

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
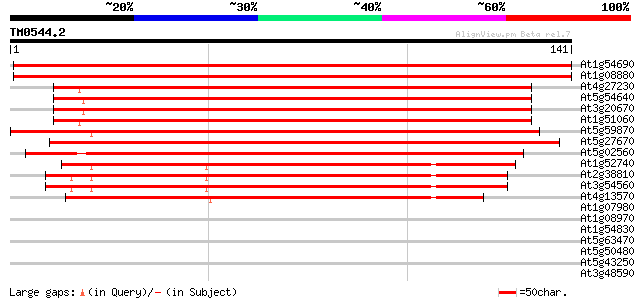
Query= TM0544.2
(141 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g54690 unknown protein 249 3e-67
At1g08880 histone H2A like protein 247 1e-66
At4g27230 histone H2A- like protein 202 4e-53
At5g54640 histone H2A (gb|AAF64418.1) 201 1e-52
At3g20670 histone H2A, putative 197 2e-51
At1g51060 putative protein 197 2e-51
At5g59870 histone H2A - like protein 179 3e-46
At5g27670 histone H2A-like protein 174 2e-44
At5g02560 unknown protein 172 4e-44
At1g52740 putative histone H2A 124 2e-29
At2g38810 histone H2A 122 8e-29
At3g54560 histone H2A.F/Z 120 2e-28
At4g13570 histone H2A.F/Z - like protein 81 2e-16
At1g07980 unknown protein 35 0.017
At1g08970 transcription factor like protein 33 0.050
At1g54830 unknown protein 32 0.15
At5g63470 transcription factor Hap5a-like protein 31 0.25
At5g50480 transcription factor Hap5a-like 31 0.25
At5g43250 unknown protein 31 0.25
At3g48590 transcription factor Hap5a 31 0.25
>At1g54690 unknown protein
Length = 142
Score = 249 bits (637), Expect = 3e-67
Identities = 128/140 (91%), Positives = 129/140 (91%)
Query: 2 SSTGGNNTKGGRGKPKAAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAV 61
S G TKGGRGKPKA KSVSRS KAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAV
Sbjct: 3 SGAGSGTTKGGRGKPKATKSVSRSSKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAV 62
Query: 62 LEYLAAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQT 121
LEYLAAEVLELAGNAARDNKK RIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQT
Sbjct: 63 LEYLAAEVLELAGNAARDNKKTRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQT 122
Query: 122 LLPKKAGKGKGEIGSASQEF 141
LLP K GK KG+IGSASQEF
Sbjct: 123 LLPSKVGKNKGDIGSASQEF 142
>At1g08880 histone H2A like protein
Length = 142
Score = 247 bits (631), Expect = 1e-66
Identities = 126/140 (90%), Positives = 129/140 (92%)
Query: 2 SSTGGNNTKGGRGKPKAAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAV 61
+ G TKGGRGKPKA KSVSRS KAGLQFPVGRIARFLK+GKYAERVGAGAPVYLSAV
Sbjct: 3 TGAGSGTTKGGRGKPKATKSVSRSSKAGLQFPVGRIARFLKSGKYAERVGAGAPVYLSAV 62
Query: 62 LEYLAAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQT 121
LEYLAAEVLELAGNAARDNKK RIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQT
Sbjct: 63 LEYLAAEVLELAGNAARDNKKTRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQT 122
Query: 122 LLPKKAGKGKGEIGSASQEF 141
LLP K GK KG+IGSASQEF
Sbjct: 123 LLPSKVGKNKGDIGSASQEF 142
>At4g27230 histone H2A- like protein
Length = 131
