

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0546.10
(175 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
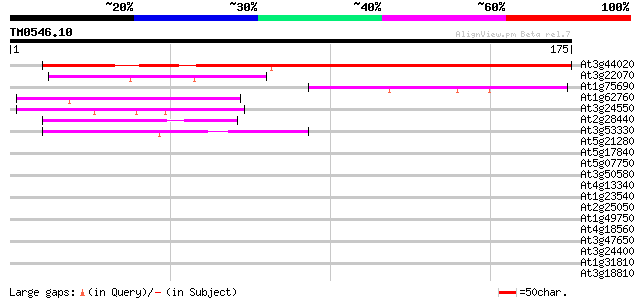
Score E
Sequences producing significant alignments: (bits) Value
At3g44020 unknown protein 156 6e-39
At3g22070 unknown protein 44 3e-05
At1g75690 unknown protein 43 1e-04
At1g62760 hypothetical protein 41 3e-04
At3g24550 protein kinase, putative 40 6e-04
At2g28440 En/Spm-like transposon protein 40 6e-04
At3g53330 putative protein 40 8e-04
At5g21280 putative protein 39 0.001
At5g17840 unknown protein 39 0.001
At5g07750 putative protein 39 0.001
At3g50580 proline-rich protein 39 0.001
At4g13340 extensin-like protein 38 0.002
At1g23540 putative serine/threonine protein kinase 38 0.002
At2g25050 hypothetical protein 38 0.003
At1g49750 unknown protein 38 0.003
At4g18560 pherophorin - like protein 37 0.004
At3g47650 putative protein 37 0.004
At3g24400 protein kinase, putative 37 0.004
At1g31810 hypothetical protein 37 0.004
At3g18810 protein kinase, putative 37 0.005
>At3g44020 unknown protein
Length = 159
Score = 156 bits (394), Expect = 6e-39
Identities = 89/167 (53%), Positives = 103/167 (61%), Gaps = 14/167 (8%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
PL S L SSSSSSS + L L P S H R +K
Sbjct: 5 PLISLQLLSSSSSSSSLKPTTKL-------LFFHKPELISHHH-----RNRNKLSRTSVH 52
Query: 71 PPRLFMIIDP--ILLLNGFGGAGFYLDTQTLLATVSVLAAIALSLFLGLKGDPVPCERCA 128
+II P + L + F G G ++DTQT + T+S++ AIALSLFLG KGDPVPCERC
Sbjct: 53 QHSANLIIHPSVLFLSSAFDGGGGFIDTQTFIVTISLVVAIALSLFLGFKGDPVPCERCG 112
Query: 129 GNGGMKCVFCNDGKMKQEMGLIDCKVCKGSGLILCKKCGGSGYSRRL 175
GNGG KCVFC +GKMK E G++DCKVCKGSGLI CKKCGGSGYSRRL
Sbjct: 113 GNGGTKCVFCLEGKMKVESGMVDCKVCKGSGLIFCKKCGGSGYSRRL 159
>At3g22070 unknown protein
Length = 146
Score = 44.3 bits (103), Expect = 3e-05
Identities = 31/71 (43%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPC-SSHSLPQPSPPPPSKSHFPS--LPRFPSKCRHKPP 69
SSS+ P SSSSSSSSSTS ++P + + P P+PPPPS + P P P P
Sbjct: 44 SSSTDPSSSSSSSSSSTSPFITPFPNPNPNPNPNPPPPSTPNPPPEFSPPPPDLDTTTAP 103
Query: 70 KPPRLFMIIDP 80
PP + I P
Sbjct: 104 PPPSTDIPIPP 114
>At1g75690 unknown protein
Length = 154
Score = 42.7 bits (99), Expect = 1e-04
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 17/98 (17%)
Query: 94 LDTQTLLATVSVLAAIALSLFLGL-----------KGDPVPCERCAGNGGMKCVFC-NDG 141
LD T++A +A++AL + + + + + PC C G G KC C G
Sbjct: 46 LDPNTVVAISVGVASVALGIGIPVFYETQIDNAAKRENTQPCFPCNGTGAQKCRLCVGSG 105
Query: 142 KMKQEMG-----LIDCKVCKGSGLILCKKCGGSGYSRR 174
+ E+G + +C C G+G + C C GSG R
Sbjct: 106 NVTVELGGGEKEVSNCINCDGAGSLTCTTCQGSGVQPR 143
>At1g62760 hypothetical protein
Length = 312
Score = 41.2 bits (95), Expect = 3e-04
Identities = 31/74 (41%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query: 3 TLSYSTLHPLSSSSL----PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLP 58
T S S+L P SSS PPSSS SS+ +SL S SL SPPPP S P
Sbjct: 21 TTSSSSLSPSSSSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSS 80
Query: 59 RFPSKCRHKPPKPP 72
PS P P
Sbjct: 81 LSPSLSPSPPSSSP 94
Score = 36.6 bits (83), Expect = 0.007
Identities = 26/56 (46%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query: 7 STLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPS 62
S+L P S S PPSSS SS+ +SL S SL SPPPP S P PS
Sbjct: 79 SSLSPSLSPS-PPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPS 133
Score = 36.6 bits (83), Expect = 0.007
Identities = 32/72 (44%), Positives = 33/72 (45%), Gaps = 18/72 (25%)
Query: 8 TLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSH-------SLPQPSPPPPSKSHFPSLPRF 60
+L P S PPSSS SS S SL SP SS SL SPPP S S PS P
Sbjct: 62 SLSPSSPPPPPPSSSPLSSLSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLS--PSSP-- 117
Query: 61 PSKCRHKPPKPP 72
PP PP
Sbjct: 118 -------PPPPP 122
Score = 36.2 bits (82), Expect = 0.009
Identities = 32/74 (43%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query: 2 ATLSYSTLHPLS-SSSLPPSSSSSSS--SSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLP 58
++LS S+ PLS S S PP SSS SS S LSP S P +PP S PS P
Sbjct: 51 SSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLSPSPPSSSPSSAPP---SSLSPSSP 107
Query: 59 RFPSKCRHKPPKPP 72
S PP PP
Sbjct: 108 PPLSLSPSSPPPPP 121
Score = 30.8 bits (68), Expect = 0.39
Identities = 25/54 (46%), Positives = 25/54 (46%), Gaps = 6/54 (11%)
Query: 5 SYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPP--PPSKSHFPS 56
