

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.5
(471 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
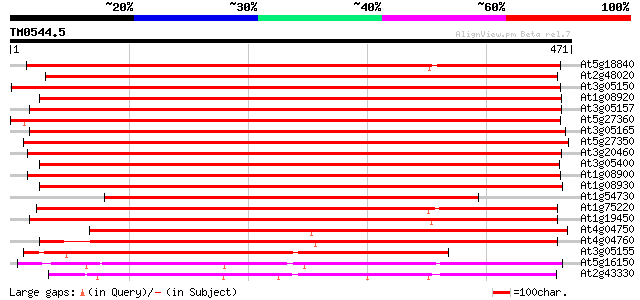
Score E
Sequences producing significant alignments: (bits) Value
At5g18840 sugar transporter - like protein 509 e-145
At2g48020 sugar transporter like protein 502 e-142
At3g05150 putative sugar transporter 465 e-131
At1g08920 putative sugar transport protein 465 e-131
At3g05157 sugar transporter like protein 464 e-131
At5g27360 sugar-porter family protein 2 (SFP2) 463 e-131
At3g05165 sugar transporter like protein 462 e-130
At5g27350 sugar transporter like protein 451 e-127
At3g20460 sugar transporter, putative 451 e-127
At3g05400 sugar transporter like protein 443 e-124
At1g08900 Beta integral like membrane protein 441 e-124
At1g08930 ERD6 protein 441 e-124
At1g54730 Unknown protein 419 e-117
At1g75220 integral membrane protein, putative 407 e-114
At1g19450 integral membrane protein, putative 407 e-114
At4g04750 putative sugar transporter 388 e-108
At4g04760 putative sugar transporter 385 e-107
At3g05155 putative protein 310 1e-84
At5g16150 sugar transporter like protein 234 6e-62
At2g43330 membrane transporter like protein 213 2e-55
>At5g18840 sugar transporter - like protein
Length = 482
Score = 509 bits (1312), Expect = e-145
Identities = 258/452 (57%), Positives = 333/452 (73%), Gaps = 7/452 (1%)
Query: 15 NDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSI 74
+DE++ + + ST VAV GS+ FG+ +GYS+PTQS+I DLNL++A+FS+FGSI
Sbjct: 30 DDEKESENNESYLMVLFSTFVAVCGSFEFGSCVGYSAPTQSSIRQDLNLSLAEFSMFGSI 89
Query: 75 LTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIG 134
LTIGAM+GA++SGK++D++GR+ AM S FCI GWL + F+K A L VGR G GIG
Sbjct: 90 LTIGAMLGAVMSGKISDFSGRKGAMRTSACFCITGWLAVFFTKGALLLDVGRFFTGYGIG 149
Query: 135 LLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQ 194
+ SYVVPVY+AEI+PKNLRGG TT++QLMI G S+++LIG+ ++W+ LAL G PC
Sbjct: 150 VFSYVVPVYIAEISPKNLRGGLTTLNQLMIVIGSSVSFLIGSLISWKTLALTGLAPCIVL 209
Query: 195 LLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASII 254
L L FIP+SPRWLAK GH KE ALQ+LRGK+AD+ EA I+ + L+ +A I
Sbjct: 210 LFGLCFIPESPRWLAKAGHEKEFRVALQKLRGKDADITNEADGIQVSIQALEILPKARIQ 269
Query: 255 GLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGF-SESIGTIAMVAVKIPVT 313
L KY S+ IGV LM+ QQF GINGI FYA+ FV AGF S +GTIA+ V++P+T
Sbjct: 270 DLVSKKYGRSVIIGVSLMVFQQFVGINGIGFYASETFVKAGFTSGKLGTIAIACVQVPIT 329
Query: 314 VLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQG---LHEWKGVSPILALVGVLVY 370
VLG +L+DKSGRRPL+++SA G LGC L +FLL+G L EW P LA+ GVL+Y
Sbjct: 330 VLGTILIDKSGRRPLIMISAGGIFLGCILTGTSFLLKGQSLLLEW---VPSLAVGGVLIY 386
Query: 371 VGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFF 430
V ++S+GMG +PWVIMSEIFPINVKG AGSLV NW +W VSY FNFLMSWSS GTF+
Sbjct: 387 VAAFSIGMGPVPWVIMSEIFPINVKGIAGSLVVLVNWSGAWAVSYTFNFLMSWSSPGTFY 446
Query: 431 IFSSICGFTVLFVAKLVPETKGRTLEEIQASL 462
++S+ T++FVAK+VPETKG+TLEEIQA +
Sbjct: 447 LYSAFAAATIIFVAKMVPETKGKTLEEIQACI 478
>At2g48020 sugar transporter like protein
Length = 463
Score = 502 bits (1293), Expect = e-142
Identities = 240/430 (55%), Positives = 320/430 (73%)
Query: 31 LSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVA 90
LST VAV GS+AFG+ GYSSP Q+AI DL+LT+A+FS+FGS+LT GAMIGAI SG +A
Sbjct: 30 LSTFVAVCGSFAFGSCAGYSSPAQAAIRNDLSLTIAEFSLFGSLLTFGAMIGAITSGPIA 89
Query: 91 DYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPK 150
D GR+ AM S FC++GWL I F+K L +GRL G G+G SYVVP+++AEI PK
Sbjct: 90 DLVGRKGAMRVSSAFCVVGWLAIIFAKGVVALDLGRLATGYGMGAFSYVVPIFIAEIAPK 149
Query: 151 NLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSPRWLAK 210
RG TT++Q++IC G+S++++IG V WR+LALIG IPC A L L FIP+SPRWLAK
Sbjct: 150 TFRGALTTLNQILICTGVSVSFIIGTLVTWRVLALIGIIPCAASFLGLFFIPESPRWLAK 209
Query: 211 VGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVG 270
VG E ++AL++LRGK AD+ +EAAEI+DY ETL+R +A ++ LFQ +Y+ S+ I G
Sbjct: 210 VGRDTEFEAALRKLRGKKADISEEAAEIQDYIETLERLPKAKMLDLFQRRYIRSVLIAFG 269
Query: 271 LMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLL 330
LM+ QQFGGINGI FY +SIF AGF +G I +++ +T L ++D++GR+PLLL
Sbjct: 270 LMVFQQFGGINGICFYTSSIFEQAGFPTRLGMIIYAVLQVVITALNAPIVDRAGRKPLLL 329
Query: 331 LSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIF 390
+SA G +GC +AA++F L+ P+LA+VG++VY+GS+S GMGA+PWV+MSEIF
Sbjct: 330 VSATGLVIGCLIAAVSFYLKVHDMAHEAVPVLAVVGIMVYIGSFSAGMGAMPWVVMSEIF 389
