

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0546.8
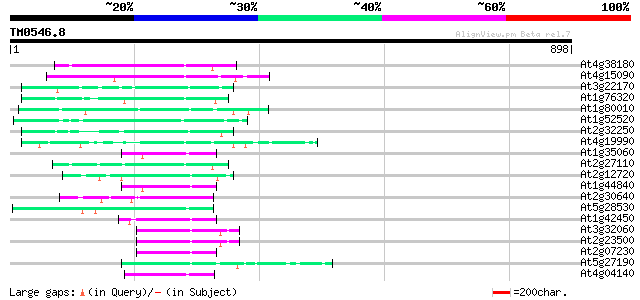
(898 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 79 1e-14
At4g15090 unknown protein 76 7e-14
At3g22170 far-red impaired response protein, putative 69 9e-12
At1g76320 putative phytochrome A signaling protein 69 1e-11
At1g80010 hypothetical protein 68 2e-11
At1g52520 F6D8.26 67 3e-11
At2g32250 Mutator-like transposase 67 4e-11
At4g19990 putative protein 58 2e-08
At1g35060 hypothetical protein 58 2e-08
At2g27110 Mutator-like transposase 57 6e-08
At2g12720 putative protein 56 8e-08
At1g44840 hypothetical protein 56 1e-07
At2g30640 Mutator-like transposase 55 1e-07
At5g28530 far-red impaired response protein (FAR1) - like 54 4e-07
At1g42450 mudrA-like protein 54 4e-07
At3g32060 hypothetical protein 54 5e-07
At2g23500 Mutator-like transposase 54 5e-07
At2g07230 Mutator-like transposase 54 5e-07
At5g27190 putative protein 52 1e-06
At4g04140 putative transposon protein 52 1e-06
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 78.6 bits (192), Expect = 1e-14
Identities = 79/314 (25%), Positives = 129/314 (40%), Gaps = 29/314 (9%)
Query: 73 LKRNRTGTKK-CNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVG--RLNPE 129
+KR RT T+ C ++ + S K W V HNH+ Q H +++
Sbjct: 139 IKRPRTITRVGCKASLSVKMQDSGK---WLVSGFVKDHNHELVPPDQVHCLRSHRQISGP 195
Query: 130 EKAMVGEMIEERVKASDILIA-IRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMK 188
K ++ + + I+ A I+++ + ++ N ++ E+Q L+
Sbjct: 196 AKTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMRNNRQKSIEGEIQLLLD 255
Query: 189 LLEEHKYAH---WSRVQDGTDV-VRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPL 244
L + + + VQ D V ++FWA P +I F F V D+TY++NRYR+P
Sbjct: 256 YLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPF 315
Query: 245 LEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEA 304
GV A++ NE + VW N L + P TD D V+ A
Sbjct: 316 APFTGVNHHGQPILFGCAFIINETEASFVWLFNTW--LAAMSAHPPVSITTDHDAVIRAA 373
Query: 305 LETVFPAARCLLCQFHVLK--------------SVTGEMKKLVKDIETREDITDRWNRLM 350
+ VFP AR C++H+LK S + K V E+ ED W L+
Sbjct: 374 IMHVFPGARHRFCKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFERCWFSLL 433
Query: 351 --YAANEVEYDKAV 362
Y + E+ +A+
Sbjct: 434 DKYELRDHEWLQAI 447
>At4g15090 unknown protein
Length = 768
Score = 76.3 bits (186), Expect = 7e-14
Identities = 85/372 (22%), Positives = 152/372 (40%), Gaps = 25/372 (6%)
Query: 60 CERGGKYKPSTSVLKRNRTGT-KKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQ 118
C R G S S +R T KK +C + +R D W + HNH+ L
Sbjct: 40 CSRYGVTPESESSGSSSRRSTVKKTDCKASMHVKR-RPDGKWIIHEFVKDHNHELLPALA 98
Query: 119 GHSYVGR---LNPEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQI----YNEKQA 171
H + R L + + + ER K + ++ + N+ + Q +K
Sbjct: 99 YHFRIQRNVKLAEKNNIDILHAVSERTKKMYVEMSRQSGGYKNIGSLLQTDVSSQVDKGR 158
Query: 172 YNRLKRG-SLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVI 230
Y L+ G S +++ ++ +E+ ++ + +R+LFWA S + F VV
Sbjct: 159 YLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVS 218