Score = 202 bits (515), Expect = 4e-53
Identities = 107/124 (86%), Positives = 109/124 (87%), Gaps = 4/124 (3%)
Query: 12 GRGKP----KAAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAA 67
GRGK A KS SRS KAGLQFPVGRIARFLKAGKYAERVGAGAPVYL+AVLEYLAA
Sbjct: 3 GRGKQLGSGAAKKSTSRSSKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLAAVLEYLAA 62
Query: 68 EVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLPKKA 127
EVLELAGNAARDNKK RIVPRHIQLAVRNDEELSKLLG VTIANGGV+PNIH LLPKKA
Sbjct: 63 EVLELAGNAARDNKKTRIVPRHIQLAVRNDEELSKLLGDVTIANGGVMPNIHNLLLPKKA 122
Query: 128 GKGK 131
G K
Sbjct: 123 GSSK 126
>At5g54640 histone H2A (gb|AAF64418.1)
Length = 130
Score = 201 bits (510), Expect = 1e-52
Identities = 106/124 (85%), Positives = 109/124 (87%), Gaps = 4/124 (3%)
Query: 12 GRGKPK----AAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAA 67
GRGK A K+ SRS KAGLQFPVGRIARFLKAGKYAERVGAGAPVYL+AVLEYLAA
Sbjct: 3 GRGKTLGSGGAKKATSRSSKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLAAVLEYLAA 62
Query: 68 EVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLPKKA 127
EVLELAGNAARDNKK RIVPRHIQLAVRNDEELSKLLG VTIANGGV+PNIH LLPKKA
Sbjct: 63 EVLELAGNAARDNKKTRIVPRHIQLAVRNDEELSKLLGDVTIANGGVMPNIHNLLLPKKA 122
Query: 128 GKGK 131
G K
Sbjct: 123 GASK 126
>At3g20670 histone H2A, putative
Length = 132
Score = 197 bits (501), Expect = 2e-51
Identities = 105/124 (84%), Positives = 107/124 (85%), Gaps = 4/124 (3%)
Query: 12 GRGKPK----AAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAA 67
GRGK A KS SRS KAGLQFPVGRIARFLK GKYA RVGAGAPVYL+AVLEYLAA
Sbjct: 3 GRGKTLGSGVAKKSTSRSSKAGLQFPVGRIARFLKNGKYATRVGAGAPVYLAAVLEYLAA 62
Query: 68 EVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLPKKA 127
EVLELAGNAARDNKK RIVPRHIQLAVRNDEELSKLLG VTIANGGV+PNIH LLPKKA
Sbjct: 63 EVLELAGNAARDNKKTRIVPRHIQLAVRNDEELSKLLGDVTIANGGVMPNIHSLLLPKKA 122
Query: 128 GKGK 131
G K
Sbjct: 123 GASK 126
>At1g51060 putative protein
Length = 132
Score = 197 bits (500), Expect = 2e-51
Identities = 103/124 (83%), Positives = 107/124 (86%), Gaps = 4/124 (3%)
Query: 12 GRGKP----KAAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAA 67
GRGK A K+ +RS KAGLQFPVGRIARFLK GKYAERVGAGAPVYL+AVLEYLAA
Sbjct: 3 GRGKTLGSGSAKKATTRSSKAGLQFPVGRIARFLKKGKYAERVGAGAPVYLAAVLEYLAA 62
Query: 68 EVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLPKKA 127
EVLELAGNAARDNKK RIVPRHIQLAVRNDEELSKLLG VTIANGGV+PNIH LLPKK
Sbjct: 63 EVLELAGNAARDNKKTRIVPRHIQLAVRNDEELSKLLGDVTIANGGVMPNIHNLLLPKKT 122
Query: 128 GKGK 131
G K
Sbjct: 123 GASK 126
>At5g59870 histone H2A - like protein