Query: 391 PINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPET 450
PIN+KG AG + T NW +W VSY FNFLMSWSS GTF I+++I ++FV +VPET
Sbjct: 390 PINIKGVAGGMATLVNWFGAWAVSYTFNFLMSWSSYGTFLIYAAINALAIVFVIAIVPET 449
Query: 451 KGRTLEEIQA 460
KG+TLE+IQA
Sbjct: 450 KGKTLEQIQA 459
>At3g05150 putative sugar transporter
Length = 463
Score = 465 bits (1197), Expect = e-131
Identities = 234/462 (50%), Positives = 317/462 (67%), Gaps = 1/462 (0%)
Query: 2 MVKTSEISIPLVFNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDL 61
M K ++ S PL+ + + + LST++AV GSY FG +GYS+PTQ IM +L
Sbjct: 1 MEKRNDKSEPLLLPENGSDVSEEASWMVYLSTIIAVCGSYEFGTCVGYSAPTQFGIMEEL 60
Query: 62 NLTVAQFSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWW 121
NL+ +QFS+FGSIL +GA++GAI SGK++D+ GR+ AM S + +GWL+I +K
Sbjct: 61 NLSYSQFSVFGSILNMGAVLGAITSGKISDFIGRKGAMRLSSVISAIGWLIIYLAKGDVP 120
Query: 122 LYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWR 181
L GR L G G G LS+VVPV++AEI+P+ LRG T++QL I G++ +LIGA VNWR
Sbjct: 121 LDFGRFLTGYGCGTLSFVVPVFIAEISPRKLRGALATLNQLFIVIGLASMFLIGAVVNWR 180
Query: 182 ILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDY 241
LAL G PC FIP+SPRWL VG + + ALQ+LRG A++ +EA EI++Y
Sbjct: 181 TLALTGVAPCVVLFFGTWFIPESPRWLEMVGRHSDFEIALQKLRGPQANITREAGEIQEY 240
Query: 242 TETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIG 301
+L +A+++ L K + + +GVGLM QQF GING++FYA IFVSAG S ++G
Sbjct: 241 LASLAHLPKATLMDLIDKKNIRFVIVGVGLMFFQQFVGINGVIFYAQQIFVSAGASPTLG 300
Query: 302 TIAMVAVKIPVTVLG-VLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSP 360
+I ++ +T LG LL+D+ GRRPLL+ SA G +GC L +FLL+ + P
Sbjct: 301 SILYSIEQVVLTALGATLLIDRLGRRPLLMASAVGMLIGCLLIGNSFLLKAHGLALDIIP 360
Query: 361 ILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFL 420
LA+ GVLVY+GS+S+GMGAIPWVIMSEIFPIN+KG+AG LVT NWL SW+VS+ FNFL
Sbjct: 361 ALAVSGVLVYIGSFSIGMGAIPWVIMSEIFPINLKGTAGGLVTVVNWLSSWLVSFTFNFL 420
Query: 421 MSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASL 462
M WS GTF+++ +C ++F+AKLVPETKGRTLEEIQA +
Sbjct: 421 MIWSPHGTFYVYGGVCVLAIIFIAKLVPETKGRTLEEIQAMM 462
>At1g08920 putative sugar transport protein
Length = 470
Score = 465 bits (1197), Expect = e-131
Identities = 223/438 (50%), Positives = 312/438 (70%)
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIV 85
TA + ST V+V GS+ FG A GYSS Q+ I+ DL L+VAQ+S+FGSI+T G MIGAI
Sbjct: 29 TAVVLFSTFVSVCGSFCFGCAAGYSSVAQTGIINDLGLSVAQYSMFGSIMTFGGMIGAIF 88
Query: 86 SGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVA 145
SGKVAD GR+ M F+Q+FCI GW+ + +K + WL +GRL G +GLLSYV+PVY+A
Sbjct: 89 SGKVADLMGRKGTMWFAQIFCIFGWVAVALAKDSMWLDIGRLSTGFAVGLLSYVIPVYIA 148
Query: 146 EITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSP 205
EITPK++RG F +QLM CG+SL Y+IG FV+WR LALIG IPC Q+++L FIP+SP
Sbjct: 149 EITPKHVRGAFVFANQLMQSCGLSLFYVIGNFVHWRNLALIGLIPCALQVVTLFFIPESP 208
Query: 206 RWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSL 265
R L K GH KE ++LQ LRG +AD+ +EA I++ ++ ++ LFQ +Y S+
Sbjct: 209 RLLGKWGHEKECRASLQSLRGDDADISEEANTIKETMILFDEGPKSRVMDLFQRRYAPSV 268
Query: 266 TIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGR 325
IGVGLM+LQQ G +G+++Y S+F GF SIG++ + + IP +LG++L++K GR
Sbjct: 269 VIGVGLMLLQQLSGSSGLMYYVGSVFDKGGFPSSIGSMILAVIMIPKALLGLILVEKMGR 328
Query: 326 RPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVI 385
RPLLL S G C L + +F + ++PI +GV+ ++ S+++GMG +PW+I
Sbjct: 329 RPLLLASTGGMCFFSLLLSFSFCFRSYGMLDELTPIFTCIGVVGFISSFAVGMGGLPWII 388
Query: 386 MSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAK 445
MSEIFP+NVK SAG+LVT NW WIV++A+NF++ W+++GTF IF +ICG ++F+
Sbjct: 389 MSEIFPMNVKVSAGTLVTLANWSFGWIVAFAYNFMLEWNASGTFLIFFTICGAGIVFIYA 448
Query: 446 LVPETKGRTLEEIQASLS 463
+VPETKGRTLE+IQASL+
Sbjct: 449 MVPETKGRTLEDIQASLT 466
>At3g05157 sugar transporter like protein
Length = 458
Score = 464 bits (1194), Expect = e-131
Identities = 219/447 (48%), Positives = 317/447 (69%)
Query: 17 EEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILT 76
E + R TA + LST VAV S+++G A GY+S ++AIM +L+L++AQFS FGS L
Sbjct: 9 ENDRDDRRITACVILSTFVAVCSSFSYGCANGYTSGAETAIMKELDLSMAQFSAFGSFLN 68
Query: 77 IGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLL 136
+G +GA+ SG++A GRR + LFCI GWL I F+K WL +GR+ +G G+GL
Sbjct: 69 LGGAVGALFSGQLAVILGRRRTLWACDLFCIFGWLSIAFAKNVLWLDLGRISLGIGVGLT 128
Query: 137 SYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLL 196
SYVVPVY+AEITPK++RG F+ L+ G+SL Y G +NWR+LA+IGA+PCF ++