Query: 231 LDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLP 290
D+TY ++PL +GV + + A + +E + VW + L P
Sbjct: 219 FDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESMETFVWLIKTW--LRAMGGRAP 276
Query: 291 KVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLM 350
KV +TD+D+ LM A+ + P R +HVL+ + ++K E+ ++N+ +
Sbjct: 277 KVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSHVMK---RHENFLLKFNKCI 333
Query: 351 YAA-NEVEYD----KAVGAMSNTGELWTYIETNWL-PLRSKFVKFEIDTCMHMGCTTTNR 404
+ + + E+D K V + W WL R K+V + G +T+ R
Sbjct: 334 FRSWTDDEFDMRWWKMVSQFGLENDEWLL----WLHEHRQKWVPTFMSDVFLAGMSTSQR 389
Query: 405 VEGAHSKLKKLL 416
E +S K +
Sbjct: 390 SESVNSFFDKYI 401
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 69.3 bits (168), Expect = 9e-12
Identities = 78/355 (21%), Positives = 140/355 (38%), Gaps = 38/355 (10%)
Query: 19 FESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAV-MTLGCERGGKYKPSTSVLKR-- 75
FES + ++ + + G+ IQ S R + C R G + R
Sbjct: 76 FESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTREFIDAKFACSRYGTKREYDKSFNRPR 135
Query: 76 ---------NRTGTKKC---NCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYV 123
N G + C +C + +R D W + HNH E+L +
Sbjct: 136 ARQSKQDPENMAGRRTCAKTDCKASMHVKR-RPDGKWVIHSFVREHNH---ELLPAQAVS 191
Query: 124 GRLNPEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTEM 183
+ AM + E + +I+++ + ++ + + + E + L L +
Sbjct: 192 EQTRKIYAAMAKQFAEYKT-----VISLKSDSKSSFEKGRTLSVETGDFKIL----LDFL 242
Query: 184 QHLMKLLEEHKYAHWSRVQDGTDV-VRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRI 242
+ L YA V G D V+++FW S H + F VV LD+TY N+Y++
Sbjct: 243 SRMQSLNSNFFYA----VDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTYVRNKYKM 298
Query: 243 PLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLM 302
PL VGV + A + +E W + I + PKV +T+ D V+
Sbjct: 299 PLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQ--APKVLITELDVVMN 356
Query: 303 EALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLMYAANEVE 357
+ +FP R L +HVL V+ + ++VK ++ ++ + +Y + + E
Sbjct: 357 SIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQ---HDNFMPKFEKCIYKSGKDE 408
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 68.9 bits (167), Expect = 1e-11
Identities = 77/361 (21%), Positives = 136/361 (37%), Gaps = 46/361 (12%)
Query: 19 FESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAV-MTLGCERGGKYKPSTSVLKRNR 77
FE+ +D + K K G+ S + + C R G + S + N
Sbjct: 3 FETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSKQQSDDAI--NP 60
Query: 78 TGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLNPEEKAMVGEM 137
+ K C + +R D W V HNHD Q H + N E
Sbjct: 61 RASPKIGCKASMHVKR-RPDGKWYVYSFVKEHNHDLLPE-QAHYFRSHRNTE-------- 110
Query: 138 IEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTE--------------M 183
VK++D + +K+ P + Y++ + R + +
Sbjct: 111 ---LVKSNDSRLRRKKNTPLTDCKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILL 167
Query: 184 QHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIP 243
+ LM++ EE+ ++ ++R++FW I + F VV +++Y ++Y++P
