLKYAHFRPPVPV 373
L E + + P ++A V L+DV ++++++ Y D +KL + + + ++ +H RP VPV
Sbjct: 328 LGNQERNNPEIPFERASVILNDVNVTITEEQYHDWIKLVEVVSRYKTYIEISHLRPMVPV 387
Query: 374 KADPRSWWKYAYRAVSDQMKKASGKMSWEQVLRYTSLRKRYIYLYASLLKSDPSQVTISG 433
PR WW++A +A Q + L YT RYI LYA+ L+ Q +
Sbjct: 388 SEAPRLWWRFAAQASLQQKR-----------LWYT----RYIQLYANFLQ----QSSDVN 428
Query: 434 NKEIEDLDHELDIELILQWRMLAHKFVEQSAEPNLSVRKQKAGNSWWSFGWTGKS---PK 490
E+ +++ +LD ++IL WR+LAH VE + +++ W+SF W ++ P+
Sbjct: 429 YPEMREIEKDLDSKVILLWRLLAHAKVESVKSKEAAEQRKLKKGGWFSFNWRTEAEDDPE 488
Query: 491 EDSE-------ELSFSEEDWNRLNKIIGYKEGDDGQSPVNSKADVMHTFLV-VHMNHNAS 542
DS E ++++W +NK++ ++ ++ M FLV V + A+
Sbjct: 489 VDSVAGGSKLMEERLTKDEWKAINKLLSHQPDEEMNLYSGKDMQNMTHFLVTVSIGQGAA 548
Query: 543 KLIGEAQDLVAELSCEDLSCSVKLYPETKVFDIKLGSYQLSSPKGLLAESAAS---FDSL 599
+++ Q V E L + K + D+ L Y LS+P+G LA+S +S ++L
Sbjct: 549 RIVDINQTEVLCGRFEQLDVTTKFRHRSTQCDVSLRFYGLSAPEGSLAQSVSSERKTNAL 608
Query: 600 VGVFKYKPFDDKVDWSMVAKASPCYMTYMKDSINQIVKFFETNATVSQTIALETAAAVQL 659
+ F P + +DW + A SPC+ T +S +++++F + + VS T+ALETAA +Q+
Sbjct: 609 MASFVNAPIGENIDWRLSATISPCHATIWTESYDRVLEFVKRSNAVSPTVALETAAVLQM 668
Query: 660 KIDEVKRTAQQQMNRALKDHARFSLDLDIAAPKITIPTDFYPDNTHATKLLLDLGNLMIR 719
K++EV R AQ+Q+ L++ +RF+LD+DI APK+ IP + ++ LLD GN +
Sbjct: 669 KLEEVTRRAQEQLQIVLEEQSRFALDIDIDAPKVRIPLRASGSSKCSSHFLLDFGNFTLT 728
Query: 720 TQDDSRQESAEDNMYLRFDLVLSDVSAFLFD--GDYHWSEISVNKLTHSTNTS------- 770
T D+R E N+Y RF + D++AF D D + + T+ S
Sbjct: 729 TM-DTRSEEQRQNLYSRFCISGRDIAAFFTDCGSDNQGCSLVMEDFTNQPILSPILEKAD 787
Query: 771 -FFPIIDRCGVILQLQQILLETPYYPSTRLAVRLPSLAFHFSPARYHRLMHVIKIF---- 825
+ +IDRCG+ + + QI + P YPSTR+++++P++ HFSP RY R+M + I
Sbjct: 788 NVYSLIDRCGMAVIVDQIKVPHPSYPSTRISIQVPNIGVHFSPTRYMRIMQLFDILYGAM 847
Query: 826 ----EEGDDGSSEFLRPWNQADLEGWLSLLTWK 854
+ D + ++PW+ DL +L WK
Sbjct: 848 KTYSQAPVDHMPDGIQPWSPTDLASDARILVWK 880
Score = 308 bits (788), Expect = 2e-83
Identities = 219/724 (30%), Positives = 354/724 (48%), Gaps = 102/724 (14%)
Query: 939 KSTDPISG--LSETSS-DHDDIESELDNQGVIDVAISERLFVTGVLDELKVCF--SYSYQ 993
+++ P+SG L +TS D D E + N D L +TG L E K+ +
Sbjct: 979 QASAPLSGDVLGQTSDGDGDFHEPQTRNMKAAD------LVITGALVETKLYLYGKIKNE 1032
Query: 994 PDQSLMKVLLNQEKRLFEFRAIGGQVEVSMKDNDIFIGTILKSLEVEDLVCYSQPSQPRY 1053
D+ + +VLL + A GG+V + ++ + + T L SL+++D + Q +Y
Sbjct: 1033 CDEQVEEVLL------LKVLASGGKVHLISSESGLTVRTKLHSLKIKDELQQQQSGSAQY 1086