Sbjct: 129 SYVVPVYIAEITPKHVRGAFSASTLLLQNSGISLIYFFGTVINWRVLAVIGALPCFIPVI 188
Query: 197 SLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGL 256
+ FIP+SPRWLAK+G +KE +++L RLRGK+ADV EAAEI+ T+ L+ +++S +
Sbjct: 189 GIYFIPESPRWLAKIGSVKEVENSLHRLRGKDADVSDEAAEIQVMTKMLEEDSKSSFCDM 248
Query: 257 FQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLG 316
FQ KY +L +G+GLM++QQ G +GI +Y+N+IF AGFSE +G++ IP ++G
Sbjct: 249 FQKKYRRTLVVGIGLMLIQQLSGASGITYYSNAIFRKAGFSERLGSMIFGVFVIPKALVG 308
Query: 317 VLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSL 376
++L+D+ GRRPLLL SA G +G L ++F LQ ++ + P+ + +LVY G +++
Sbjct: 309 LILVDRWGRRPLLLASAVGMSIGSLLIGVSFTLQEMNLFPEFIPVFVFINILVYFGFFAI 368
Query: 377 GMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSIC 436
G+G +PW+IMSEIFPIN+K SAGS+V +W W VSY FNF+ WS+ GTF+IF+ +
Sbjct: 369 GIGGLPWIIMSEIFPINIKVSAGSIVALTSWTTGWFVSYGFNFMFEWSAQGTFYIFAMVG 428
Query: 437 GFTVLFVAKLVPETKGRTLEEIQASLS 463
G ++LF+ LVPETKG++LEE+QASL+
Sbjct: 429 GLSLLFIWMLVPETKGQSLEELQASLT 455
>At5g27360 sugar-porter family protein 2 (SFP2)
Length = 478
Score = 463 bits (1192), Expect = e-131
Identities = 228/465 (49%), Positives = 327/465 (70%), Gaps = 3/465 (0%)
Query: 1 MMVKTSEISIP---LVFNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAI 57
MMV E SI L ++ + TA + LST +AV GS++FG ++GY+S + I
Sbjct: 4 MMVVEKERSIEERLLQLKNQNDDSECRITACVILSTFIAVCGSFSFGVSLGYTSGAEIGI 63
Query: 58 MPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSK 117
M DL+L++AQFS F S+ T+GA IGA+ SGK+A GRR M S L CI+GW I F+K
Sbjct: 64 MKDLDLSIAQFSAFASLSTLGAAIGALFSGKMAIILGRRKTMWVSDLLCIIGWFSIAFAK 123
Query: 118 VAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAF 177
WL GR+ G G+GL+SYVVPVY+AEI+PK++RG FT +QL+ G+++ Y G F
Sbjct: 124 DVMWLNFGRISSGIGLGLISYVVPVYIAEISPKHVRGTFTFTNQLLQNSGLAMVYFSGNF 183
Query: 178 VNWRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAE 237
+NWRILAL+GA+PCF Q++ L F+P+SPRWLAKVG KE +++L RLRG NAD+ +EA++
Sbjct: 184 LNWRILALLGALPCFIQVIGLFFVPESPRWLAKVGSDKELENSLLRLRGGNADISREASD 243
Query: 238 IRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFS 297
I T+ ++ +++S LFQ KY +L +G+GLM++QQF G + ++ YA++I AGFS
Sbjct: 244 IEVMTKMVENDSKSSFCDLFQRKYRYTLVVGIGLMLIQQFSGSSAVLSYASTILRKAGFS 303
Query: 298 ESIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKG 357
+IG+ + IP ++GV+L+DK GRRPLLL S +G C+ L +AF LQ +
Sbjct: 304 VTIGSTLLGLFMIPKAMIGVILVDKWGRRPLLLTSVSGMCITSMLIGVAFTLQKMQLLPE 363
Query: 358 VSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAF 417
++P+ + V +Y+G+Y++G+G +PWVIMSEIFP+N+K +AGS+VT +W S IV+YAF
Sbjct: 364 LTPVFTFICVTLYIGTYAIGLGGLPWVIMSEIFPMNIKVTAGSIVTLVSWSSSSIVTYAF 423
Query: 418 NFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASL 462
NFL+ WS+ GTF++F ++ G +LF+ LVPETKG +LEEIQASL
Sbjct: 424 NFLLEWSTQGTFYVFGAVGGLALLFIWLLVPETKGLSLEEIQASL 468
>At3g05165 sugar transporter like protein
Length = 467
Score = 462 bits (1189), Expect = e-130
Identities = 219/450 (48%), Positives = 317/450 (69%)
Query: 17 EEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILT 76
+ + R TA + LST VAV ++++G A GY+S ++AIM +L+L++AQFS FGS L
Sbjct: 18 QNDRDDRRITACVILSTFVAVCSAFSYGCAAGYTSGAETAIMKELDLSMAQFSAFGSFLN 77
Query: 77 IGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLL 136
+G +GA+ SG++A GRR + FC+ GWL I F+K +WL +GR+ +G G+GL+
Sbjct: 78 VGGAVGALFSGQLAVILGRRRTLWACDFFCVFGWLSIAFAKNVFWLDLGRISLGIGVGLI 137
Query: 137 SYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLL 196
SYVVPVY+AEITPK++RG FT +QL+ G+SL Y G +NWR++A+IGAIPC Q +
Sbjct: 138 SYVVPVYIAEITPKHVRGAFTASNQLLQNSGVSLIYFFGTVINWRVMAVIGAIPCILQTI 197
Query: 197 SLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGL 256
+ FIP+SPRWLAK+ KE +S+L RLRGK+ DV EAAEI+ T+ L+ +++S +
Sbjct: 198 GIFFIPESPRWLAKIRLSKEVESSLHRLRGKDTDVSGEAAEIQVMTKMLEEDSKSSFSDM 257
Query: 257 FQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLG 316
FQ KY +L +G+GLM++QQ G +GI +Y+N+IF AGFSE +G++ IP ++G
Sbjct: 258 FQKKYRRTLVVGIGLMLIQQLSGASGITYYSNAIFRKAGFSERLGSMIFGVFVIPKALVG 317
Query: 317 VLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSL 376
++L+D+ GRRPLLL SA G +G L ++F LQ ++ + PI V +LVY G ++
Sbjct: 318 LILVDRWGRRPLLLASAVGMSIGSLLIGVSFTLQQMNVLPELIPIFVFVNILVYFGCFAF 377
Query: 377 GMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSIC 436
G+G +PWVIMSEIFPIN+K SAG++V +W W VSYAFNF+ WS+ GTF+IF+++
Sbjct: 378 GIGGLPWVIMSEIFPINIKVSAGTIVALTSWTSGWFVSYAFNFMFEWSAQGTFYIFAAVG 437
Query: 437 GFTVLFVAKLVPETKGRTLEEIQASLSSHS 466