Sbjct: 168 EFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVP 227
Query: 244 LLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLME 303
L+ VGV + + ++ VW + L+ PKV +TD++ +
Sbjct: 228 LVLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSW--LVAMGGQKPKVMLTDQNNAIKA 285
Query: 304 ALETVFPAARCLLCQFHVLKSVTGE-----------MKKLVKDIE---TREDITDRWNRL 349
A+ V P R C +HVL + MKKL K I + E+ RW +L
Sbjct: 286 AIAAVLPETRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKL 345
Query: 350 M 350
+
Sbjct: 346 I 346
>At1g80010 hypothetical protein
Length = 696
Score = 68.2 bits (165), Expect = 2e-11
Identities = 95/441 (21%), Positives = 172/441 (38%), Gaps = 51/441 (11%)
Query: 14 TTSQAFESSKDLVNWAKAVGKEHGYVIIIQKS---DYGSDKRRAVMTLGCERGGKYKPST 70
T FES D ++ + +E G+ I ++ S +KR AV+ C+ K +
Sbjct: 67 TPGMEFESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGAVLCCNCQGFKLLKDAH 126
Query: 71 SVLKRNRTGTKKCNCPFRLRARRSNKDKMWTV-LVHSGIHNHDTAEVLQGH-------SY 122
S K RTG C RLR ++ K+ V L H+ + A + H S
Sbjct: 127 SRRKETRTG---CQAMIRLRLIHFDRWKVDQVKLDHNHSFDPQRAHNSKSHKKSSSSASP 183
Query: 123 VGRLNPEEK--AMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSL 180
+ NPE V + R A D A+ + T + + + + RG
Sbjct: 184 ATKTNPEPPPHVQVRTIKLYRTLALDTPPALGTSLSSGETSDLSLDHFQSSRRLELRGGF 243
Query: 181 TEMQHL---MKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKT 237
+Q ++L + DG+ +R++FW + ++ F V++ D+T +
Sbjct: 244 RALQDFFFQIQLSSPNFLYLMDLADDGS--LRNVFWIDARARAAYSHFGDVLLFDTTCLS 301
Query: 238 NRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDK 297
N Y +PL+ VG+ T + + ++ + VW ++ P++F+T++
Sbjct: 302 NAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCMLGRP--PQIFITEQ 359
Query: 298 DQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLMYAANEVE 357
+ + A+ VFP A L HVL ++ + V ++ + NR++Y +VE
Sbjct: 360 CKAMRTAVSEVFPRAHHRLSLTHVLHNIC----QSVVQLQDSDLFPMALNRVVYGCLKVE 415
Query: 358 ----------------YDKAVGAMSNTGELW--TYIETNWL------PLRSKFVKFEIDT 393
++ + M ELW Y++ +L PL + F
Sbjct: 416 EFETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFSG 475
Query: 394 CMHMGCTTTNRVEGAHSKLKK 414
+H + +EG S L K
Sbjct: 476 YVHENTSLREFLEGYESFLDK 496
>At1g52520 F6D8.26
Length = 703
Score = 67.4 bits (163), Expect = 3e-11
Identities = 88/386 (22%), Positives = 152/386 (38%), Gaps = 33/386 (8%)
Query: 7 VDLTKAF---TTSQAFESSKDLVNWAKAVGKEHGYVIIIQKSDY---GSDKRRAVMTLGC 60
VD K F FES D N+ E G+ + ++ S + +K AV+
Sbjct: 76 VDERKEFDAPAVGMEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGAVLCCSS 135
Query: 61 ERGGKYKPSTSVLKRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGH 120
+ + V K RTG CP +R R+ + K W V+ + HNH L G
Sbjct: 136 QGFKRINDVNRVRKETRTG-----CPAMIRMRQVDS-KRWRVVEVTLDHNH-----LLGC 184
Query: 121 SYVGRLNPEEKAMVGEMIEER-VKASDILIAIRKHNPT-NLTRIKQIYNEKQAYN--RLK 176
+ + K + + + + +K + N N T K+ N + + LK
Sbjct: 185 KLYKSVKRKRKCVSSPVSDAKTIKLYRACVVDNGSNVNPNSTLNKKFQNSTGSPDLLNLK 244
Query: 177 RGSLTEMQHLM--KLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDST 234
RG + + L + + V D + R++FWA S + F V+ +DS+