Query: 1054 LARSF---------------------IGAADEKSLFYDTMRENVESSGLIPTE--SDDKF 1090
LA S +G AD++ + D + E L PTE + D
Sbjct: 1087 LAYSVLKNEDIQESLGTCDSFDKEMPVGHADDEDAYTDALPE-----FLSPTEPGTPDMD 1141
Query: 1091 YEAPETLADSDVYM---QSPGGTSEYPSSSSNEIKFNYSSLEPPKFSRIIGLLPTDAPST 1147
+ DSD ++ + GG E +S + D
Sbjct: 1142 MIQCSMMMDSDEHVGLEDTEGGFHEKDTSQGKSL--------------------CDEVFY 1181
Query: 1148 STKEHELNDTLESFVKAQIIIYDQNSTRYNNIDKQVIVTLATLTFFCRRPTILAIMEFIN 1207
+ E +D FV + +S YN ID Q+ + ++ L FFC RPT++A++ F
Sbjct: 1182 EVQGGEFSD----FVSVVFLTRSSSSHDYNGIDTQMSIRMSKLEFFCSRPTVVALIGF-- 1235
Query: 1208 SINIENGNLATSSDSSSTSMVKND------LSNDLDVLHVTTVEEHAVKGLLGKGKSRVM 1261
D S+ S ++ND + D T E ++GLLG GK RV+
Sbjct: 1236 -----------GFDLSTASYIENDKDANTLVPEKSDSEKETNDESGRIEGLLGYGKDRVV 1284
Query: 1262 FSLTLKMAQAQILLMKENETKLACLSQESLLAEIKVFPSSFSIKAALGNLKISDDSLSSS 1321
F L + + + L KE+ ++LA QE + +IKV PSS S++ LGN K+ D SL S
Sbjct: 1285 FYLNMNVDNVTVFLNKEDGSQLAMFVQERFVLDIKVHPSSLSVEGTLGNFKLCDKSLDSG 1344
Query: 1322 HFYYWACDMRNPGGRSFVELEFTSFSCDDEDYEGYDFSLFGELSEVRIVYLNRFLQEIVG 1381
+ + W CD+R+PG S ++ +F+S+S D+DYEGYD+SL G+LS VRIV+L RF+QE+
Sbjct: 1345 NCWSWLCDIRDPGVESLIKFKFSSYSAGDDDYEGYDYSLSGKLSAVRIVFLYRFVQEVTA 1404
Query: 1382 YFMGLVPNSPRSVVKVTDQVTNSEKWFSASDIEGSPAVKFDLSLRKPIILMPRNRDSLDF 1441
YFMGL V+K+ D+V E +++G+ AVK DLSL PII++PR+ S D+
Sbjct: 1405 YFMGLATPHSEEVIKLVDKVGGFEWLIQKDEMDGATAVKLDLSLDTPIIVVPRDSLSKDY 1464
Query: 1442 LRLDIVHITVRNTFQWIGGSKSEINAVHLETLMVQVEDINLNVGTGTDLGESIIKDVNGL 1501
++LD+ + V N W G + + AV ++ L ++ +N++VG +G+ +I++ GL
Sbjct: 1465 IQLDLGQLEVSNEISWHGCPEKDATAVRVDVLHAKILGLNMSVGINGSIGKPMIREGQGL 1524
Query: 1502 SVIIHRSLRDLSHQFPSIEIIIKMEELKAAMSNKEYQIITECAVSNFSEVPDIPSPLNQY 1561
+ + RSLRD+ + P++ + +K++ L A MS+KEY II C N E P +P
Sbjct: 1525 DIFVRRSLRDVFKKVPTLSVEVKIDFLHAVMSDKEYDIIVSCTSMNLFEEPKLP------ 1578
Query: 1562 SSKTLNGATDDIVPEVTSGADSVTTD---VEASVLLKIWVSINLVELSLYTGISRDASLA 1618
G++ ++ AD V + + + + + V IN L L ++ ++SLA
Sbjct: 1579 --PDFRGSSSGPKAKMRLLADKVNLNSQMIMSRTVTILAVDINYALLELRNSVNEESSLA 1636
Query: 1619 TVQV 1622
V V
Sbjct: 1637 HVAV 1640
>At4g17130 hypothetical protein
Length = 747
Score = 142 bits (357), Expect = 2e-33
Identities = 76/144 (52%), Positives = 101/144 (69%), Gaps = 2/144 (1%)
Query: 1479 DINLNVGTGTDLGESIIKDVNGLSVIIHRSLRDLSHQFPSIEIIIKMEELKAAMSNKEYQ 1538
DINLNVG+G ++GESII+DV G+SV I+RSLRDL HQ PSIE+ I+++EL+AA+SN+EYQ
Sbjct: 2 DINLNVGSGAEIGESIIQDVKGVSVTINRSLRDLLHQIPSIEVSIEIDELRAALSNREYQ 61