S S P SS S P SS S SS L LSP S P P PP P S PS
Sbjct: 84 SLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSS----PPPPPPSSSPLSSLSPS 133
>At3g24550 protein kinase, putative
Length = 652
Score = 40.0 bits (92), Expect = 6e-04
Identities = 33/81 (40%), Positives = 40/81 (48%), Gaps = 10/81 (12%)
Query: 3 TLSYSTLHPLSSSSLPPSSSSSS---SSSTSLHLSPCSS---HSLPQPSPP----PPSKS 52
T S +T P ++SS PP+++ SS S ST+ P SS SLP PSPP PP
Sbjct: 20 TNSTTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLPPPSPPGSLTPPLPQ 79
Query: 53 HFPSLPRFPSKCRHKPPKPPR 73
PS P PS P PR
Sbjct: 80 PSPSAPITPSPPSPTTPSNPR 100
Score = 32.0 bits (71), Expect = 0.17
Identities = 29/74 (39%), Positives = 30/74 (40%), Gaps = 19/74 (25%)
Query: 7 STLHPLSSS---SLPPSSSSSSSSSTSLHLSPCSSHSLPQPSP-----PPPSKSHFPSLP 58
ST P SS SLPP S S L+P LPQPSP P P PS P
Sbjct: 51 STSPPPSSPLPPSLPPPSPPGS-------LTP----PLPQPSPSAPITPSPPSPTTPSNP 99
Query: 59 RFPSKCRHKPPKPP 72
R P PP P
Sbjct: 100 RSPPSPNQGPPNTP 113
Score = 32.0 bits (71), Expect = 0.17
Identities = 23/62 (37%), Positives = 31/62 (49%), Gaps = 7/62 (11%)
Query: 15 SSLPPSSSSSSSSSTSLHLS----PCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
S PPS ++S+++T + P ++ S P PSP S S PS P PS PP
Sbjct: 12 SPSPPSPPTNSTTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSL---PPPS 68
Query: 71 PP 72
PP
Sbjct: 69 PP 70
Score = 31.6 bits (70), Expect = 0.23
Identities = 28/82 (34%), Positives = 33/82 (40%), Gaps = 17/82 (20%)
Query: 5 SYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLP-QPSPP----------PPSKSH 53
S ++ P SS LPPS S + P S S P PSPP PPS +
Sbjct: 48 STNSTSPPPSSPLPPSLPPPSPPGSLTPPLPQPSPSAPITPSPPSPTTPSNPRSPPSPNQ 107
Query: 54 FP------SLPRFPSKCRHKPP 69
P S PR PS + PP
Sbjct: 108 GPPNTPSGSTPRTPSNTKPSPP 129
Score = 26.6 bits (57), Expect = 7.3
Identities = 18/45 (40%), Positives = 19/45 (42%), Gaps = 16/45 (35%)
Query: 35 PCSSH---SLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRLFM 76
P +H SLP P PP P PR P PP PP FM
Sbjct: 207 PSDNHVVTSLPPPKPPSP--------PRKP-----PPPPPPPAFM 238
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 40.0 bits (92), Expect = 6e-04
Identities = 24/61 (39%), Positives = 28/61 (45%), Gaps = 5/61 (8%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P + S LPPSSS ++S S SP PSPPPP P P PS + P
Sbjct: 111 PEADSPLPPSSSPEANSPQSPASSPKPESLADSPSPPPP-----PPQPESPSSPSYPEPA 165
Query: 71 P 71
P
Sbjct: 166 P 166
Score = 33.5 bits (75), Expect = 0.060
Identities = 28/82 (34%), Positives = 33/82 (40%), Gaps = 22/82 (26%)
Query: 11 PLSSSSLPPSSSS--------SSSSSTSLHLSPCSSHSLPQPSPP----------PPSKS 52
P S LPPSSS SSS L+P SS + P PP PPS S
Sbjct: 63 PEEDSPLPPSSSPEEDSPLAPSSSPEVDSPLAPSSSPEVDSPQPPSSSPEADSPLPPSSS 122
Query: 53 HFPSLPRFPSKCRHKPPKPPRL 74
+ P+ P+ PKP L
Sbjct: 123 PEANSPQSPA----SSPKPESL 140
Score = 32.0 bits (71), Expect = 0.17
Identities = 27/72 (37%), Positives = 32/72 (43%), Gaps = 17/72 (23%)
Query: 7 STLHPLSS----SSLPPSSSSSSS-----SSTSLHLSPCSSHSLPQPS--------PPPP 49
S L P SS S PPSSS + SS+ SP S S P+P PPPP
Sbjct: 91 SPLAPSSSPEVDSPQPPSSSPEADSPLPPSSSPEANSPQSPASSPKPESLADSPSPPPPP 150
Query: 50 SKSHFPSLPRFP 61
+ PS P +P
Sbjct: 151 PQPESPSSPSYP 162
Score = 26.6 bits (57), Expect = 7.3
Identities = 22/52 (42%), Positives = 23/52 (43%), Gaps = 6/52 (11%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPS 62
P + S L PSSS S LSP SS SP PPS S P PS
Sbjct: 39 PSTDSPLSPSSSPEEDSP----LSPSSSPEED--SPLPPSSSPEEDSPLAPS 84
Score = 26.2 bits (56), Expect = 9.5
Identities = 22/72 (30%), Positives = 27/72 (36%), Gaps = 6/72 (8%)
Query: 7 STLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPS-KSHFPSLPRFPSKCR 65
S L P SS + + SSS P SS P PP S +++ P P K
Sbjct: 79 SPLAPSSSPEVDSPLAPSSSPEVDSPQPPSSSPEADSPLPPSSSPEANSPQSPASSPKPE 138
Query: 66 H-----KPPKPP 72
PP PP
Sbjct: 139 SLADSPSPPPPP 150
>At3g53330 putative protein
Length = 310
Score = 39.7 bits (91), Expect = 8e-04
Identities = 28/90 (31%), Positives = 37/90 (41%), Gaps = 13/90 (14%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPS-------PPPPSKSHFPSLPRFPSK 63
PL+ + PP S + S + SP +PS PPPPSK+H PS PS
Sbjct: 116 PLAITPSPPPPSKTHERSRPITPSPPPPSKTHEPSRPNTPPPPPPPSKTHEPSRRITPS- 174
Query: 64 CRHKPPKPPRLFMIIDPILLLNGFGGAGFY 93
P PP + I + +GG Y
Sbjct: 175 -----PPPPSKILPFGKIYRVGDYGGWSVY 199
>At5g21280 putative protein
Length = 302
Score = 39.3 bits (90), Expect = 0.001