G + +F+ LVPETKG++LEE+QASL+ S
Sbjct: 438 GMSFIFIWMLVPETKGQSLEELQASLTGTS 467
>At5g27350 sugar transporter like protein
Length = 474
Score = 451 bits (1160), Expect = e-127
Identities = 225/458 (49%), Positives = 318/458 (69%)
Query: 12 LVFNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIF 71
L ++ + TA + LST VAV GS++FG A GY+S ++ +M DL+L++AQFS F
Sbjct: 14 LQLKNKNDDSECRITACVILSTFVAVCGSFSFGVATGYTSGAETGVMKDLDLSIAQFSAF 73
Query: 72 GSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGC 131
GS T+GA IGA+ G +A GRR M S CI GWL I F+K L GR++ G
Sbjct: 74 GSFATLGAAIGALFCGNLAMVIGRRGTMWVSDFLCITGWLSIAFAKEVVLLNFGRIISGI 133
Query: 132 GIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPC 191
G GL SYVVPVY+AEITPK++RG FT +QL+ G+++ Y G F+ WR LAL+GA+PC
Sbjct: 134 GFGLTSYVVPVYIAEITPKHVRGTFTFSNQLLQNAGLAMIYFCGNFITWRTLALLGALPC 193
Query: 192 FAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEA 251
F Q++ L F+P+SPRWLAKVG KE +++L RLRG++AD+ +EA+EI+ T+ ++ +++
Sbjct: 194 FIQVIGLFFVPESPRWLAKVGSDKELENSLFRLRGRDADISREASEIQVMTKMVENDSKS 253
Query: 252 SIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIP 311
S LFQ KY +L +G+GLM++QQF G ++ YA++IF AGFS +IGT + IP
Sbjct: 254 SFSDLFQRKYRYTLVVGIGLMLIQQFSGSAAVISYASTIFRKAGFSVAIGTTMLGIFVIP 313
Query: 312 VTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYV 371
++G++L+DK GRRPLL+ SA G + C L +AF LQ + ++PIL+ + V++Y+
Sbjct: 314 KAMIGLILVDKWGRRPLLMTSAFGMSMTCMLLGVAFTLQKMQLLSELTPILSFICVMMYI 373
Query: 372 GSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFI 431
+Y++G+G +PWVIMSEIFPIN+K +AGS+VT ++ S IV+YAFNFL WS+ GTFFI
Sbjct: 374 ATYAIGLGGLPWVIMSEIFPINIKVTAGSIVTLVSFSSSSIVTYAFNFLFEWSTQGTFFI 433
Query: 432 FSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTKR 469
F+ I G +LF+ LVPETKG +LEEIQ SL +R
Sbjct: 434 FAGIGGAALLFIWLLVPETKGLSLEEIQVSLIHQPDER 471
>At3g20460 sugar transporter, putative
Length = 488
Score = 451 bits (1160), Expect = e-127
Identities = 208/448 (46%), Positives = 313/448 (69%)
Query: 16 DEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSIL 75
+ + ++ P T L +T A+ G++++G A G++SP Q+ IM LNL++A+FS FG++L
Sbjct: 40 NNDGEEDGPVTLILLFTTFTALCGTFSYGTAAGFTSPAQTGIMAGLNLSLAEFSFFGAVL 99
Query: 76 TIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGL 135
TIG ++GA +SGK+AD GRR A+G S FC+ GWL+I FS+ W L +GRL +G G+
Sbjct: 100 TIGGLVGAAMSGKLADVFGRRGALGVSNSFCMAGWLMIAFSQATWSLDIGRLFLGVAAGV 159
Query: 136 LSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQL 195
SYVVPVY+ EI PK +RG F+ ++ L++C +++TYL+G+ ++W+ LALI +PC +
Sbjct: 160 ASYVVPVYIVEIAPKKVRGTFSAINSLVMCASVAVTYLLGSVISWQKLALISTVPCVFEF 219
Query: 196 LSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIG 255
+ L FIP+SPRWL++ G +KES+ +LQRLRG N D+ +EAAEI+ Y + LQ E
Sbjct: 220 VGLFFIPESPRWLSRNGRVKESEVSLQRLRGNNTDITKEAAEIKKYMDNLQEFKEDGFFD 279
Query: 256 LFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVL 315
LF +Y +T+G+GL++LQQ GG++G FY +SIF +GF ++G + V+ +VL
Sbjct: 280 LFNPRYSRVVTVGIGLLVLQQLGGLSGYTFYLSSIFKKSGFPNNVGVMMASVVQSVTSVL 339
Query: 316 GVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYS 375
G++++DK GRR LL ++ CLG + L+FL Q + +PI +GVLV++ S +
Sbjct: 340 GIVIVDKYGRRSLLTVATIMMCLGSLITGLSFLFQSYGLLEHYTPISTFMGVLVFLTSIT 399
Query: 376 LGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSI 435
+G+G IPWV++SE+ PIN+KGSAG+L +W +W VSY FNFL WSS+G FFI++ I
Sbjct: 400 IGIGGIPWVMISEMTPINIKGSAGTLCNLTSWSSNWFVSYTFNFLFQWSSSGVFFIYTMI 459
Query: 436 CGFTVLFVAKLVPETKGRTLEEIQASLS 463
G +LFV K+VPET+GR+LEEIQA+++
Sbjct: 460 SGVGILFVMKMVPETRGRSLEEIQAAIT 487
>At3g05400 sugar transporter like protein
Length = 462
Score = 443 bits (1139), Expect = e-124
Identities = 215/436 (49%), Positives = 303/436 (69%)
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIV 85
T L ST + V S+ FGAAIGY++ T S+IM DL+L++AQFS+FGS+ T G MIGAI
Sbjct: 23 TPLLIFSTFIIVSASFTFGAAIGYTADTMSSIMSDLDLSLAQFSLFGSLSTFGGMIGAIF 82
Query: 86 SGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVA 145
S K A G ++ + + LFCI GWL I+ +K WL +GR LVG G+GL+SYVVPVY+A
Sbjct: 83 SAKAASAFGHKMTLWVADLFCITGWLAISLAKDIIWLDMGRFLVGIGVGLISYVVPVYIA 142
Query: 146 EITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSP 205
EITPK++RG FT +QL+ CG+++ Y G F++WR LA+IG+IPC+ Q++ L FIP+SP
Sbjct: 143 EITPKHVRGAFTFSNQLLQNCGVAVVYYFGNFLSWRTLAIIGSIPCWIQVIGLFFIPESP 202
Query: 206 RWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSL 265
RWLAK G KE + LQ+LRG+ D+ EA EI+ E ++ + +I LF+ +Y L