Length = 150
Score = 179 bits (455), Expect = 3e-46
Identities = 94/136 (69%), Positives = 107/136 (78%), Gaps = 3/136 (2%)
Query: 1 MSSTGGNNTKGGRGKPKAA---KSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVY 57
M STG G KP A KSVS+S KAGLQFPVGRI RFLK G+YA+R+G GAPVY
Sbjct: 1 MESTGKVKKAFGGRKPPGAPKTKSVSKSMKAGLQFPVGRITRFLKKGRYAQRLGGGAPVY 60
Query: 58 LSAVLEYLAAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPN 117
++AVLEYLAAEVLELAGNAARDNKK+RI+PRH+ LA+RNDEEL KLL VTIA+GGVLPN
Sbjct: 61 MAAVLEYLAAEVLELAGNAARDNKKSRIIPRHLLLAIRNDEELGKLLSGVTIAHGGVLPN 120
Query: 118 IHQTLLPKKAGKGKGE 133
I+ LLPKK+ E
Sbjct: 121 INSVLLPKKSATKPAE 136
>At5g27670 histone H2A-like protein
Length = 150
Score = 174 bits (440), Expect = 2e-44
Identities = 91/128 (71%), Positives = 103/128 (80%)
Query: 11 GGRGKPKAAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVL 70
GGR KSVS+S KAGLQFPVGRIAR+LK G+YA R G+GAPVYL+AVLEYLAAEVL
Sbjct: 15 GGRKGGDRKKSVSKSVKAGLQFPVGRIARYLKKGRYALRYGSGAPVYLAAVLEYLAAEVL 74
Query: 71 ELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLPKKAGKG 130
ELAGNAARDNKKNRI PRH+ LA+RNDEEL +LL VTIA+GGVLPNI+ LLPKK+
Sbjct: 75 ELAGNAARDNKKNRINPRHLCLAIRNDEELGRLLHGVTIASGGVLPNINPVLLPKKSTAS 134
Query: 131 KGEIGSAS 138
+ AS
Sbjct: 135 SSQAEKAS 142
>At5g02560 unknown protein
Length = 153
Score = 172 bits (437), Expect = 4e-44
Identities = 90/125 (72%), Positives = 102/125 (81%), Gaps = 2/125 (1%)
Query: 5 GGNNTKGGRGKPKAAKSVSRSQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEY 64
G + G G K K VSRS K+GLQFPVGRI R+LK G+Y++RVG GAPVYL+AVLEY
Sbjct: 10 GAAGRRSGGGPKK--KPVSRSVKSGLQFPVGRIGRYLKKGRYSKRVGTGAPVYLAAVLEY 67
Query: 65 LAAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLP 124
LAAEVLELAGNAARDNKKNRI+PRH+ LAVRNDEEL LL VTIA+GGVLPNI+ LLP
Sbjct: 68 LAAEVLELAGNAARDNKKNRIIPRHVLLAVRNDEELGTLLKGVTIAHGGVLPNINPILLP 127
Query: 125 KKAGK 129
KK+ K
Sbjct: 128 KKSEK 132
>At1g52740 putative histone H2A
Length = 134
Score = 124 bits (310), Expect = 2e-29
Identities = 69/122 (56%), Positives = 86/122 (69%), Gaps = 9/122 (7%)
Query: 14 GKPKAA-------KSVSRSQKAGLQFPVGRIARFLKAGKYAE-RVGAGAPVYLSAVLEYL 65
GKP + K ++RS +AGLQFPVGR+ R LK A RVGA A VY +A+LEYL
Sbjct: 12 GKPSGSDKDKDKKKPITRSSRAGLQFPVGRVHRLLKTRSTAHGRVGATAAVYTAAILEYL 71
Query: 66 AAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIHQTLLPK 125
AEVLELAGNA++D K RI PRH+QLA+R DEEL L+ TIA GGV+P+IH++L+ K
Sbjct: 72 TAEVLELAGNASKDLKVKRISPRHLQLAIRGDEELDTLIKG-TIAGGGVIPHIHKSLINK 130
Query: 126 KA 127
A
Sbjct: 131 SA 132
>At2g38810 histone H2A
Length = 136