Identities = 22/52 (42%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query: 12 LSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPP-SKSHFPSLPRFPS 62
+ +SS SSSSSS+SS+S + P + H PSPPP + + S P+ PS
Sbjct: 153 IQTSSTTSSSSSSSTSSSSSSIFPTNPHIYSSPSPPPTFTTATSDSAPQLPS 204
>At5g17840 unknown protein
Length = 154
Score = 39.3 bits (90), Expect = 0.001
Identities = 21/61 (34%), Positives = 28/61 (45%), Gaps = 8/61 (13%)
Query: 119 GDPVPCERCAGNGGMKCVFCND-----GKMKQEMGLI---DCKVCKGSGLILCKKCGGSG 170
GD C C G + C C+ + + G+I C C GSG I+CK CGG G
Sbjct: 91 GDNQRCTCCEAKGALLCSTCSGTGLYVDSIMESQGIIVKVRCLGCGGSGNIMCKLCGGRG 150
Query: 171 Y 171
+
Sbjct: 151 H 151
>At5g07750 putative protein
Length = 1289
Score = 39.3 bits (90), Expect = 0.001
Identities = 22/62 (35%), Positives = 28/62 (44%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P+ ++ PP S++ + L P S S P P PPPPS P P P PP
Sbjct: 913 PVKTAPPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPP 972
Query: 71 PP 72
PP
Sbjct: 973 PP 974
Score = 37.7 bits (86), Expect = 0.003
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 7/71 (9%)
Query: 9 LHPLSSSSLPPSSSSSS-------SSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP 61
LH +SS+ PP ++ S++ S+ P S+ P P PPPP P P P
Sbjct: 903 LHGISSAPSPPVKTAPPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSYGSPPPPPPP 962
Query: 62 SKCRHKPPKPP 72
PP PP
Sbjct: 963 PPSYGSPPPPP 973
Score = 35.4 bits (80), Expect = 0.016
Identities = 25/70 (35%), Positives = 32/70 (45%), Gaps = 6/70 (8%)
Query: 7 STLHPLSSSSLPPSSSS-SSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCR 65
S+L P S PP S +S + T LH S + S P P PPPP P+ + +
Sbjct: 613 SSLPPASPHQAPPPLPSLTSEAKTVLHSSQAVA-SPPPPPPPPP----LPTYSHYQTSQL 667
Query: 66 HKPPKPPRLF 75
PP PP F
Sbjct: 668 PPPPPPPPPF 677
Score = 33.9 bits (76), Expect = 0.046
Identities = 22/64 (34%), Positives = 23/64 (35%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRLFMI 77
PP S S P S S P P PPPP P P P PP PP F
Sbjct: 946 PPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSH 1005
Query: 78 IDPI 81
+ I
Sbjct: 1006 VSSI 1009
Score = 33.9 bits (76), Expect = 0.046
Identities = 21/62 (33%), Positives = 22/62 (34%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P S PP S S P S+ P P PPPP P P P PP
Sbjct: 938 PSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPP 997
Query: 71 PP 72
PP
Sbjct: 998 PP 999
Score = 33.1 bits (74), Expect = 0.078
Identities = 23/64 (35%), Positives = 24/64 (36%), Gaps = 2/64 (3%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSH--SLPQPSPPPPSKSHFPSLPRFPSKCRHKP 68
P S PP S S P SH S+P P PPPP P P P P
Sbjct: 977 PGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPPPPPPPMHGGAP 1036
Query: 69 PKPP 72
P PP
Sbjct: 1037 PPPP 1040
Score = 32.7 bits (73), Expect = 0.10
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 10/71 (14%)
Query: 4 LSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPP--SKSHFPSLPRFP 61
L Y+++ P S + P SS+ S + +P P PPPP S +H P P
Sbjct: 887 LPYTSIAPSPSVKILPLHGISSAPSPPVKTAP--------PPPPPPPFSNAHSVLSPPPP 938
Query: 62 SKCRHKPPKPP 72
S PP PP
Sbjct: 939 SYGSPPPPPPP 949
Score = 31.2 bits (69), Expect = 0.30
Identities = 27/79 (34%), Positives = 34/79 (42%), Gaps = 12/79 (15%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCR-HKPPKPPRLFM 76
PP S +S+ H CS+ S P P PPPP P P +K + P PP +
Sbjct: 837 PPPWKSLYASTFETH-EACSTSSSPPPPPPPP-----PFSPLNTTKANDYILPPPPLPYT 890
Query: 77 IIDP-----ILLLNGFGGA 90
I P IL L+G A
Sbjct: 891 SIAPSPSVKILPLHGISSA 909
Score = 30.8 bits (68), Expect = 0.39
Identities = 23/67 (34%), Positives = 27/67 (39%), Gaps = 11/67 (16%)
Query: 8 TLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKC--R 65
TL P PP +S +S +L LP P PPPP KS + S C
Sbjct: 807 TLLPPPPPPPPPPFASVRRNSETL---------LPPPPPPPPWKSLYASTFETHEACSTS 857
Query: 66 HKPPKPP 72
PP PP
Sbjct: 858 SSPPPPP 864
Score = 30.8 bits (68), Expect = 0.39
Identities = 20/55 (36%), Positives = 23/55 (41%), Gaps = 6/55 (10%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
PP+ S S+ L S LP P PPPP F S+ R PP PP
Sbjct: 768 PPAYYSVGQKSSDLQTS-----QLPSPPPPPPPPP-FASVRRNSETLLPPPPPPP 816
Score = 30.4 bits (67), Expect = 0.51
Identities = 19/55 (34%), Positives = 19/55 (34%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
PP S SS P P P PPPP P P P PP PP
Sbjct: 999 PPPPFSHVSSIPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPP 1053
Score = 30.4 bits (67), Expect = 0.51