Sbjct: 245 RGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQL-RNVFWADAFSKVSCSYFGDVIFIDSS 303
Query: 235 YKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFV 294
Y + ++ IPL+ GV T ++ ++ E + W LK + P+ V
Sbjct: 304 YISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWL---LKVWLSVMKRSPQTIV 360
Query: 295 TDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLMYAAN 354
TD+ + L A+ VFP + H+++ + ++ L R+ T +
Sbjct: 361 TDRCKPLEAAISQVFPRSHQRFSLTHIMRKIPEKLGGLHNYDAVRKAFT---KAVYETLK 417
Query: 355 EVEYDKAVGAMSNTGELWTYIETNWL 380
VE++ A G M + + IE WL
Sbjct: 418 VVEFEAAWGFMVHN---FGVIENEWL 440
>At2g32250 Mutator-like transposase
Length = 684
Score = 67.0 bits (162), Expect = 4e-11
Identities = 72/361 (19%), Positives = 136/361 (36%), Gaps = 62/361 (17%)
Query: 19 FESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAV-MTLGCERGGKYKPSTSVLKRNR 77
FES + + + + G+ I I+ S + + + + C R G + + + N
Sbjct: 44 FESKEAAYYFYREYARSVGFGITIKASRRSKRSGKFIDVKIACSRFGTKREKATAI--NP 101
Query: 78 TGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLNPEEKAMVGEM 137
K C L +R +D+ W + HNH+
Sbjct: 102 RSCPKTGCKAGLHMKRK-EDEKWVIYNFVKEHNHE------------------------- 135
Query: 138 IEERVKASDILIAIR-KHNPTNLTRIK---QIYNEKQAYNRLKRGSLTEMQHLMKLLEEH 193
+ D +++R K+ P IK Q+ E++ L ++H M++ ++
Sbjct: 136 ----ICPDDFYVSVRGKNKPAGALAIKKGLQLALEEEDLKLL-------LEHFMEMQDKQ 184
Query: 194 KYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTST 253
++ D VR++FW + H + F VV+ D+ Y N YRIP +GV+
Sbjct: 185 PGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHH 244
Query: 254 NLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAAR 313
+ A + + W + + P V +TD+D++L + + VFP R
Sbjct: 245 RQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQ--APGVMITDQDKLLSDIVVEVFPDVR 302
Query: 314 CLLCQFHVLKSVTGEMKKLVKDIE--------------TREDITDRWNRLM--YAANEVE 357
+ C + VL ++ + V + T E RW+ ++ + NE E
Sbjct: 303 HIFCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMIGKFELNENE 362
Query: 358 Y 358
+
Sbjct: 363 W 363
>At4g19990 putative protein
Length = 672
Score = 58.2 bits (139), Expect = 2e-08
Identities = 109/512 (21%), Positives = 181/512 (35%), Gaps = 113/512 (22%)
Query: 19 FESSKDLVNWAKAVGKEHGYVIIIQKSD-----------------YGSDKRRAVMTLGCE 61
FES ++ + K G+ II+ S YGS K LG +
Sbjct: 27 FESKEEAFEFYKEYANSVGFTTIIKASRRSRMTGKFIDAKFVCTRYGSKKEDIDTGLGTD 86
Query: 62 RGGKYKPSTSVLKRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNH-------DTA 114
G P R + K +C L +R +D W V HNH D+
Sbjct: 87 --GFNIPQARKRGRINRSSSKTDCKAFLHVKR-RQDGRWVVRSLVKEHNHEIFTGQADSL 143
Query: 115 EVLQGHSYVGRLNPEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNR 174
L G + +LN G +++E + RK ++ R+ +
Sbjct: 144 RELSGRRKLEKLN-------GAIVKE--------VKSRKLEDGDVERLLNFF-------- 180
Query: 175 LKRGSLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDST 234
T+MQ L R++FW + F VV +D+T
Sbjct: 181 ------TDMQSL----------------------RNIFWVDAKGRFDYTCFSDVVSIDTT 212
Query: 235 YKTNRYRIPLLEMVGVTSTNLTYSIAFA-YMQNEQKDEVVWAVNRLKDLIINEDNLPKVF 293
+ N Y++PL+ GV + F + +E K VW L P+V