Score = 122 bits (305), Expect = 8e-29
Identities = 73/130 (56%), Positives = 90/130 (69%), Gaps = 15/130 (11%)
Query: 10 KGGRG----KPKAA---------KSVSRSQKAGLQFPVGRIARFLKAGKYAE-RVGAGAP 55
KGG+G K AA KS+SRS +AG+QFPVGRI R LK A RVGA A
Sbjct: 4 KGGKGLLAAKTTAAAANKDSVKKKSISRSSRAGIQFPVGRIHRQLKQRVSAHGRVGATAA 63
Query: 56 VYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVL 115
VY +++LEYL AEVLELAGNA++D K RI PRH+QLA+R DEEL L+ TIA GGV+
Sbjct: 64 VYTASILEYLTAEVLELAGNASKDLKVKRITPRHLQLAIRGDEELDTLIKG-TIAGGGVI 122
Query: 116 PNIHQTLLPK 125
P+IH++L+ K
Sbjct: 123 PHIHKSLVNK 132
>At3g54560 histone H2A.F/Z
Length = 136
Score = 120 bits (301), Expect = 2e-28
Identities = 72/130 (55%), Positives = 89/130 (68%), Gaps = 15/130 (11%)
Query: 10 KGGRG----KPKAA---------KSVSRSQKAGLQFPVGRIARFLKAGKYAE-RVGAGAP 55
KGG+G K AA K +SRS +AG+QFPVGRI R LK A RVGA A
Sbjct: 4 KGGKGLVAAKTMAANKDKDKDKKKPISRSARAGIQFPVGRIHRQLKTRVSAHGRVGATAA 63
Query: 56 VYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVL 115
VY +++LEYL AEVLELAGNA++D K RI PRH+QLA+R DEEL L+ TIA GGV+
Sbjct: 64 VYTASILEYLTAEVLELAGNASKDLKVKRITPRHLQLAIRGDEELDTLIKG-TIAGGGVI 122
Query: 116 PNIHQTLLPK 125
P+IH++L+ K
Sbjct: 123 PHIHKSLINK 132
>At4g13570 histone H2A.F/Z - like protein
Length = 118
Score = 80.9 bits (198), Expect = 2e-16
Identities = 49/106 (46%), Positives = 64/106 (60%), Gaps = 2/106 (1%)
Query: 15 KPKAAKSVSRSQKAGLQFPVGRIARFLKAGKYAER-VGAGAPVYLSAVLEYLAAEVLELA 73
K A ++V G F V RI + LK A VGA VY++++LEYL EVL+LA
Sbjct: 14 KISAFENVRMIMVEGEMFQVARIHKQLKNRVSAHSSVGATDVVYMTSILEYLTTEVLQLA 73
Query: 74 GNAARDNKKNRIVPRHIQLAVRNDEELSKLLGSVTIANGGVLPNIH 119
N ++D K RI PRH+QLA+R DEEL L+ TI G V+P+IH
Sbjct: 74 ENTSKDLKVKRITPRHLQLAIRGDEELDTLIKG-TIIGGSVIPHIH 118
>At1g07980 unknown protein
Length = 206
Score = 34.7 bits (78), Expect = 0.017
Identities = 21/83 (25%), Positives = 40/83 (47%)
Query: 25 SQKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNR 84
++ A ++FP+ RI R +++ A ++ A ++ E E A +++ +KK
Sbjct: 103 AEDAKIKFPMNRIRRIMRSDNSAPQIMQDAVFLVNKATEMFIERFSEEAYDSSVKDKKKF 162
Query: 85 IVPRHIQLAVRNDEELSKLLGSV 107
I +H+ V ND+ L SV
Sbjct: 163 IHYKHLSSVVSNDQRYEFLADSV 185
>At1g08970 transcription factor like protein
Length = 231
Score = 33.1 bits (74), Expect = 0.050
Identities = 25/100 (25%), Positives = 43/100 (43%), Gaps = 2/100 (2%)
Query: 33 PVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHIQL 92
P+ RI + +KA + + A APV + E E+ + N +NK+ + I
Sbjct: 82 PLARIKKIMKADEDVRMISAEAPVVFARACEMFILELTLRSWNHTEENKRRTLQKNDIAA 141
Query: 93 AVRNDEELSKLLGSVTIAN--GGVLPNIHQTLLPKKAGKG 130