Query: 1539 IITECAVSNFSEVPDIPSPLNQYSSKTLNGATDDIVPEVTSGADSVTTDVEASVLLKIWV 1598
I+TECA SN SE+P PL+ + + + E T+ A + TD S +K+ V
Sbjct: 62 ILTECAQSNISELPHAVPPLSGDVVTSSRNLHETLTSEDTNAAQTEKTDTWIS--MKVSV 119
Query: 1599 SINLVELSLYTGISRDASLATVQV 1622
INLVEL LY G +RD LA VQ+
Sbjct: 120 VINLVELCLYAGTARDTPLAAVQI 143
>At4g21490 NADH dehydrogenase like protein
Length = 478
Score = 125 bits (315), Expect = 2e-28
Identities = 69/123 (56%), Positives = 86/123 (69%), Gaps = 4/123 (3%)
Query: 820 HVIKIFEEGDDGSSEFLRPWNQADLEGWLSLLTWKVMVSFICASLTIGGPTGVEFAASLH 879
H +K E+ S + + +A L G L+ K M+ F+ GGPTGVEFA+ LH
Sbjct: 173 HFLKEVEDAQRIRSTVIDSFEKASLPG-LNEQERKRMLHFVVVG---GGPTGVEFASELH 228
Query: 880 DFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQRDGIDVKTGSVVVK 939
DFVNEDLV+LYP K++V+ITLLEA DHIL+MFDKRIT FA++KF RDGIDVK GS+VVK
Sbjct: 229 DFVNEDLVKLYPKAKNLVQITLLEAADHILTMFDKRITEFAEEKFTRDGIDVKLGSMVVK 288
Query: 940 STD 942
D
Sbjct: 289 VND 291
>At2g20800 putative NADH-ubiquinone oxireductase
Length = 582
Score = 117 bits (292), Expect = 7e-26
Identities = 56/80 (70%), Positives = 70/80 (87%), Gaps = 1/80 (1%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVEF+A LHDF+ +D+ ++YP +++ KITLLEAGDHIL+MFDKRITAFA++KFQR
Sbjct: 234 GGPTGVEFSAELHDFLVQDVAKIYPKVQEFTKITLLEAGDHILNMFDKRITAFAEEKFQR 293
Query: 927 DGIDVKTGSVVVKST-DPIS 945
DGID+KTGS+VV T D IS
Sbjct: 294 DGIDLKTGSMVVGVTADEIS 313
>At4g05020 unknown protein
Length = 582
Score = 114 bits (286), Expect = 4e-25
Identities = 56/73 (76%), Positives = 61/73 (82%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVEFAA LHDFV EDLV LYP K V+ITLLEA DHIL+MFDKRIT FA++KF R
Sbjct: 230 GGPTGVEFAAELHDFVTEDLVSLYPRAKGSVRITLLEAADHILTMFDKRITEFAEEKFSR 289
Query: 927 DGIDVKTGSVVVK 939
DGIDVK GS+V K
Sbjct: 290 DGIDVKLGSMVTK 302
>At4g28220 putative NADH dehydrogenase
Length = 571
Score = 110 bits (274), Expect = 9e-24
Identities = 48/76 (63%), Positives = 65/76 (85%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVEFAA LHDF+ ED+ ++YP +K++VKITL+++GDHIL+ FD+RI++FA+ KF R
Sbjct: 222 GGPTGVEFAAELHDFIIEDITKIYPSVKELVKITLIQSGDHILNTFDERISSFAEQKFTR 281
Query: 927 DGIDVKTGSVVVKSTD 942
DGIDV+TG V+ TD
Sbjct: 282 DGIDVQTGMRVMSVTD 297
>At5g24740 VPS13 - like protein
Length = 3306
Score = 89.4 bits (220), Expect = 2e-17
Identities = 43/112 (38%), Positives = 70/112 (62%), Gaps = 5/112 (4%)