Sbjct: 213 FIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAW--LKAMHGCRPRVI 270
Query: 294 VTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEM-KKLVKDIETREDITDRWNRLMYA 352
+T DQ+L EA+ VFP++R + L G+M +KL I + + D N +Y
Sbjct: 271 LTKHDQMLKEAVLEVFPSSRHCFYMWDTL----GQMPEKLGHVIRLEKKLVDEINDAIYG 326
Query: 353 ANEVE-----YDKAVGAMSNTGELWTYI----ETNWLPLRSKFVKFEIDTCMHMGCTTTN 403
+ + E + + V +W W+P+ K V G T
Sbjct: 327 SCQSEDFEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVSL-------AGMCTAQ 379
Query: 404 RVEGAHSKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLN--DPFY 461
R + +S L K + + A+ K +I + + +E ++ + L PF
Sbjct: 380 RSDSVNSGLDKYI--QRKTTFKAFLEQYKKMIQERYEEEEKSEIETLYKQPGLKSPSPFG 437
Query: 462 KHLRGFVSRAALHIIVKEKTQIGMIGVGGIYC 493
K + +R +K Q+ ++ GG+ C
Sbjct: 438 KQMAEVYTREMF-----KKFQVEVL--GGVAC 462
>At1g35060 hypothetical protein
Length = 873
Score = 58.2 bits (139), Expect = 2e-08
Identities = 40/156 (25%), Positives = 68/156 (42%), Gaps = 6/156 (3%)
Query: 179 SLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRS----LFWAHPDSIHLFNEFPHVVILDST 234
+L E HL+KL H D D + +F A SI F V+++D T
Sbjct: 472 NLAEYLHLLKLTNPGTITHIETEPDIEDERKERFLYMFLAFGASIQGFKHLRRVLVVDGT 531
Query: 235 YKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFV 294
+ +Y+ LL G + Y +AFA + +E D W +L+ +I DN +
Sbjct: 532 HLKGKYKGVLLTASGQDANFQVYPLAFAVVDSENDDAWTWFFTKLERII--ADNNTLTIL 589
Query: 295 TDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMK 330
+D+ + + ++ VFP A C H+ +++ K
Sbjct: 590 SDRHESIKVGVKKVFPQAHHGACIIHLCRNIQARFK 625
>At2g27110 Mutator-like transposase
Length = 851
Score = 56.6 bits (135), Expect = 6e-08
Identities = 62/298 (20%), Positives = 108/298 (35%), Gaps = 27/298 (9%)
Query: 69 STSVLKRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLN- 127
S+S + R ++ C+ R+ + K W V H H A H R +
Sbjct: 94 SSSSKRSKRRLSESCDAMVRIELQGHEK---WVVTKFVKEHTHGLASSNMLHCLRPRRHF 150
Query: 128 -PEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHL 186
EK+ E + S ++ N N K+ R L ++
Sbjct: 151 ANSEKSSYQEGVNV---PSGMMYVSMDANSRGARNASMATNTKRTIGRDAHNLL---EYF 204
Query: 187 MKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLE 246
++ E+ ++ D + + ++FWA S + F V LD+ Y+ N++R+P
Sbjct: 205 KRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAP 264
Query: 247 MVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALE 306
GV A + +E +W L D P VTD+D+ + A
Sbjct: 265 FTGVNHHGQAILFGCALILDESDTSFIWLFKTF--LTAMRDQPPVSLVTDQDRAIQIAAG 322
Query: 307 TVFPAARCLLCQFHVLK--------------SVTGEMKKLVKDIETREDITDRWNRLM 350
VFP AR + ++ VL+ S E+ + ET E+ W+ ++
Sbjct: 323 QVFPGARHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSWSSVI 380
>At2g12720 putative protein
Length = 819
Score = 56.2 bits (134), Expect = 8e-08
Identities = 67/294 (22%), Positives = 117/294 (39%), Gaps = 38/294 (12%)
Query: 85 CPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLNPEEKAMVGEMIEER--- 141
C +R+RA R D + V ++ H E GR ++G + +
Sbjct: 301 CLWRVRACRQGNDPNFYVYIYDSEHTCSVRE------RSGRSRQATPDVLGVLYRDYLGD 354
Query: 142 ----VKASDILIAIRKHNPTNLTRIKQIYNEKQAYNRLKR--------GSLTEMQHLMKL 189