AV + L+ V + VL +I + +P+ A G
Sbjct: 142 AVTRTDIFDFLVDIVPREDLRDEVLGSIPRGTVPEAAAAG 181
>At1g54830 unknown protein
Length = 217
Score = 31.6 bits (70), Expect = 0.15
Identities = 17/62 (27%), Positives = 28/62 (44%)
Query: 33 PVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHIQL 92
P+ RI + +KA + + A APV + E E+ + N +NK+ + I
Sbjct: 72 PLARIKKIMKADEDVRMISAEAPVVFARACEMFILELTLRSWNHTEENKRRTLQKNDIAA 131
Query: 93 AV 94
AV
Sbjct: 132 AV 133
>At5g63470 transcription factor Hap5a-like protein
Length = 250
Score = 30.8 bits (68), Expect = 0.25
Identities = 16/64 (25%), Positives = 29/64 (45%)
Query: 31 QFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHI 90
Q P+ RI + +KA + + A AP+ + E E+ + A +NK+ + I
Sbjct: 78 QLPLARIKKIMKADEDVRMISAEAPILFAKACELFILELTIRSWLHAEENKRRTLQKNDI 137
Query: 91 QLAV 94
A+
Sbjct: 138 AAAI 141
>At5g50480 transcription factor Hap5a-like
Length = 202
Score = 30.8 bits (68), Expect = 0.25
Identities = 20/77 (25%), Positives = 33/77 (41%)
Query: 31 QFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHI 90
Q P+ RI + +KA V A AP+ + E ++ + A +NK++ + I
Sbjct: 54 QLPLARIKKIMKADPDVHMVSAEAPIIFAKACEMFIVDLTMRSWLKAEENKRHTLQKSDI 113
Query: 91 QLAVRNDEELSKLLGSV 107
AV + LL V
Sbjct: 114 SNAVASSFTYDFLLDVV 130
>At5g43250 unknown protein
Length = 130
Score = 30.8 bits (68), Expect = 0.25
Identities = 15/74 (20%), Positives = 35/74 (47%)
Query: 31 QFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHI 90
+FP+GR+ + +K K ++ + A ++ E + E + + K+ + H+
Sbjct: 11 EFPIGRVKKIMKLDKDINKINSEALHVITYSTELFLHFLAEKSAVVTAEKKRKTVNLDHL 70
Query: 91 QLAVRNDEELSKLL 104
++AV+ + S L
Sbjct: 71 RIAVKRHQPTSDFL 84
>At3g48590 transcription factor Hap5a
Length = 234
Score = 30.8 bits (68), Expect = 0.25
Identities = 16/64 (25%), Positives = 29/64 (45%)
Query: 31 QFPVGRIARFLKAGKYAERVGAGAPVYLSAVLEYLAAEVLELAGNAARDNKKNRIVPRHI 90
Q P+ RI + +KA + + A AP+ + E E+ + A +NK+ + I
Sbjct: 65 QLPLARIKKIMKADEDVRMISAEAPILFAKACELFILELTIRSWLHAEENKRRTLQKNDI 124
Query: 91 QLAV 94
A+
Sbjct: 125 AAAI 128
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,018,539
Number of Sequences: 26719
Number of extensions: 115069
Number of successful extensions: 288
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 252
Number of HSP's gapped (non-prelim): 43
length of query: 141
length of database: 11,318,596
effective HSP length: 89
effective length of query: 52
effective length of database: 8,940,605
effective search space: 464911460
effective search space used: 464911460
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0544.2