Query: 9 LLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGFLGSVKLKV 68
LL YLG Y++ + K+ LKIS+W G+V L+N++L PEA L+LP+ +K G +G + +K+
Sbjct: 12 LLLGYLGRYIKDIQKDQLKISLWNGEVLLENVELNPEAFEYLQLPIALKQGRVGKLSIKI 71
Query: 69 PWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQE----AKKIRIEEMEL 116
PW +L +DPV + ++ +F+ A E S D V++ KK ++ EL
Sbjct: 72 PWKKLHRDPVTIMIEDVFICASQRNDQE-WSSDVVEKREFAGKKAKLASAEL 122
Score = 47.0 bits (110), Expect = 9e-05
Identities = 115/602 (19%), Positives = 214/602 (35%), Gaps = 81/602 (13%)
Query: 282 KFGTKDGKPADRLLQKHSYVLEPVTGKGNYSKLLLNEVADSKQ-----PLQKAVVNLDDV 336
+F K K A L K S + G +Y + +V S + P L DV
Sbjct: 109 EFAGKKAKLASAELAKLSRRVFDNPGGNSYMSYITAKVNRSGELYDDLPQYSISAELTDV 168
Query: 337 TISLSKDGYRDIMKLADNFAAFNQRLKYAHFRP-PVPVKADP----RSWWKYAYRAVSDQ 391
++L++ + I+ L D R +Y +RP + P + WW YA +V
Sbjct: 169 VVTLNEFQLQQILILLDYLQTSQLRERYGRYRPCSTSLSRKPPGWQKLWWHYAQNSVLSD 228
Query: 392 MKKASGKMSWEQVLRYTSLRKRYIYLYASLLKSDPSQVTISGNKEIEDLDHELDIELILQ 451
S ++ E++ + + + Y A+ E+++ EL + +
Sbjct: 229 SIDKSIRLGLEELEKKSDIDDILSYRSAA-------------EGEMQEACSELTVNMGAT 275
Query: 452 WRMLAHKFVEQSA---EPN--LSVRKQKAGNSWWSFGWTGKSPKEDSEELS--FSEE--- 501
+ K EQSA EP+ ++ K + +W S G G EDS + S S+E
Sbjct: 276 GATRSEK--EQSAPEKEPSDDPTLNKSRGWLNWLSRGMLGAGGTEDSSQFSGVVSDEVVK 333
Query: 502 DWNRLNKIIGYKEGDDGQSPVNSKADVMHTFLVVHMNHNASKLIGEAQDLVAELSCEDLS 561
D ++ K S V + + G + ++ EL +
Sbjct: 334 DIHKATKFYPLSSSPRNTSATGKICTCSIRLNVRKFSATLQHISGSSSQVITELDILGVI 393
Query: 562 CSVKLYPETKVFDIKLGSYQLSSP---KGLLA-----------ESAASFDSLVGVFKYKP 607
K + ++ + + S +L P K +L E+ S+ + + + K
Sbjct: 394 VECKSWKDSTAMILSVISGRLVYPHNGKEILTMKRVCSQSDTLETKPSYGARLELSK--- 450
Query: 608 FDDKVDWSMVAKASPCYMTYMKDSINQIVKFF--------ETNATVSQTIALETAAAVQL 659
D V S+ A P Y I + FF + +S LE+ +
Sbjct: 451 -DHDVALSLKATLQPLEAAYDGGFILAVSNFFSGSRYFKLQHERILSSLNGLESETRLAA 509
Query: 660 KIDEVKRTAQQQMNRALKDHARFSLDLDIAAPKITIPTDFYPDNTHATKLLLDLGNLMIR 719
K + + L + DLDI ++ P ++ L+L+ ++
Sbjct: 510 KAEYL-----------LSSRNKVKWDLDIKDLTLSFPGRLVESESYNLVLVLESLSITSS 558
Query: 720 TQDD----SRQESAEDNMYLRFDLVLSDVSAFLFDGDYHWSEISV----NKLTHSTNTSF 771
+ D R +S D++ + + AF Y +I + KL
Sbjct: 559 STDALSQIPRLQSDVDHVVNSLQSSVEALDAFQVKDLYDHFDIKICNLEMKLMKIHPFQE 618
Query: 772 FPIIDRCGVILQLQQ-ILLETPYYPSTRLAVRLPSLAFHFSPARYHRLMHVIKIFEEGDD 830
P++++ ++++ I+ E + LP HFSP+ + +M VI+ + D
Sbjct: 619 LPLVEKSSLLIKFASCIIPEESILKQLEVEATLPMFNVHFSPSIFKGVMSVIEYLDIQDH 678