Identities = 24/78 (30%), Positives = 35/78 (44%), Gaps = 9/78 (11%)
Query: 4 LSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSP---CSSHSLP--QPSPPPPSKSHF---- 54
L +S+ P S + LPP S S + L+ CS+ P P+PPPP +++
Sbjct: 717 LPFSSERPNSGTVLPPPPSPPWKSVYASALAIPAICSTSQAPTSSPTPPPPPPAYYSVGQ 776
Query: 55 PSLPRFPSKCRHKPPKPP 72
S S+ PP PP
Sbjct: 777 KSSDLQTSQLPSPPPPPP 794
Score = 30.4 bits (67), Expect = 0.51
Identities = 13/30 (43%), Positives = 15/30 (49%)
Query: 43 QPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
QP PPPP + P P P + PP PP
Sbjct: 1074 QPPPPPPMRGGAPPPPPPPMRGGAPPPPPP 1103
Score = 30.0 bits (66), Expect = 0.66
Identities = 21/62 (33%), Positives = 23/62 (36%), Gaps = 10/62 (16%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P S PP S P S+ P P PPPP SH S+P PP
Sbjct: 964 PSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPF-SHVSSIP---------PPP 1013
Query: 71 PP 72
PP
Sbjct: 1014 PP 1015
Score = 29.6 bits (65), Expect = 0.86
Identities = 21/64 (32%), Positives = 22/64 (33%), Gaps = 2/64 (3%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP--SKCRHKP 68
P S PP S S P + P P PPPP P P P S P
Sbjct: 951 PSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSSIP 1010
Query: 69 PKPP 72
P PP
Sbjct: 1011 PPPP 1014
Score = 28.5 bits (62), Expect = 1.9
Identities = 12/29 (41%), Positives = 14/29 (47%)
Query: 44 PSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P PPPP + P P P + PP PP
Sbjct: 1087 PPPPPPMRGGAPPPPPPPMRGGAPPPPPP 1115
Score = 28.5 bits (62), Expect = 1.9
Identities = 21/57 (36%), Positives = 23/57 (39%), Gaps = 14/57 (24%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRL 74
PP SS +S T L P P PPPP S P+ S PP PP L
Sbjct: 675 PPFSSERPNSGTVLP---------PPPPPPPPFSSERPN-----SGTVLPPPPPPPL 717
Score = 28.5 bits (62), Expect = 1.9
Identities = 13/31 (41%), Positives = 14/31 (44%)
Query: 42 PQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P P PPPP P P P +PP PP
Sbjct: 1049 PPPPPPPPMHGGAPPPPPPPMFGGAQPPPPP 1079
Score = 28.5 bits (62), Expect = 1.9
Identities = 13/31 (41%), Positives = 13/31 (41%)
Query: 42 PQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P P PPPP P P P PP PP
Sbjct: 1036 PPPPPPPPMHGGAPPPPPPPPMHGGAPPPPP 1066
Score = 27.7 bits (60), Expect = 3.3
Identities = 13/33 (39%), Positives = 15/33 (45%)
Query: 42 PQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRL 74
P P PPP ++ P P P PP PP L
Sbjct: 1148 PPPPPPPGGRAPGPPPPPGPRPPGGGPPPPPML 1180
Score = 27.3 bits (59), Expect = 4.3
Identities = 12/29 (41%), Positives = 13/29 (44%)
Query: 44 PSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P PPPP + P P P PP PP
Sbjct: 1099 PPPPPPMRGGAPPPPPPPMHGGAPPPPPP 1127
Score = 27.3 bits (59), Expect = 4.3
Identities = 12/29 (41%), Positives = 13/29 (44%)
Query: 44 PSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P PPPP P P P + PP PP
Sbjct: 1111 PPPPPPMHGGAPPPPPPPMRGGAPPPPPP 1139
Score = 26.6 bits (57), Expect = 7.3
Identities = 14/41 (34%), Positives = 16/41 (38%)
Query: 35 PCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRLF 75
P + P P PPP P P P PP PP +F
Sbjct: 1030 PMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPMF 1070
Score = 26.2 bits (56), Expect = 9.5
Identities = 19/58 (32%), Positives = 19/58 (32%)
Query: 15 SSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
S L P S S P P P PPP S P P P PP PP
Sbjct: 931 SVLSPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPP 988
Score = 26.2 bits (56), Expect = 9.5
Identities = 16/62 (25%), Positives = 19/62 (29%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P+ + PP P H P PPPP P P + PP
Sbjct: 1030 PMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAPPPP 1089
Query: 71 PP 72
PP
Sbjct: 1090 PP 1091
>At3g50580 proline-rich protein
Length = 265
Score = 39.3 bits (90), Expect = 0.001
Identities = 26/68 (38%), Positives = 29/68 (42%), Gaps = 6/68 (8%)
Query: 11 PLSSSSLPPSSSSSSSSSTSL-----HLSPCSSHSLPQ-PSPPPPSKSHFPSLPRFPSKC 64
P S PP + S S SL H +P S S P PS PPP+ PS P P
Sbjct: 93 PAPKKSPPPPTPKKSPSPPSLTPFVPHPTPKKSPSPPPTPSLPPPAPKKSPSTPSLPPPT 152
Query: 65 RHKPPKPP 72
K P PP
Sbjct: 153 PKKSPPPP 160
Score = 35.0 bits (79), Expect = 0.021
Identities = 22/64 (34%), Positives = 27/64 (41%), Gaps = 4/64 (6%)
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRF----PSKCRHKP 68
S+ S P S S S+ S +P P+ SPPPP+ PS P P K