VK I I I KH R+K Y++ R R S E+ + +
Sbjct: 355 VGPDVKPKSIGIIITKH-----FRVKMSYSKSYKTLRFARELTLGTPDSSFEELPSYLYM 409
Query: 190 LEEHKYAHWSRVQ-DGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMV 248
+ +R+Q D + +F SI F+ VV++D T+ Y+ LL +
Sbjct: 410 IRRANPGTVARLQIDESGRFNYMFIVFGASIAGFHYMRRVVVVDGTFLHGSYKGTLLTAL 469
Query: 249 GVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETV 308
+ +AF + E D W +LK +I + +L ++D+ + + +A+ V
Sbjct: 470 AQDGNFQIFPLAFGVVDTENDDSWRWLFTQLKVVIPDATDL--AIISDRHKSIGKAIGEV 527
Query: 309 FPAARCLLCQFHVLKSVTGEMKK-----LVKDIETREDITDRWNRLMYAANEVE 357
+P A +C +H+ K++ + K+ LVK + D N A NE+E
Sbjct: 528 YPLAARGICTYHLYKNILVKFKRKDLFPLVKKAARCYRLNDFTN----AFNEIE 577
>At1g44840 hypothetical protein
Length = 926
Score = 55.8 bits (133), Expect = 1e-07
Identities = 37/155 (23%), Positives = 69/155 (43%), Gaps = 6/155 (3%)
Query: 180 LTEMQHLMKLLEEHKYAHWSRVQDGTDVVRS----LFWAHPDSIHLFNEFPHVVILDSTY 235
L E HL+KL H +D D + +F A SI F ++++D T+
Sbjct: 522 LAEYLHLLKLTNPGTITHIETERDVEDESKERFLYMFLAFGASIAGFRHLRRILVVDGTH 581
Query: 236 KTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVT 295
+Y+ LL G + Y + FA + +E + W +L+ +I + L ++
Sbjct: 582 LKGKYKGVLLTSSGQDANFQVYPLGFAVVDSENDESWTWFFTKLERIIADSKTL--TILS 639
Query: 296 DKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMK 330
D+ ++ A++ VFP A C H+ +++ + K
Sbjct: 640 DRHSSILVAVKRVFPQANHGACIIHLCRNIQTKYK 674
>At2g30640 Mutator-like transposase
Length = 754
Score = 55.5 bits (132), Expect = 1e-07
Identities = 68/267 (25%), Positives = 115/267 (42%), Gaps = 32/267 (11%)
Query: 80 TKKCN---CPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLNPEEKAMVGE 136
T KCN CP+R+ + +T+ G H G S++G + V +
Sbjct: 218 TAKCNSEGCPWRIHCAKLPGLPTFTIRTIHGSHT------CGGISHLGH-HQASVQWVAD 270
Query: 137 MIEERVKAS------DILIAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKLL 190
+ ER+K + +IL I + + LT KQ + K+ RGS E L+
Sbjct: 271 AVTERLKVNPHCKPKEILEQIHQVHGITLT-YKQAWRGKERIMAAVRGSYEEDYRLLPRY 329
Query: 191 EEH---------KYAHWSRVQDGTDVVRSLFWAHPDSIHLF-NEFPHVVILDSTYKTNRY 240
+ H S V DG+ + F + SI F N ++ LD T ++Y
Sbjct: 330 CDQIRRTNPGSVAVVHGSPV-DGS--FQQFFISFQASICGFLNACRPLIGLDRTVLKSKY 386
Query: 241 RIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLI-INEDNLPKVFV-TDKD 298
LL G + +AFA + E W ++ L+ L+ +N +N+PK+ + + +D
Sbjct: 387 VGTLLLATGFDGEGAVFPLAFAIISEENDSSWQWFLSELRQLLEVNSENMPKLTILSSRD 446
Query: 299 QVLMEALETVFPAARCLLCQFHVLKSV 325
Q +++ ++T FP A LC + +SV
Sbjct: 447 QSIVDGVDTNFPTAFHGLCVHCLTESV 473
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 53.9 bits (128), Expect = 4e-07
Identities = 69/345 (20%), Positives = 131/345 (37%), Gaps = 26/345 (7%)
Query: 5 PSVDLTKAFT--TSQAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCER 62
P+ FT Q F + + + ++ G+ I +S + C R
Sbjct: 60 PTSQYDTVFTPYVGQIFTTDDEAFEYYSTFARKSGFSIRKARSTESQNLGVYRRDFVCYR 119
Query: 63 GGKYKP--STSVLKRNRTGTKKCNCPFRLRARRSNKDKM--WTVLVHSGIHNHDTAE--- 115
G +P +V + +C C +L + D + W V S +HNH+ E
Sbjct: 120 SGFNQPRKKANVEHPRERKSVRCGCDGKLYLTKEVVDGVSHWYVSQFSNVHNHELLEDDQ 179