Sbjct: 203 RWLAKKGRDKECEEVLQKLRGRKYDIVPEACEIKISVEASKKNSNINIRSLFEKRYAHQL 262
Query: 266 TIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGR 325
TIG+GLM+LQQ G GI Y +++F AGF IG + + + +P +++G++L+D+ GR
Sbjct: 263 TIGIGLMLLQQLCGTAGISSYGSTLFKLAGFPARIGMMVLSLIVVPKSLMGLILVDRWGR 322
Query: 326 RPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVI 385
RPLL+ SA G CL C A+AF ++ + ++PI +G+L + +++GMGA+PW+I
Sbjct: 323 RPLLMTSALGLCLSCITLAVAFGVKDVPGIGKITPIFCFIGILSFTMMFAIGMGALPWII 382
Query: 386 MSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAK 445
MSEIFP+++K AGSLVT NW WI +YAFNF++ WS +GTF I + ICG T++F
Sbjct: 383 MSEIFPMDIKVLAGSLVTIANWFTGWIANYAFNFMLVWSPSGTFIISAIICGATIVFTWC 442
Query: 446 LVPETKGRTLEEIQAS 461
LVPET+ TLEEIQ S
Sbjct: 443 LVPETRRLTLEEIQLS 458
>At1g08900 Beta integral like membrane protein
Length = 462
Score = 441 bits (1134), Expect = e-124
Identities = 217/447 (48%), Positives = 311/447 (69%)
Query: 16 DEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSIL 75
++++++ FT+ L LST V V GS+ +G A+ YSSP QS IM +L L+VA +S F S++
Sbjct: 12 NKQEEEASSFTSGLLLSTSVVVAGSFCYGCAMSYSSPAQSKIMEELGLSVADYSFFTSVM 71
Query: 76 TIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGL 135
T+G MI A+ SGK++ GRR M S + CI GWL + F+ L GRL +G G+GL
Sbjct: 72 TLGGMITAVFSGKISALVGRRQTMWISDVCCIFGWLAVAFAHDIIMLNTGRLFLGFGVGL 131
Query: 136 LSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQL 195
+SYVVPVY+AEITPK RGGF+ +QL+ C G+SL + G F +WR LAL+ AIP Q+
Sbjct: 132 ISYVVPVYIAEITPKTFRGGFSYSNQLLQCLGISLMFFTGNFFHWRTLALLSAIPSAFQV 191
Query: 196 LSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIG 255
+ L FIP+SPRWLA G +E + +L++LRG+N+D+ +EAAEIR+ E ++++++ I
Sbjct: 192 ICLFFIPESPRWLAMYGQDQELEVSLKKLRGENSDILKEAAEIRETVEISRKESQSGIRD 251
Query: 256 LFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVL 315
LF SL IG+GLM+LQQF G I YA IF AGF IGT + + IP +++
Sbjct: 252 LFHIGNAHSLIIGLGLMLLQQFCGSAAISAYAARIFDKAGFPSDIGTTILAVILIPQSIV 311
Query: 316 GVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYS 375
+L +D+ GRRPLL++S+ G C+ F L++ LQ E++ + ++ +VG++ YV S+
Sbjct: 312 VMLTVDRWGRRPLLMISSIGMCICSFFIGLSYYLQKNGEFQKLCSVMLIVGLVGYVSSFG 371
Query: 376 LGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSI 435
+G+G +PWVIMSEIFP+NVK +AGSLVT NW +WI+ Y+FNF++ WS++GT+FIFS +
Sbjct: 372 IGLGGLPWVIMSEIFPVNVKITAGSLVTMSNWFFNWIIIYSFNFMIQWSASGTYFIFSGV 431
Query: 436 CGFTVLFVAKLVPETKGRTLEEIQASL 462
T++F+ LVPETKGRTLEEIQ SL
Sbjct: 432 SLVTIVFIWTLVPETKGRTLEEIQTSL 458
>At1g08930 ERD6 protein
Length = 496
Score = 441 bits (1133), Expect = e-124
Identities = 212/439 (48%), Positives = 304/439 (68%)
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIV 85
TA++ LST VAV GS+ G +G+SS Q+ I DL+L+VA++S+FGSILT+G +IGA+
Sbjct: 55 TASVFLSTFVAVSGSFCTGCGVGFSSGAQAGITKDLSLSVAEYSMFGSILTLGGLIGAVF 114
Query: 86 SGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVA 145
SGKVAD GR+ M F + FCI GWL + ++ A WL GRLL+G G+G+ SYV+PVY+A
Sbjct: 115 SGKVADVLGRKRTMLFCEFFCITGWLCVALAQNAMWLDCGRLLLGIGVGIFSYVIPVYIA 174
Query: 146 EITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSP 205
EI PK++RG F +QLM CG+SL ++IG F+ WR+L ++G +PC + L FIP+SP
Sbjct: 175 EIAPKHVRGSFVFANQLMQNCGISLFFIIGNFIPWRLLTVVGLVPCVFHVFCLFFIPESP 234
Query: 206 RWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSL 265
RWLAK+G KE S+LQRLRG + D+ +EA IRD + + E + LFQ +Y L
Sbjct: 235 RWLAKLGRDKECRSSLQRLRGSDVDISREANTIRDTIDMTENGGETKMSELFQRRYAYPL 294
Query: 266 TIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGR 325
IGVGLM LQQ G +G+ +YA+S+F GF +IGT + + +P +L +L+DK GR
Sbjct: 295 IIGVGLMFLQQLCGSSGVTYYASSLFNKGGFPSAIGTSVIATIMVPKAMLATVLVDKMGR 354
Query: 326 RPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVI 385
R LL+ S + L L ++++ Q ++PI +GVL ++ S+++GMG +PW+I
Sbjct: 355 RTLLMASCSAMGLSALLLSVSYGFQSFGILPELTPIFTCIGVLGHIVSFAMGMGGLPWII 414
Query: 386 MSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAK 445
M+EIFP+NVK SAG+LVT NWL WI++Y FNF++ W+++G F IFS + +++F+
Sbjct: 415 MAEIFPMNVKVSAGTLVTVTNWLFGWIITYTFNFMLEWNASGMFLIFSMVSASSIVFIYF 474
Query: 446 LVPETKGRTLEEIQASLSS 464
LVPETKGR+LEEIQA L++
Sbjct: 475 LVPETKGRSLEEIQALLNN 493
>At1g54730 Unknown protein
Length = 332
Score = 419 bits (1078), Expect = e-117
Identities = 206/314 (65%), Positives = 250/314 (79%)
Query: 80 MIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYV 139
MIGA +SG++AD GRR MGFS++FCILGWL I SKVA WL VGR LVG G+G+ S+V
Sbjct: 1 MIGAAMSGRIADMIGRRATMGFSEMFCILGWLAIYLSKVAIWLDVGRFLVGYGMGVFSFV 60