Sbjct: 66 SNGSAPAISISPSTPIPSTPSTPSPPPPAPKKSPPPPTPKKSPSPPSLTPFVPHPTPKKS 125
Query: 69 PKPP 72
P PP
Sbjct: 126 PSPP 129
Score = 30.8 bits (68), Expect = 0.39
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 7/58 (12%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKP 68
P + SLPP + S S+ SL P+ SPPPP SH S P P + P
Sbjct: 128 PPPTPSLPPPAPKKSPSTPSL------PPPTPKKSPPPP-PSHHSSSPSNPPHHQQNP 178
Score = 28.1 bits (61), Expect = 2.5
Identities = 20/61 (32%), Positives = 25/61 (40%), Gaps = 2/61 (3%)
Query: 16 SLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPP--PPSKSHFPSLPRFPSKCRHKPPKPPR 73
S+ PS+ S+ ST P S P P+P P S P +P K PP P
Sbjct: 74 SISPSTPIPSTPSTPSPPPPAPKKSPPPPTPKKSPSPPSLTPFVPHPTPKKSPSPPPTPS 133
Query: 74 L 74
L
Sbjct: 134 L 134
Score = 26.6 bits (57), Expect = 7.3
Identities = 20/65 (30%), Positives = 24/65 (36%), Gaps = 4/65 (6%)
Query: 8 TLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHK 67
+L P P S S + + +P S S P PP P KS P PS
Sbjct: 112 SLTPFVPHPTPKKSPSPPPTPSLPPPAPKKSPSTPSLPPPTPKKSPPPP----PSHHSSS 167
Query: 68 PPKPP 72
P PP
Sbjct: 168 PSNPP 172
>At4g13340 extensin-like protein
Length = 760
Score = 38.1 bits (87), Expect = 0.002
Identities = 22/56 (39%), Positives = 25/56 (44%)
Query: 17 LPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
LPP S S + +P + S P PSPPPP S P P P PP PP
Sbjct: 402 LPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPP 457
Score = 34.7 bits (78), Expect = 0.027
Identities = 27/82 (32%), Positives = 34/82 (40%), Gaps = 7/82 (8%)
Query: 6 YSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQP--SPPPPSKSHFPS-LPRFPS 62
YS P SS PP S + + P HS P P SPPPP ++ S P S
Sbjct: 510 YSPPPPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSS 569
Query: 63 KCRHKPP----KPPRLFMIIDP 80
H PP PP ++ + P
Sbjct: 570 PPPHSPPPPHSPPPPIYPYLSP 591
Score = 33.1 bits (74), Expect = 0.078
Identities = 26/73 (35%), Positives = 30/73 (40%), Gaps = 11/73 (15%)
Query: 11 PLSSSSLPPSSSSSSSSSTS----LHLSP---CSSHSLPQP----SPPPPSKSHFPSLPR 59
P+ SS PP S+ S +H SP HS P P SPPPPS + LP
Sbjct: 686 PVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQSPPPPSPEYEGPLPP 745
Query: 60 FPSKCRHKPPKPP 72
PP PP
Sbjct: 746 VIGVSYASPPPPP 758
Score = 32.0 bits (71), Expect = 0.17
Identities = 22/61 (36%), Positives = 24/61 (39%), Gaps = 1/61 (1%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P S PP SS H SP HS P P PPP + S P P+ PP
Sbjct: 546 PPPQFSPPPPEPYYYSSPPPPHSSP-PPHSPPPPHSPPPPIYPYLSPPPPPTPVSSPPPT 604
Query: 71 P 71
P
Sbjct: 605 P 605
Score = 31.2 bits (69), Expect = 0.30
Identities = 24/69 (34%), Positives = 27/69 (38%), Gaps = 5/69 (7%)
Query: 9 LHPLSSSSLPPSSSSSSSSSTSLHLSPCSS-----HSLPQPSPPPPSKSHFPSLPRFPSK 63
L P S S PP + S+ T P S P P PPPP S P P P
Sbjct: 402 LPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPP 461
Query: 64 CRHKPPKPP 72
+ PP PP
Sbjct: 462 PVYSPPPPP 470
Score = 30.4 bits (67), Expect = 0.51
Identities = 24/65 (36%), Positives = 26/65 (39%), Gaps = 10/65 (15%)
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPS----PPPPSKSHFPSLPRFPSK-CRHK 67
SS PP SS +H S S P P PPPS H+ S P PS C
Sbjct: 649 SSPPPPPVYYSSPPPPPPVHYS-----SPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEES 703
Query: 68 PPKPP 72
PP P
Sbjct: 704 PPPAP 708
Score = 30.4 bits (67), Expect = 0.51
Identities = 14/31 (45%), Positives = 15/31 (48%)
Query: 42 PQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P P PPPP S P P P + PP PP
Sbjct: 471 PPPPPPPPVYSPPPPSPPPPPPPVYSPPPPP 501
Score = 30.4 bits (67), Expect = 0.51
Identities = 16/44 (36%), Positives = 19/44 (42%), Gaps = 6/44 (13%)
Query: 35 PCSSHSLPQPSP------PPPSKSHFPSLPRFPSKCRHKPPKPP 72
PC +S P P P PPP ++ S P P PP PP
Sbjct: 623 PCIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPPPP 666
Score = 30.0 bits (66), Expect = 0.66
Identities = 19/63 (30%), Positives = 27/63 (42%), Gaps = 3/63 (4%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRLFMI 77
PP + SS T ++ SP +P PPPP + P P P + P PP ++
Sbjct: 593 PPPTPVSSPPPTPVY-SPPPPPPCIEPPPPPPCIEYSPPPP--PPVVHYSSPPPPPVYYS 649
Query: 78 IDP 80
P
Sbjct: 650 SPP 652
Score = 29.3 bits (64), Expect = 1.1
Identities = 21/60 (35%), Positives = 26/60 (43%), Gaps = 3/60 (5%)
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
SS PP SS + SP + SPPPP + H+ S P PS + P PP
Sbjct: 639 SSPPPPPVYYSSPPPPPVYYSSPPPPPPVHYSSPPPP-EVHYHSPP--PSPVHYSSPPPP 695
Score = 29.3 bits (64), Expect = 1.1