Query: 116 --VLQGHSYVGRLNPEEKAMVGE----------MIE-ERVKASDILIAIRKHNPTNLTRI 162
+L + + + + E ++ + ++E E+ S L I K +
Sbjct: 180 VRLLPAYRKIQQSDQERILLLSKAGFPVNRIVKLLELEKGVVSGQLPFIEKDVRNFVRAC 239
Query: 163 KQIYNEKQAYNRLKRGSLT-EMQHLMKLLEEHKYAH-WSRVQDGTDVVRSLFWAHPDSIH 220
K+ E A+ KR S T E+ K L E + D V ++ WA+ DS+
Sbjct: 240 KKSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVR 299
Query: 221 LFNEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLK 280
++ F VV+ D++Y++ Y + L G+ + + +Q+E WA+
Sbjct: 300 GYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFV 359
Query: 281 DLIINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSV 325
+ P+ +TD D L +A+ P ++ H++ +
Sbjct: 360 RFM--RGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKL 402
>At1g42450 mudrA-like protein
Length = 601
Score = 53.9 bits (128), Expect = 4e-07
Identities = 40/168 (23%), Positives = 73/168 (42%), Gaps = 19/168 (11%)
Query: 174 RLKRGSLTEMQHLMKL-----------LEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLF 222
R GS T +Q KL L+ KY G + R LF+A P ++H +
Sbjct: 419 RSAMGSYTLIQPYFKLPLETNPNSLVALDTEKY------NKGVERFRYLFFALPAAVHGY 472
Query: 223 NEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDL 282
V+++ T+ RY L+ + + I F + +E D W + RL +
Sbjct: 473 AYMRKVMVIVGTHLRGRYGGCLIAASAQDANFQVFPITFGIVNSENDDAWTWFMERLTEA 532
Query: 283 IINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMK 330
I ++ +L VFV+D+ + ++ V+P + C H+++++ K
Sbjct: 533 IPDDPDL--VFVSDRHSSIYASIRKVYPMSSHAACVIHLMRNIVSMFK 578
>At3g32060 hypothetical protein
Length = 487
Score = 53.5 bits (127), Expect = 5e-07
Identities = 46/173 (26%), Positives = 78/173 (44%), Gaps = 10/173 (5%)
Query: 203 DGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFA 262
D D + +F A SI F V+I+D+T+ N Y L+ Y IAFA
Sbjct: 241 DENDKFQYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFA 300
Query: 263 YMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVL 322
+ E W +LK ++ + L VF+TD++ L++A+ V+ AA C +H+
Sbjct: 301 VLDGENDASWEWFFEKLKTVVPDTSEL--VFMTDRNASLIKAIRNVYTAAHHGYCIWHLS 358
Query: 323 KSVTGEMKKLVKDI-------ETREDITDRWNRLMYAANEVEYDKAVGAMSNT 368
++V G +D+ +R + +NR Y ++ Y KA + +T
Sbjct: 359 QNVKGHATHTNRDVLAWKFQELSRVYVVADFNR-AYDGFKLRYPKATKYLEDT 410
>At2g23500 Mutator-like transposase
Length = 784
Score = 53.5 bits (127), Expect = 5e-07
Identities = 46/173 (26%), Positives = 78/173 (44%), Gaps = 10/173 (5%)
Query: 203 DGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFA 262
D D + +F A SI F V+I+D+T+ N Y L+ Y IAFA
Sbjct: 371 DENDKFQYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFA 430
Query: 263 YMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVL 322
+ E W +LK ++ + L VF+TD++ L++A+ V+ AA C +H+
Sbjct: 431 VLDGENDASWEWFFEKLKTVVPDTSEL--VFMTDRNASLIKAIRNVYTAAHHGYCIWHLS 488
Query: 323 KSVTGEMKKLVKDI-------ETREDITDRWNRLMYAANEVEYDKAVGAMSNT 368
++V G +D+ +R + +NR Y ++ Y KA + +T
Sbjct: 489 QNVKGHATHTNRDVLAWKFQELSRVYVVADFNR-AYDGFKLRYPKATKYLEDT 540
>At2g07230 Mutator-like transposase
Length = 590
Score = 53.5 bits (127), Expect = 5e-07