Query: 140 VPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLP 199
VPVY+AEITPK LRGGFTT+HQL+IC G+S+TYL+G+F+ WRILALIG IPC Q++ L
Sbjct: 61 VPVYIAEITPKGLRGGFTTVHQLLICLGVSVTYLLGSFIGWRILALIGMIPCVVQMMGLF 120
Query: 200 FIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQW 259
IP+SPRWLAKVG +E + ALQRLRG++AD+ E+ EI+DYT L +E SI+ LFQ
Sbjct: 121 VIPESPRWLAKVGKWEEFEIALQRLRGESADISYESNEIKDYTRRLTDLSEGSIVDLFQP 180
Query: 260 KYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLL 319
+Y SL +GVGLM+LQQFGG+NGI FYA+SIF SAG S IG IAMV V+IP+T LGVLL
Sbjct: 181 QYAKSLVVGVGLMVLQQFGGVNGIAFYASSIFESAGVSSKIGMIAMVVVQIPMTTLGVLL 240
Query: 320 MDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMG 379
MDKSGRRPLLL+SA GTC+GCFL L+F LQ + + G + LAL GVLVY GS+SLGMG
Sbjct: 241 MDKSGRRPLLLISATGTCIGCFLVGLSFSLQFVKQLSGDASYLALTGVLVYTGSFSLGMG 300
Query: 380 AIPWVIMSEIFPIN 393
IPWVIMSE+ I+
Sbjct: 301 GIPWVIMSEVSNIH 314
>At1g75220 integral membrane protein, putative
Length = 487
Score = 407 bits (1046), Expect = e-114
Identities = 201/446 (45%), Positives = 299/446 (66%), Gaps = 11/446 (2%)
Query: 23 RPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIG 82
R + ++ L+ LG FG GYSSPTQ+AI DL LTV+++S+FGS+ +GAM+G
Sbjct: 42 RDSSISVLACVLIVALGPIQFGFTCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVG 101
Query: 83 AIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPV 142
AI SG++A+Y GR+ ++ + + I+GWL I+F+K +LY+GRLL G G+G++SY VPV
Sbjct: 102 AIASGQIAEYIGRKGSLMIAAIPNIIGWLCISFAKDTSFLYMGRLLEGFGVGIISYTVPV 161
Query: 143 YVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIP 202
Y+AEI P+N+RGG +++QL + G+ L YL+G FV WRILA++G +PC + L FIP
Sbjct: 162 YIAEIAPQNMRGGLGSVNQLSVTIGIMLAYLLGLFVPWRILAVLGILPCTLLIPGLFFIP 221
Query: 203 DSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYM 262
+SPRWLAK+G E +++LQ LRG D+ E EI+ + ++ + L + +Y
Sbjct: 222 ESPRWLAKMGMTDEFETSLQVLRGFETDITVEVNEIKRSVASSTKRNTVRFVDLKRRRYY 281
Query: 263 TSLTIGVGLMILQQFGGINGIVFYANSIFVSAGF-SESIGTIAMVAVKIPVTVLGVLLMD 321
L +G+GL++LQQ GGING++FY+++IF SAG S + T + A+++ T + L+D
Sbjct: 282 FPLMVGIGLLVLQQLGGINGVLFYSSTIFESAGVTSSNAATFGVGAIQVVATAISTWLVD 341
Query: 322 KSGRRPLLLLSAAGTCLGCFLAALAFLLQ-------GLHEWKGVSPILALVGVLVYVGSY 374
K+GRR LL +S+ G + + A AF L+ ++ W IL++VGV+ V +
Sbjct: 342 KAGRRLLLTISSVGMTISLVIVAAAFYLKEFVSPDSDMYSWLS---ILSVVGVVAMVVFF 398
Query: 375 SLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSS 434
SLGMG IPW+IMSEI P+N+KG AGS+ T NW SW+++ N L++WSS GTF ++
Sbjct: 399 SLGMGPIPWLIMSEILPVNIKGLAGSIATLANWFFSWLITMTANLLLAWSSGGTFTLYGL 458
Query: 435 ICGFTVLFVAKLVPETKGRTLEEIQA 460
+C FTV+FV VPETKG+TLEE+Q+
Sbjct: 459 VCAFTVVFVTLWVPETKGKTLEELQS 484
>At1g19450 integral membrane protein, putative
Length = 488
Score = 407 bits (1046), Expect = e-114
Identities = 202/449 (44%), Positives = 302/449 (66%), Gaps = 5/449 (1%)
Query: 17 EEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILT 76
E Q R + ++ L+ LG FG GYSSPTQ+AI DL LTV+++S+FGS+
Sbjct: 37 ESSQVIRDSSISVLACVLIVALGPIQFGFTCGYSSPTQAAITKDLGLTVSEYSVFGSLSN 96
Query: 77 IGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLL 136
+GAM+GAI SG++A+Y GR+ ++ + + I+GWL I+F+K +LY+GRLL G G+G++
Sbjct: 97 VGAMVGAIASGQIAEYVGRKGSLMIAAIPNIIGWLSISFAKDTSFLYMGRLLEGFGVGII 156
Query: 137 SYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLL 196
SY VPVY+AEI P+ +RG +++QL + G+ L YL+G FV WRILA++G +PC +
Sbjct: 157 SYTVPVYIAEIAPQTMRGALGSVNQLSVTIGIMLAYLLGLFVPWRILAVLGVLPCTLLIP 216
Query: 197 SLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGL 256
L FIP+SPRWLAK+G + +++LQ LRG D+ E EI+ + +++ + L
Sbjct: 217 GLFFIPESPRWLAKMGLTDDFETSLQVLRGFETDITVEVNEIKRSVASSSKRSAVRFVDL 276
Query: 257 FQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGF-SESIGTIAMVAVKIPVTVL 315
+ +Y L +G+GL+ LQQ GGING++FY+++IF SAG S ++ T + V++ T +
Sbjct: 277 KRRRYYFPLMVGIGLLALQQLGGINGVLFYSSTIFESAGVTSSNVATFGVGVVQVVATGI 336
Query: 316 GVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGL----HEWKGVSPILALVGVLVYV 371
L+DK+GRR LL++S+ G + + A+AF L+ + ++++VGV+ V
Sbjct: 337 ATWLVDKAGRRLLLMISSIGMTISLVIVAVAFYLKEFVSPDSNMYNILSMVSVVGVVAMV 396
Query: 372 GSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFI 431
S SLGMG IPW+IMSEI P+N+KG AGS+ T NW SW+V+ N L++WSS GTF +
Sbjct: 397 ISCSLGMGPIPWLIMSEILPVNIKGLAGSIATLLNWFVSWLVTMTANMLLAWSSGGTFTL 456
Query: 432 FSSICGFTVLFVAKLVPETKGRTLEEIQA 460
++ +CGFTV+FV+ VPETKG+TLEEIQA
Sbjct: 457 YALVCGFTVVFVSLWVPETKGKTLEEIQA 485