Identities = 21/60 (35%), Positives = 25/60 (41%), Gaps = 7/60 (11%)
Query: 16 SLPPSSSSSSSSSTSLHLSPCSSHSLPQP---SPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
S PP + S S S+ P LP P SPPPP+ P P+ PP PP
Sbjct: 377 SRPPVNCGSFSCGRSVSPRPPVVTPLPPPSLPSPPPPA----PIFSTPPTLTSPPPPSPP 432
Score = 28.5 bits (62), Expect = 1.9
Identities = 18/55 (32%), Positives = 22/55 (39%), Gaps = 1/55 (1%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
PP SS ++ S + SPPPP H+ S P P H PP P
Sbjct: 633 PPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPPPPVHYSSPPP-PEVHYHSPPPSP 686
Score = 28.1 bits (61), Expect = 2.5
Identities = 15/41 (36%), Positives = 19/41 (45%), Gaps = 6/41 (14%)
Query: 35 PCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRLF 75
P +S P PSPPPP + P P PP PP ++
Sbjct: 476 PPPVYSPPPPSPPPPPPPVYSPPPPPP------PPPPPPVY 510
Score = 27.7 bits (60), Expect = 3.3
Identities = 22/67 (32%), Positives = 23/67 (33%), Gaps = 5/67 (7%)
Query: 6 YSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCR 65
YS P PP S S P S P P PPPP P P + S
Sbjct: 464 YSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSS--- 520
Query: 66 HKPPKPP 72
PP PP
Sbjct: 521 --PPPPP 525
Score = 27.7 bits (60), Expect = 3.3
Identities = 17/59 (28%), Positives = 20/59 (33%)
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKP 71
SS PP SS +H + SPPPP + P H PP P
Sbjct: 659 SSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPP 717
Score = 26.9 bits (58), Expect = 5.6
Identities = 14/35 (40%), Positives = 15/35 (42%), Gaps = 4/35 (11%)
Query: 42 PQPSPPPPSKSHFPSLPRFPSK----CRHKPPKPP 72
P SPPPP P P P+ C PP PP
Sbjct: 508 PVYSPPPPPVYSSPPPPPSPAPTPVYCTRPPPPPP 542
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 38.1 bits (87), Expect = 0.002
Identities = 25/66 (37%), Positives = 29/66 (43%), Gaps = 18/66 (27%)
Query: 7 STLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRH 66
S L PL+ S PPS SS ST PSPPPP+ + PS P +
Sbjct: 69 SILPPLTDSPPPPSDSSPPVDST--------------PSPPPPTSNESPS----PPEDSE 110
Query: 67 KPPKPP 72
PP PP
Sbjct: 111 TPPAPP 116
Score = 35.8 bits (81), Expect = 0.012
Identities = 27/77 (35%), Positives = 33/77 (42%), Gaps = 11/77 (14%)
Query: 7 STLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLP---------QPSPPPPSKSHFP-- 55
S L P+ SS P + SSS+ S +P LP SPPPPS S P
Sbjct: 30 SALPPVDSSPPSPPADSSSTPPLSEPSTPPPDSQLPPLPSILPPLTDSPPPPSDSSPPVD 89
Query: 56 SLPRFPSKCRHKPPKPP 72
S P P ++ P PP
Sbjct: 90 STPSPPPPTSNESPSPP 106
Score = 32.3 bits (72), Expect = 0.13
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 8/68 (11%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSP-------CSSHSLPQPSPPPPSKSHFPSLPRFPSK 63
P + ++ PP + S +S+ + SP S+ L +PS PPP S P LP
Sbjct: 15 PPADTAPPPETPSENSALPPVDSSPPSPPADSSSTPPLSEPSTPPPD-SQLPPLPSILPP 73
Query: 64 CRHKPPKP 71
PP P
Sbjct: 74 LTDSPPPP 81
Score = 30.0 bits (66), Expect = 0.66
Identities = 22/64 (34%), Positives = 29/64 (44%), Gaps = 4/64 (6%)
Query: 13 SSSSLPPSSSSSSS---SSTSLHLSPCSSHSLPQPSPP-PPSKSHFPSLPRFPSKCRHKP 68
S+ + PP S S SS S ++ P + S P SPP PP+ S P P + P
Sbjct: 119 SNDNNPPPSQDLQSPPPSSPSPNVGPTNPESPPLQSPPAPPASDPTNSPPASPLDPTNPP 178
Query: 69 PKPP 72
P P
Sbjct: 179 PIQP 182
Score = 27.7 bits (60), Expect = 3.3
Identities = 22/72 (30%), Positives = 29/72 (39%), Gaps = 14/72 (19%)
Query: 13 SSSSLPPSSSSSSSS------------STSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRF 60
S+ S PP +S+ S S + S +P S L P P PS + P+ P
Sbjct: 90 STPSPPPPTSNESPSPPEDSETPPAPPNESNDNNPPPSQDLQSPPPSSPSPNVGPTNPES 149
Query: 61 PSKCRHKPPKPP 72
P PP PP
Sbjct: 150 PP--LQSPPAPP 159
>At2g25050 hypothetical protein
Length = 742
Score = 37.7 bits (86), Expect = 0.003
Identities = 30/88 (34%), Positives = 36/88 (40%), Gaps = 24/88 (27%)
Query: 8 TLHPLSSSSL------------------PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPP 49
TLHPL+SS PPS+ ++ +SS L P S P P PPPP
Sbjct: 519 TLHPLTSSQPKKASPQCPQSPTPVHSNGPPSAEAAVTSSPLPPLKPLRILSRPPPPPPPP 578
Query: 50 SKSHFPSLPRFPSKCRHK-----PPKPP 72
S S P PS + PP PP
Sbjct: 579 PISSLRSTPS-PSSTSNSIATQGPPPPP 605
Score = 37.4 bits (85), Expect = 0.004
Identities = 21/56 (37%), Positives = 28/56 (49%), Gaps = 4/56 (7%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPR 73
PP SS S+ S S + ++ P P PPPP +SH +L P PP PP+
Sbjct: 578 PPISSLRSTPSPSSTSNSIATQGPPPPPPPPPLQSHRSALSSSPL----PPPLPPK 629
Score = 28.5 bits (62), Expect = 1.9