Identities = 32/127 (25%), Positives = 60/127 (47%), Gaps = 2/127 (1%)
Query: 204 GTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAY 263
G + R LF+A ++ + V+++D T+ RY L+ + + IAF
Sbjct: 227 GLERFRYLFFALDAAVKGYAYMRKVIVIDGTHLRGRYGGCLIAASAQDANFQVFPIAFGI 286
Query: 264 MQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLK 323
+ +E D W + RL D I N+ +L VFV+D+ + ++ V+P + C H+ +
Sbjct: 287 VNSENDDAWTWFMERLTDAIPNDPDL--VFVSDRHSSIYASMRKVYPMSSHAACVVHLKR 344
Query: 324 SVTGEMK 330
++ K
Sbjct: 345 NIVSIFK 351
>At5g27190 putative protein
Length = 779
Score = 52.4 bits (124), Expect = 1e-06
Identities = 79/348 (22%), Positives = 141/348 (39%), Gaps = 34/348 (9%)
Query: 179 SLTEMQHLMKLLEEHKYAHWSRVQ-DGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKT 237
S T++ + +L+E S V+ D + LF A I F VVI+D+T+
Sbjct: 343 SYTDLYSYLHVLKEVNPGTISYVEVDAQQKFKYLFVALGACIEGFKVMRKVVIVDATFLK 402
Query: 238 NRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDK 297
Y L+ + Y IA A + E W N+LK +I + L VFV+D+
Sbjct: 403 TVYGGMLVFATAQDPNHHNYIIASAVIDRENDASWSWFFNKLKTVIPDVPGL--VFVSDR 460
Query: 298 DQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLMYAANEVE 357
Q +++++ VFP AR C +H+ ++V +K +E+ + + + E
Sbjct: 461 HQSIIKSIMQVFPNARHGHCVWHLSQNVKVRVK------TEKEEAAANFIACAHVYTQFE 514
Query: 358 YDKAVG----AMSNTG---ELWTYIETNWLPLRSKFVKFEIDTCMHMGCTTTNRV-EGAH 409
+ + N G + W +E NW + ++ IDT C + N E A
Sbjct: 515 FTREYARFRRRFPNVGPYLDRWAPVE-NWAMCYFEGDRYNIDT--SNACESLNSTFERAR 571
Query: 410 SKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYKHLRGFVS 469
L D+ + + W ++ +++ K + + V E+ ++ L
Sbjct: 572 KYFLLPLLDAIIEKISEWFNKHRKESVKYSKTRG----LVPVVENEIHS-----LCPIAK 622
Query: 470 RAALHIIVKEKTQIGMIGVGGI-YCECVLRRTHGLPCACELMKFESVP 516
+ + + Q +IG GGI Y +LR+T C+C + P
Sbjct: 623 TMPVTELNSFERQYSVIGEGGISYYVDLLRKT----CSCRRFDVDKYP 666
>At4g04140 putative transposon protein
Length = 946
Score = 52.4 bits (124), Expect = 1e-06
Identities = 37/145 (25%), Positives = 67/145 (45%), Gaps = 4/145 (2%)
Query: 185 HLMKLLEEHKYAHWSRVQD--GTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRI 242
H+++L H D G D + +F + S+ VV++D T+ +Y
Sbjct: 475 HVLQLANPGTVYHLETELDDIGDDRFKYVFLSLGASVKRLKYIRRVVVVDGTHLFGKYLG 534
Query: 243 PLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLM 302
LL + + IAFA + +E W +N+L ++I +D FV+D++Q +
Sbjct: 535 CLLTASCQDANFQIFPIAFAVVDSETDHSWTWFMNKLSEII--KDGPDLTFVSDRNQSIF 592
Query: 303 EALETVFPAARCLLCQFHVLKSVTG 327
+++ VFP A C H+ ++V G
Sbjct: 593 KSVGLVFPQAHHGACLVHIRRNVKG 617
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,263,098
Number of Sequences: 26719
Number of extensions: 870773
Number of successful extensions: 2606
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 2472
Number of HSP's gapped (non-prelim): 134
length of query: 898
length of database: 11,318,596
effective HSP length: 108
effective length of query: 790
effective length of database: 8,432,944
effective search space: 6662025760
effective search space used: 6662025760
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0546.8