>At4g04750 putative sugar transporter
Length = 461
Score = 388 bits (996), Expect = e-108
Identities = 195/403 (48%), Positives = 274/403 (67%), Gaps = 2/403 (0%)
Query: 68 FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRL 127
FS FGSILT+G ++GA++ GK+AD GR + + + ++GWL I F+K L +GRL
Sbjct: 54 FSFFGSILTVGLILGALICGKLADLVGRVYTIWITNILVLIGWLAIAFAKDVRLLDLGRL 113
Query: 128 LVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIG 187
L G +G+ SY+ P+Y++E+ P+NLRG +++ QL + G+S Y +G V WR LA++G
Sbjct: 114 LQGISVGISSYLGPIYISELAPRNLRGAASSLMQLFVGVGLSAFYALGTAVAWRSLAILG 173
Query: 188 AIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQR 247
+IP L L FIP+SPRWLAKVG KE + L LRG +DV EAA I +YT+ +++
Sbjct: 174 SIPSLVVLPLLFFIPESPRWLAKVGREKEVEGVLLSLRGAKSDVSDEAATILEYTKHVEQ 233
Query: 248 QTEAS--IIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAM 305
Q S LFQ KY LTIGV L+ + Q GG+NG FY ++IF S G S IG I
Sbjct: 234 QDIDSRGFFKLFQRKYALPLTIGVVLISMPQLGGLNGYTFYTDTIFTSTGVSSDIGFILT 293
Query: 306 VAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALV 365
V++ VLGVLL+D SGRR LLL S AG LGC A++F LQ + W+ +PI+AL+
Sbjct: 294 SIVQMTGGVLGVLLVDISGRRSLLLFSQAGMFLGCLATAISFFLQKNNCWETGTPIMALI 353
Query: 366 GVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSS 425
V+VY GSY LGMG IPW+I SEI+P++VKG+AG++ + SW+V+Y+FNFL+ WSS
Sbjct: 354 SVMVYFGSYGLGMGPIPWIIASEIYPVDVKGAAGTVCNLVTSISSWLVTYSFNFLLQWSS 413
Query: 426 TGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTK 468
TGTF +F+++ G +F AKLVPETKG++LEEIQ++ + +++
Sbjct: 414 TGTFMMFATVMGLGFVFTAKLVPETKGKSLEEIQSAFTDSTSE 456
>At4g04760 putative sugar transporter
Length = 457
Score = 385 bits (990), Expect = e-107
Identities = 198/438 (45%), Positives = 282/438 (64%), Gaps = 24/438 (5%)
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIV 85
T L+ +V G++AFG +S+FGSILT+G ++GA++
Sbjct: 28 TRPFVLAFIVGSCGAFAFGCI---------------------YSLFGSILTVGLILGALI 66
Query: 86 SGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVA 145
GK+ D GR + + + ++GW I F+K W L +GRLL G IG+ Y+ PVY+
Sbjct: 67 CGKLTDLVGRVKTIWITNILFVIGWFAIAFAKGVWLLDLGRLLQGISIGISVYLGPVYIT 126
Query: 146 EITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSP 205
EI P+NLRG ++ QL G+S+ Y +G V WR LA++G IP L L FIP+SP
Sbjct: 127 EIAPRNLRGAASSFAQLFAGVGISVFYALGTIVAWRNLAILGCIPSLMVLPLLFFIPESP 186
Query: 206 RWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIG---LFQWKYM 262
RWLAKVG E ++ L LRG+ +DV EAAEI +YTE +++Q + G LFQ KY
Sbjct: 187 RWLAKVGREMEVEAVLLSLRGEKSDVSDEAAEILEYTEHVKQQQDIDDRGFFKLFQRKYA 246
Query: 263 TSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDK 322
SLTIGV L+ L Q GG+NG FY +SIF+S G S G I+ V++ +LG +L+D
Sbjct: 247 FSLTIGVVLIALPQLGGLNGYSFYTDSIFISTGVSSDFGFISTSVVQMFGGILGTVLVDV 306
Query: 323 SGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIP 382
SGRR LLL+S AG LGC A++F L+ H W+ +P+LAL V+VY GSY GMG+IP
Sbjct: 307 SGRRTLLLVSQAGMFLGCLTTAISFFLKENHCWETGTPVLALFSVMVYFGSYGSGMGSIP 366
Query: 383 WVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLF 442
W+I SEI+P++VKG+AG++ + + +W+V+Y+F++L+ WSSTGTF +F+++ G +F
Sbjct: 367 WIIASEIYPVDVKGAAGTMCNLVSSISAWLVAYSFSYLLQWSSTGTFLMFATVAGLGFVF 426
Query: 443 VAKLVPETKGRTLEEIQA 460
+AKLVPETKG++LEEIQ+
Sbjct: 427 IAKLVPETKGKSLEEIQS 444
>At3g05155 putative protein
Length = 418
Score = 310 bits (793), Expect = 1e-84
Identities = 165/386 (42%), Positives = 236/386 (60%), Gaps = 35/386 (9%)
Query: 12 LVFNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAA------------------------- 46
L+ EE PF L +T + V S++FG A
Sbjct: 13 LLIRKEESANTTPF---LVFTTFIIVSASFSFGVANWVLSRNIPVFKLFRYSGMYLNYFD 69
Query: 47 ---IGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQ 103
+G+++ T ++IM DL+L++ QFS+FGS+LT G MIGA+ S +AD G ++ + ++
Sbjct: 70 DNQLGHTAGTMASIMEDLDLSITQFSVFGSLLTFGGMIGALFSATIADSFGCKMTLWITE 129
Query: 104 LFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLM 163
+FCI GWL I +K WL +GR VG G+GLLSYVVPVY+AEITPK +RG FT +QL+
Sbjct: 130 VFCISGWLAIALAKNIIWLDLGRFFVGIGVGLLSYVVPVYIAEITPKTVRGTFTFSNQLL 189
Query: 164 ICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQR 223
CG++ Y +G F++WRI+ALIG +PC QL+ L F+P+SPRWLAK G +E + LQ+
Sbjct: 190 QNCGVATAYYLGNFMSWRIIALIGILPCLIQLVGLFFVPESPRWLAKEGRDEECEVVLQK 249
Query: 224 LRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGI 283
LRG AD+ +E EI +++ S+ LF+ KY LTIG+GLM+LQQ G G+
Sbjct: 250 LRGDEADIVKETQEI---LISVEASANISMRSLFKKKYTHQLTIGIGLMLLQQLSGSAGL 306