Identities = 28/89 (31%), Positives = 32/89 (35%), Gaps = 27/89 (30%)
Query: 11 PLSSSSLPPSSSSSSSSSTSL-------------HLSPCSSHSLP------------QPS 45
P+SS PS SS+S+S + H S SS LP P
Sbjct: 579 PISSLRSTPSPSSTSNSIATQGPPPPPPPPPLQSHRSALSSSPLPPPLPPKKLLATTNPP 638
Query: 46 PPPPSKSHFPSLPRFP--SKCRHKPPKPP 72
PPPP H S P S PP PP
Sbjct: 639 PPPPPPLHSNSRMGAPTSSLVLKSPPVPP 667
Score = 27.3 bits (59), Expect = 4.3
Identities = 15/44 (34%), Positives = 22/44 (49%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP 61
PP S+S + + L S P P+P P S+SH ++P P
Sbjct: 643 PPLHSNSRMGAPTSSLVLKSPPVPPPPAPAPLSRSHNGNIPPVP 686
>At1g49750 unknown protein
Length = 494
Score = 37.7 bits (86), Expect = 0.003
Identities = 16/39 (41%), Positives = 19/39 (48%)
Query: 35 PCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPR 73
PC P P PPPPS P P+ P + PP PP+
Sbjct: 69 PCPPPPSPPPCPPPPSPPPSPPPPQLPPPPQLPPPAPPK 107
Score = 28.1 bits (61), Expect = 2.5
Identities = 20/55 (36%), Positives = 21/55 (37%), Gaps = 11/55 (20%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
PPS S + C P P PPPPS P P PS PP PP
Sbjct: 47 PPSPSPEPEPEPA----DCPPPPPPPPCPPPPSP---PPCPPPPS----PPPSPP 90
>At4g18560 pherophorin - like protein
Length = 637
Score = 37.4 bits (85), Expect = 0.004
Identities = 22/56 (39%), Positives = 23/56 (40%), Gaps = 1/56 (1%)
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPP-PPSKSHFPSLPRFPSKCRHKPPKPP 72
PP S S ST P S+P P PP PP P P SK PP PP
Sbjct: 269 PPKRSISLGDSTENRADPPPQKSIPPPPPPPPPPLLQQPPPPPSVSKAPPPPPPPP 324
>At3g47650 putative protein
Length = 136
Score = 37.4 bits (85), Expect = 0.004
Identities = 36/132 (27%), Positives = 49/132 (36%), Gaps = 27/132 (20%)
Query: 54 FPSLPRFPSKCRHKPPKPPRLFMIIDPILLLNGFGGAGFYLDTQTLLATV---SVLAAIA 110
F S P F + K PKP F D L + + LL T S A
Sbjct: 8 FSSPPTFCFQSPSKNPKPSHFFSTNDNTSSL---------VQKRELLQTSRSQSFEVKAA 58
Query: 111 LSLFLGLKGDPVPCERCAGNGGMKCVFCNDGKMKQEMGLID-----------CKVCKGSG 159
+ G K + + C C G G + C C G + LID C +C+G
Sbjct: 59 NNNPQGTKPNSLVCANCEGEGCVACSQCKGGGVN----LIDHFNGQFKAGALCWLCRGKK 114
Query: 160 LILCKKCGGSGY 171
+LC C G+G+
Sbjct: 115 EVLCGDCNGAGF 126
>At3g24400 protein kinase, putative
Length = 694
Score = 37.4 bits (85), Expect = 0.004
Identities = 29/77 (37%), Positives = 35/77 (44%), Gaps = 8/77 (10%)
Query: 11 PLSSSSLPPSSS-------SSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSK 63
PL+ S LPPS + S S S L SP S L SPPPPS + + PR P
Sbjct: 118 PLTPSPLPPSPTTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPP 177
Query: 64 CRHKPPKPPRLFMIIDP 80
P PPR+ + P
Sbjct: 178 -PSTPTPPPRVGSLSPP 193
Score = 33.9 bits (76), Expect = 0.046
Identities = 26/65 (40%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPP---PSKSHFPS-LPRFPSKCRH 66
PL+ S LPPS ++ S T SP + PSPPP PS PS LP P+
Sbjct: 77 PLTPSPLPPSPTTPSPPLTP---SPTTPSPPLTPSPPPAITPSPPLTPSPLPPSPTTPSP 133
Query: 67 KPPKP 71
PP P
Sbjct: 134 PPPSP 138
Score = 32.3 bits (72), Expect = 0.13
Identities = 28/81 (34%), Positives = 36/81 (43%), Gaps = 14/81 (17%)
Query: 4 LSYSTLHPLSSSSLPPSSSSSSSSSTSLHLS---------PCSSHSLP----QPSPPPPS 50
L+ S L P ++ PP + S ++ S L S P + LP PSPPPPS
Sbjct: 78 LTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSPLPPSPTTPSPPPPS 137
Query: 51 KSHFPSLPRFPSKCRHKPPKP 71
S PS P PS P +P
Sbjct: 138 PS-IPSPPLTPSPPPSSPLRP 157
Score = 30.0 bits (66), Expect = 0.66
Identities = 21/61 (34%), Positives = 26/61 (42%), Gaps = 8/61 (13%)
Query: 15 SSLPPSSSSSSSSSTSLHLSPCSSHSLPQP---SPPPPSKSHFPSLPRFPSKCRHKPPKP 71
SS PP + S L + P PQP +PPPP + P+LP P P P
Sbjct: 2 SSAPPPGGTPSPPPQPLPIPPP-----PQPLPVTPPPPPTALPPALPPPPPPTALPPALP 56
Query: 72 P 72
P
Sbjct: 57 P 57
Score = 29.6 bits (65), Expect = 0.86
Identities = 26/73 (35%), Positives = 32/73 (43%), Gaps = 13/73 (17%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSP----------PPPSKSHFPSLPRF 60
PL+ S P + S + + L SP ++ S P PSP PPPS PS P
Sbjct: 104 PLTPSPPPAITPSPPLTPSPLPPSP-TTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPP 162
Query: 61 PSKCRHKPPKPPR 73
PS P PPR
Sbjct: 163 PSPA--TPSTPPR 173
Score = 28.1 bits (61), Expect = 2.5
Identities = 21/62 (33%), Positives = 24/62 (37%), Gaps = 4/62 (6%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P + +PPS+ S T L P S PSPP PS P PS P
Sbjct: 61 PTTVPPIPPSTPSPPPPLTPSPLPP----SPTTPSPPLTPSPTTPSPPLTPSPPPAITPS 116
Query: 71 PP 72
PP
Sbjct: 117 PP 118
Score = 28.1 bits (61), Expect = 2.5