Query: 831 GS 832
G+
Sbjct: 679 GT 680
>At1g07180 unknown protein
Length = 510
Score = 62.4 bits (150), Expect = 2e-09
Identities = 32/77 (41%), Positives = 49/77 (63%), Gaps = 3/77 (3%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVEF+ L DF+ +D+ + Y +KD +++TL+EA D ILS FD R+ +A + +
Sbjct: 249 GGPTGVEFSGELSDFIMKDVRQRYSHVKDDIRVTLIEARD-ILSSFDDRLRHYAIKQLNK 307
Query: 927 DGIDVKTGSVVVKSTDP 943
G+ + G +VK P
Sbjct: 308 SGVKLVRG--IVKEVKP 322
>At2g29990 putative NADH dehydrogenase (ubiquinone oxidoreductase)
Length = 508
Score = 59.3 bits (142), Expect = 2e-08
Identities = 30/70 (42%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVEF+ L DF+ +D+ + Y +KD + +TL+EA D ILS FD R+ +A + +
Sbjct: 247 GGPTGVEFSGELSDFIMKDVRQRYAHVKDDIHVTLIEARD-ILSSFDDRLRRYAIKQLNK 305
Query: 927 DGIDVKTGSV 936
G+ G V
Sbjct: 306 SGVRFVRGIV 315
>At3g19190 hypothetical protein
Length = 1814
Score = 42.0 bits (97), Expect = 0.003
Identities = 35/166 (21%), Positives = 77/166 (46%), Gaps = 11/166 (6%)
Query: 6 VAYLLQRYLGNYVRG-LNKEALKISVWKGDVELKNMQLKPEALN-ALKLPVKVKAGFLGS 63
V +LL++ LG + G ++ + L I + G ++L ++ + + LN P+ +K G +GS
Sbjct: 24 VKFLLKKKLGKLILGDIDLDQLDIQLRDGTIQLSDLAINVDYLNDKFDAPLVIKEGSIGS 83
Query: 64 VKLKVPWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQEAKKIRIEEMELKL---WE 120
+ +K+PW G V +D + L+ P + S + + R + ++L
Sbjct: 84 LLVKMPWKTNG---CQVEVDELELVLAPRLESNKSSSNEASTSASTREDLHNIRLEIGKH 140
Query: 121 KSQQLKSEMKTINPSSQGSAK---KLFSTIIGNLKLSISNIHIRYE 163
+++ L + K+ + K K+ + + + I N+ I ++
Sbjct: 141 ENEMLMNAAKSASIDVHEGVKTVAKIVKWFLTSFHVKIKNLIIAFD 186
>At2g14510 putative receptor-like protein kinase
Length = 868
Score = 34.3 bits (77), Expect = 0.61
Identities = 35/129 (27%), Positives = 58/129 (44%), Gaps = 13/129 (10%)
Query: 1051 PRYLARSFIGAADEKSLFYDTMRENVESSGLIPTESDDKFYE---APETLADSDVY-MQS 1106
P Y ++++ + F + E++ +I +S DK Y A LA+ D+ +
Sbjct: 733 PEYYLKNWLTEKSDVYSFGIVLLESITGQPVIE-QSRDKSYIVEWAKSMLANGDIESIMD 791
Query: 1107 PGGTSEYPSSSSNEIKFNYSSLEPPKFSRIIGLLPTDAPSTSTKEHELNDTLESFVKAQI 1166
P +Y SSSS + +LE I T P+ + HELN+ LE + +I
Sbjct: 792 PNLHQDYDSSSS------WKALELAMLC--INPSSTQRPNMTRVAHELNECLEIYNLTKI 843
Query: 1167 IIYDQNSTR 1175
DQNS++
Sbjct: 844 RSQDQNSSK 852
>At2g36570 putative receptor-like protein kinase
Length = 672
Score = 33.5 bits (75), Expect = 1.0
Identities = 25/100 (25%), Positives = 45/100 (45%), Gaps = 14/100 (14%)