Query: 284 VFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLA 343
+Y S+F AGF IG + V +P +LG++L+++ GRRPLL+ SA G CLGC
Sbjct: 307 GYYTGSVFDLAGFPSRIGMTVLSIVVVPKAILGLILVERWGRRPLLMASAFGLCLGCISL 366
Query: 344 ALAFLLQGLHEWK-GVSPILALVGVL 368
ALAF L+G+ V+P LA +G++
Sbjct: 367 ALAFGLKGVPGVNVNVTPTLAFIGIM 392
>At5g16150 sugar transporter like protein
Length = 546
Score = 234 bits (598), Expect = 6e-62
Identities = 151/470 (32%), Positives = 244/470 (51%), Gaps = 27/470 (5%)
Query: 7 EISIPLVFNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNL--- 63
E +IPL + PF VA LG+ FG +G + + DL +
Sbjct: 90 EEAIPLRSEGKSSGTVLPFVG-------VACLGAILFGYHLGVVNGALEYLAKDLGIAEN 142
Query: 64 TVAQFSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLY 123
TV Q I S+L GA +G+ G +AD GR + +G + ++ +
Sbjct: 143 TVLQGWIVSSLLA-GATVGSFTGGALADKFGRTRTFQLDAIPLAIGAFLCATAQSVQTMI 201
Query: 124 VGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVN---- 179
VGRLL G GIG+ S +VP+Y++EI+P +RG +++QL IC G+ + G +
Sbjct: 202 VGRLLAGIGIGISSAIVPLYISEISPTEIRGALGSVNQLFICIGILAALIAGLPLAANPL 261
Query: 180 -WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEI 238
WR + + IP + + F P+SPRWL + G + E++ A++ L GK V +
Sbjct: 262 WWRTMFGVAVIPSVLLAIGMAFSPESPRWLVQQGKVSEAEKAIKTLYGKERVVEL----V 317
Query: 239 RDYTETLQ--RQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGF 296
RD + + Q + EA LF +Y +++G L + QQ GIN +V+Y+ S+F SAG
Sbjct: 318 RDLSASGQGSSEPEAGWFDLFSSRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGI 377
Query: 297 SESIGTIAMV-AVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEW 355
+ A+V A + T + LMDK GR+ LLL S G L L +L+F + L +
Sbjct: 378 QSDVAASALVGASNVFGTAVASSLMDKMGRKSLLLTSFGGMALSMLLLSLSFTWKALAAY 437
Query: 356 KGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVS- 414
G LA+VG ++YV S+SLG G +P +++ EIF ++ A +L +W+ ++++
Sbjct: 438 SGT---LAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWISNFVIGL 494
Query: 415 YAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSS 464
Y + + + + + F+ +C VL++A V ETKGR+LEEI+ +L+S
Sbjct: 495 YFLSVVTKFGISSVYLGFAGVCVLAVLYIAGNVVETKGRSLEEIELALTS 544
>At2g43330 membrane transporter like protein
Length = 509
Score = 213 bits (542), Expect = 2e-55
Identities = 146/453 (32%), Positives = 226/453 (49%), Gaps = 37/453 (8%)
Query: 33 TLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFG----SILTIGAMIGAIVSGK 88
T+ A +G FG G S I D + V Q S S+ +GAMIGA G
Sbjct: 35 TVTAGIGGLLFGYDTGVISGALLYIKDDFEV-VKQSSFLQETIVSMALVGAMIGAAAGGW 93
Query: 89 VADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEIT 148
+ DY GR+ A F+ + G +V+ + + L GRLLVG G+G+ S PVY+AE +
Sbjct: 94 INDYYGRKKATLFADVVFAAGAIVMAAAPDPYVLISGRLLVGLGVGVASVTAPVYIAEAS 153
Query: 149 PKNLRGGFTTMHQLMICCGMSLTYLI-GAFV----NWRILALIGAIPCFAQLLSLPFIPD 203
P +RGG + + LMI G L+YL+ AF WR + + +P Q + + F+P+
Sbjct: 154 PSEVRGGLVSTNVLMITGGQFLSYLVNSAFTQVPGTWRWMLGVSGVPAVIQFILMLFMPE 213
Query: 204 SPRWLAKVGHLKESDSALQRL----RGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQW 259
SPRWL E+ L R R ++ H AAE E QR+ + +F+
Sbjct: 214 SPRWLFMKNRKAEAIQVLARTYDISRLEDEIDHLSAAE----EEEKQRKRTVGYLDVFRS 269
Query: 260 KYM-TSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES----IGTIAMVAVKIPVTV 314
K + + G GL QQF GIN +++Y+ +I AGF + ++ + A+ TV
Sbjct: 270 KELRLAFLAGAGLQAFQQFTGINTVMYYSPTIVQMAGFHSNQLALFLSLIVAAMNAAGTV 329
Query: 315 LGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQ-------GLHEWKGVSPILALVGV 367
+G+ +D GR+ L L S G + + +++F Q GL+ W LA++G+
Sbjct: 330 VGIYFIDHCGRKKLALSSLFGVIISLLILSVSFFKQSETSSDGGLYGW------LAVLGL 383
Query: 368 LVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTG 427
+Y+ ++ GMG +PW + SEI+P +G G + NW+ + IV+ F + + TG
Sbjct: 384 ALYIVFFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWISNLIVAQTFLTIAEAAGTG 443
Query: 428 -TFFIFSSICGFTVLFVAKLVPETKGRTLEEIQ 459
TF I + I V+FV VPET+G T E++
Sbjct: 444 MTFLILAGIAVLAVIFVIVFVPETQGLTFSEVE 476
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,737,423
Number of Sequences: 26719
Number of extensions: 395542
Number of successful extensions: 1607
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 1334
Number of HSP's gapped (non-prelim): 113
length of query: 471
length of database: 11,318,596
effective HSP length: 103
effective length of query: 368
effective length of database: 8,566,539
effective search space: 3152486352
effective search space used: 3152486352
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0544.5