Identities = 25/67 (37%), Positives = 28/67 (41%), Gaps = 10/67 (14%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQP-----SPPPPSKSHFPSLPRFPSKCR 65
PL SS PP S ++ S T P S P P SPPPP+ PS R PS
Sbjct: 154 PLRPSSPPPPSPATPS--TPPRSPPPPSTPTPPPRVGSLSPPPPAS---PSGGRSPSTPS 208
Query: 66 HKPPKPP 72
P P
Sbjct: 209 TTPGSSP 215
>At1g31810 hypothetical protein
Length = 1201
Score = 37.4 bits (85), Expect = 0.004
Identities = 23/63 (36%), Positives = 29/63 (45%), Gaps = 1/63 (1%)
Query: 11 PLSSSSLPPS-SSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPP 69
PL S S+PP + P SS S+P PS PPP PS +K + +PP
Sbjct: 581 PLPSRSIPPPLAQPPPPRPPPPPPPPPSSRSIPSPSAPPPPPPPPPSFGSTGNKRQAQPP 640
Query: 70 KPP 72
PP
Sbjct: 641 PPP 643
Score = 35.8 bits (81), Expect = 0.012
Identities = 26/71 (36%), Positives = 34/71 (47%), Gaps = 7/71 (9%)
Query: 2 ATLSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP 61
+T S+S P LP S S+ T+LH P + P P PPPP S S+P P
Sbjct: 537 STTSFSPSQPPPPPPLP--SFSNRDPLTTLH-QPINKTPPPPPPPPPPLPSR--SIP--P 589
Query: 62 SKCRHKPPKPP 72
+ PP+PP
Sbjct: 590 PLAQPPPPRPP 600
Score = 33.9 bits (76), Expect = 0.046
Identities = 25/73 (34%), Positives = 29/73 (39%), Gaps = 7/73 (9%)
Query: 10 HPLSSSSLPPSSSSSSSSSTSLHLSP-------CSSHSLPQPSPPPPSKSHFPSLPRFPS 62
+PL+ S PPSS + P +S S QP PPPP F S F
Sbjct: 463 NPLNLPSDPPSSGDHVTLLPPPPPPPPPPLFTSTTSFSPSQPPPPPPPPPLFMSTTSFSP 522
Query: 63 KCRHKPPKPPRLF 75
PP PP LF
Sbjct: 523 SQPPPPPPPPPLF 535
Score = 33.1 bits (74), Expect = 0.078
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 4/66 (6%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPR----FPSKCRH 66
P + + PP +S S S+ + P S+ P P PP + S+ P P PS R
Sbjct: 652 PAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISNAPKPPAPPPLPPSSTRL 711
Query: 67 KPPKPP 72
P PP
Sbjct: 712 GAPPPP 717
Score = 32.7 bits (73), Expect = 0.10
Identities = 20/62 (32%), Positives = 25/62 (40%), Gaps = 10/62 (16%)
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P S+ PP S ST + P P PPPP + P+ +KC PP
Sbjct: 613 PSPSAPPPPPPPPPSFGSTGN-----KRQAQPPPPPPPPPPTRIPA-----AKCAPPPPP 662
Query: 71 PP 72
PP
Sbjct: 663 PP 664
Score = 32.0 bits (71), Expect = 0.17
Identities = 20/59 (33%), Positives = 26/59 (43%), Gaps = 2/59 (3%)
Query: 18 PPSSSSSSSSSTSLH--LSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRL 74
PP ++ S++ L P S+ P PPPP S P+ P P PP PP L
Sbjct: 687 PPKANISNAPKPPAPPPLPPSSTRLGAPPPPPPPPLSKTPAPPPPPLSKTPVPPPPPGL 745
Score = 29.3 bits (64), Expect = 1.1
Identities = 23/75 (30%), Positives = 30/75 (39%), Gaps = 6/75 (8%)
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
S++S PS L +S S P PPPP + PS +PP PP
Sbjct: 495 STTSFSPSQPPPPPPPPPLFMSTTSFSPSQPPPPPPPPPLFTSTTSFSPS----QPPPPP 550
Query: 73 RL--FMIIDPILLLN 85
L F DP+ L+
Sbjct: 551 PLPSFSNRDPLTTLH 565
Score = 27.7 bits (60), Expect = 3.3
Identities = 21/65 (32%), Positives = 26/65 (39%), Gaps = 2/65 (3%)
Query: 12 LSSSSLPPSSSSSSSSSTSLHLSPCSSHS--LPQPSPPPPSKSHFPSLPRFPSKCRHKPP 69
+S++ PP+ SST L P P+PPPP S P P P R
Sbjct: 692 ISNAPKPPAPPPLPPSSTRLGAPPPPPPPPLSKTPAPPPPPLSKTPVPPPPPGLGRGTSS 751
Query: 70 KPPRL 74
PP L
Sbjct: 752 GPPPL 756
>At3g18810 protein kinase, putative
Length = 700
Score = 37.0 bits (84), Expect = 0.005
Identities = 22/57 (38%), Positives = 24/57 (41%), Gaps = 2/57 (3%)
Query: 15 SSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKP 71
S PPS SSS + S P S S P P PPP P+ PS PP P
Sbjct: 13 SPTPPSPSSSDNQQQSS--PPPSDSSSPSPPAPPPPDDSSNGSPQPPSSDSQSPPSP 67
Score = 29.6 bits (65), Expect = 0.86
Identities = 17/40 (42%), Positives = 20/40 (49%), Gaps = 1/40 (2%)
Query: 34 SPCSSHSLPQPSPPPP-SKSHFPSLPRFPSKCRHKPPKPP 72
SP SS + Q SPPP S S P P P + P+PP
Sbjct: 18 SPSSSDNQQQSSPPPSDSSSPSPPAPPPPDDSSNGSPQPP 57
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,041,440
Number of Sequences: 26719
Number of extensions: 271090
Number of successful extensions: 6169
Number of sequences better than 10.0: 453
Number of HSP's better than 10.0 without gapping: 248
Number of HSP's successfully gapped in prelim test: 218
Number of HSP's that attempted gapping in prelim test: 2948
Number of HSP's gapped (non-prelim): 2008
length of query: 175
length of database: 11,318,596
effective HSP length: 93
effective length of query: 82
effective length of database: 8,833,729
effective search space: 724365778
effective search space used: 724365778
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0546.10