Query: 1141 PTDAPSTSTKEHE-----LNDTLESFVKAQII--IYDQNSTRYNNIDKQVIVTLATLTFF 1193
P A S + +E E L + S VK + ++D RY NI+++++ L +
Sbjct: 565 PRSAASVAVEEEEEAVVDLPKWVRSVVKEEWTAEVFDPELLRYKNIEEEMVAML-HIGLA 623
Query: 1194 C------RRPTILAIMEFINSINIENGNLATSSDSSSTSM 1227
C +RPT+ +++ + I +E + D S SM
Sbjct: 624 CVVPQPEKRPTMAEVVKMVEEIRVEQSPVGEDFDESRNSM 663
>At3g53030 serine protein kinase - like
Length = 529
Score = 32.7 bits (73), Expect = 1.8
Identities = 21/75 (28%), Positives = 39/75 (52%), Gaps = 13/75 (17%)
Query: 878 LHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQRDGIDVKTGSVV 937
LH ++N + P +KD E D + + +KR + +++ ++VK G+VV
Sbjct: 462 LHPWINSGPRSIKPSLKD-------ENSDKLDTEKNKR------ENEEQEAVEVKMGNVV 508
Query: 938 VKSTDPISGLSETSS 952
+ S D G+S++SS
Sbjct: 509 ISSLDSKPGMSQSSS 523
>At3g28344 P-glycoprotein, 5' partial
Length = 626
Score = 32.7 bits (73), Expect = 1.8
Identities = 53/213 (24%), Positives = 84/213 (38%), Gaps = 22/213 (10%)
Query: 913 DKRITAFADDKFQRDGIDVKTGSVVVKSTDPISGLSETSSDHDDIESELDNQGVIDVAIS 972
D ITA A F+ I V TG V+ + + L++ S + + LD ID
Sbjct: 319 DGYITAKA--LFETFMILVSTGRVIADAGSMTTDLAKGSDAVGSVFAVLDRYTSIDPEDP 376
Query: 973 ---ERLFVTGVLDELKVCFSYSYQPDQSLMKVLLNQEKRLFEFRAIGGQVEVSMKDNDIF 1029
E +TG ++ L V FSY +PD + K N ++ E ++ V S
Sbjct: 377 DGYETERITGQVEFLDVDFSYPTRPDVIIFK---NFSIKIEEGKSTA-IVGPSGSGKSTI 432
Query: 1030 IGTI---------LKSLEVEDLVCYSQPSQPRYLARSFIGAADEKSLFYDTMRENVESSG 1080
IG I + ++ D+ Y S R++A + E +LF T+REN+ G
Sbjct: 433 IGLIERFYDPLKGIVKIDGRDIRSYHLRSLRRHIAL----VSQEPTLFAGTIRENIIYGG 488
Query: 1081 LIPTESDDKFYEAPETLADSDVYMQSPGGTSEY 1113
+ + + EA + D G Y
Sbjct: 489 VSDKIDEAEIIEAAKAANAHDFITSLTEGYDTY 521
>At1g53090 phytochrome A supressor spa1, putative
Length = 794
Score = 32.7 bits (73), Expect = 1.8
Identities = 39/160 (24%), Positives = 68/160 (42%), Gaps = 19/160 (11%)
Query: 913 DKRITAFADDKFQRDGIDVKTGSVVVKSTDPISGLSETSSDHDDIESELDNQGVIDV-AI 971
+K+I F + +DG D+ V + S +SG+ S + S + +GV+ V +
Sbjct: 505 NKKIKIFECESIIKDGRDIHYPVVELASRSKLSGICWNSYIKSQVASS-NFEGVVQVWDV 563
Query: 972 SERLFVTGVLDELKVCFSYSYQPDQSLMKVLLNQEKRLFEFRAIGGQVEVSMKDNDIFIG 1031
+ VT + + K +S Y + + L + G V++ + + IG
Sbjct: 564 ARNQLVTEMKEHEKRVWSIDYS----------SADPTLLASGSDDGSVKLWSINQGVSIG 613
Query: 1032 TILKSLEVEDLVCYSQPSQP-RYLARSFIGAADEKSLFYD 1070
TI ++ C PS+ R LA G+AD K +YD
Sbjct: 614 TIKTK---ANICCVQFPSETGRSLA---FGSADHKVYYYD 647
>At4g33400 Dem -like protein
Length = 645
Score = 32.0 bits (71), Expect = 3.0
Identities = 27/128 (21%), Positives = 54/128 (42%), Gaps = 25/128 (19%)
Query: 995 DQSLMKVLLNQEKRLFEFRAIGGQVEVSMKDNDIFIGTILKSL-EVEDLVCYSQPSQPRY 1053
D+ +K +Q + F +A+ S +D +F+ + L E V +++ ++ +
Sbjct: 154 DEMQLKAYKDQRRVDFVAKAVWAMKFASSEDFSVFVSSYNNCLFENNHGVEFNEANKAKI 213
Query: 1054 LARSFIGAADEKSLFYDTMRENVESSGLIPTESDDKFYEAPETLADSDVYMQSPGGTSEY 1113
+ FIG A+ P +DD +E + D+ +QSPGG S
Sbjct: 214 YGKDFIGWAN-------------------PEAADDSMWEDAD-----DILLQSPGGGSAT 249
Query: 1114 PSSSSNEI 1121
P+ + ++
Sbjct: 250 PARDTQDL 257
>At5g10280 putative transcription factor MYB92
Length = 334
Score = 31.6 bits (70), Expect = 3.9
Identities = 24/95 (25%), Positives = 44/95 (46%), Gaps = 7/95 (7%)
Query: 1071 TMRENVESSGLIPTESDD---KFYEAPETLADSDVYMQSPGGTSEYPSSSSNEIKFNYSS 1127
T+ N+E S + P DD KF E L S +++ P T+ P+ ++++ FN
Sbjct: 239 TLNSNMEPSSVFPQNLDDNHFKFSTQRENLPVSPIWLSDPSSTT--PAHVNDDLIFNQYG 296
Query: 1128 LEP--PKFSRIIGLLPTDAPSTSTKEHELNDTLES 1160
+E + G + S + +H L+D++ S
Sbjct: 297 IEDVNSNITSSSGQESGASASAAWPDHLLDDSIFS 331
>At5g55760 transcription regulator Sir2-like protein
Length = 473
Score = 31.2 bits (69), Expect = 5.2
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query: 1517 PSIEIIIKMEEL-KAAMSNKEYQIITECAVSNFSEVPDIPSPLN----QYSSKTLNGATD 1571
PS + K+EEL K +K + T +S +PD P Q K L A+
Sbjct: 26 PSHLLQCKIEELAKLIQKSKHLVVFTGAGISTSCGIPDFRGPKGIWTLQREGKDLPKASL 85
Query: 1572 DIVPEVTSGADSVTTDVEASVLLKIWVSINLVELSLYTGISRD 1614
+ S ++E + +LK +S N+ L L +GI R+
Sbjct: 86 PFHRAMPSMTHMALVELERAGILKFVISQNVDGLHLRSGIPRE 128
>At5g02390 putative protein
Length = 835
Score = 31.2 bits (69), Expect = 5.2
Identities = 19/54 (35%), Positives = 25/54 (46%), Gaps = 5/54 (9%)
Query: 487 KSPKEDSEELSFSEEDWNRLNKIIGYKEGDDGQSPVNSKADVMHTFLVVHMNHN 540
K K D EE F ++ G+ E DD SP SKA + ++HMN N
Sbjct: 251 KKEKSDDEEAWFDP----KMRHTKGFSENDDDTSPRRSKA-CLDALNLIHMNRN 299
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,019,356
Number of Sequences: 26719
Number of extensions: 1596739
Number of successful extensions: 4514
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 4458
Number of HSP's gapped (non-prelim): 54
length of query: 1631
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1518
effective length of database: 8,299,349
effective search space: 12598411782
effective search space used: 12598411782
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0550.1


