

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.10
(306 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
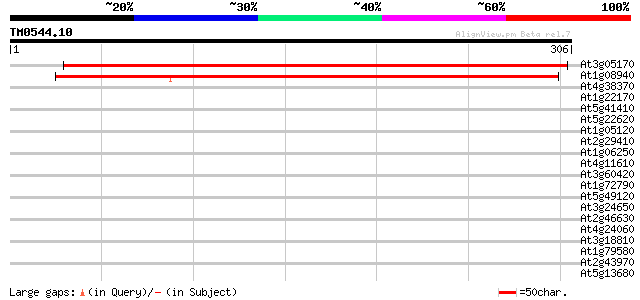
Score E
Sequences producing significant alignments: (bits) Value
At3g05170 unknown protein 412 e-115
At1g08940 unknown protein 368 e-102
At4g38370 unknown protein 39 0.005
At1g22170 phosphoglycerate mutase like protein 33 0.15
At5g41410 homeotic protein BEL1 homolog 32 0.33
At5g22620 phosphoglycerate mutase like protein 32 0.43
At1g05120 hypothetical protein, 3' partial 32 0.56
At2g29410 putative zinc transporter 31 0.96
At1g06250 lipase like protein 31 0.96
At4g11610 phosphoribosylanthranilate transferase like protein 30 1.3
At3g60420 unknown protein 30 1.3
At1g72790 unknown protein 30 1.3
At5g49120 putative protein 30 1.6
At3g24650 abscisic acid-insensitive protein 3 30 1.6
At2g46630 putative extensin 30 1.6
At4g24060 unknown protein 29 2.8
At3g18810 protein kinase, putative 29 2.8
At1g79580 unknown protein 29 2.8
At2g43970 unknown protein 29 3.6
At5g13680 putative protein 28 4.8
>At3g05170 unknown protein
Length = 316
Score = 412 bits (1059), Expect = e-115
Identities = 198/276 (71%), Positives = 232/276 (83%), Gaps = 1/276 (0%)
Query: 30 VLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISST-SSTD 88
+LPKRIILVRHGES+GNLD +AYTTTPDHKI LT G+ QA+ AGAR+ +ISS SS +
Sbjct: 7 LLPKRIILVRHGESEGNLDTAAYTTTPDHKIQLTDSGLLQAQEAGARLHALISSNPSSPE 66
Query: 89 WRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQ 148
WRVYFYVSPY RTRSTLREIGRSFS+ R++GVREECRIREQDFGNFQV+ER+ A K+ R+
Sbjct: 67 WRVYFYVSPYDRTRSTLREIGRSFSRRRVIGVREECRIREQDFGNFQVKERMRATKKVRE 126
Query: 149 RFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVF 208
RFGRFF+RFP+GESAADVFDRVSSFLESLWRDIDMNRL+ +PS++LN +IVSHGL SRVF
Sbjct: 127 RFGRFFYRFPEGESAADVFDRVSSFLESLWRDIDMNRLHINPSHELNFVIVSHGLTSRVF 186
Query: 209 LMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPDMIADQKW 268
LMKWFKW+VEQF+ LNN GN E RVM+LG GG+YSLA+HHT+ E+ WGLSP+MIADQKW
Sbjct: 187 LMKWFKWSVEQFEGLNNPGNSEIRVMELGQGGDYSLAIHHTEEELATWGLSPEMIADQKW 246
Query: 269 RAHATKGAWHDQGPLSLDTFFDHLHDSDNEDNDDQT 304
RA+A KG W + FFDH+ DSD E + T
Sbjct: 247 RANAHKGEWKEDCKWYFGDFFDHMADSDKECETEAT 282
>At1g08940 unknown protein
Length = 281
Score = 368 bits (944), Expect = e-102
Identities = 177/279 (63%), Positives = 223/279 (79%), Gaps = 5/279 (1%)
Query: 26 HKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTS 85
++ +LPKRIIL+RHGES GN+D AY TTPDHKI LT +G QAR AG ++R +IS+ S
Sbjct: 3 NEKKMLPKRIILMRHGESAGNIDAGAYATTPDHKIPLTEEGRAQAREAGKKMRALISTQS 62
Query: 86 S----TDWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERIN 141
+WRVYFYVSPY RTR+TLRE+G+ FS+ R++GVREECRIREQDFGNFQV+ER+
Sbjct: 63 GGACGENWRVYFYVSPYERTRTTLREVGKGFSRKRVIGVREECRIREQDFGNFQVEERMR 122
Query: 142 ALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSH 201
+KETR+RFGRFF+RFP+GESAADV+DRVSSFLES+WRD+DMNR DPS++LNL+IVSH
Sbjct: 123 VVKETRERFGRFFYRFPEGESAADVYDRVSSFLESMWRDVDMNRHQVDPSSELNLVIVSH 182
Query: 202 GLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPD 261
GL SRVFL KWFKWTV +F+ LNNFGN EFRVM+LG+ GEY+ A+HH++ EM +WG+S D
Sbjct: 183 GLTSRVFLTKWFKWTVAEFERLNNFGNCEFRVMELGASGEYTFAIHHSEEEMLDWGMSKD 242
Query: 262 MIADQKWRAHATK-GAWHDQGPLSLDTFFDHLHDSDNED 299
MI DQK R + +D L L+ +FD L +D+E+
Sbjct: 243 MIDDQKDRVDGCRVTTSNDSCSLHLNEYFDLLDVTDDEE 281
>At4g38370 unknown protein
Length = 225
Score = 38.5 bits (88), Expect = 0.005
Identities = 47/182 (25%), Positives = 80/182 (43%), Gaps = 31/182 (17%)
Query: 34 RIILVRHGESQGN---LDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWR 90
R ++RHG+S N L S+ + L P G+ QAR+AG + ++ +
Sbjct: 11 RYWVLRHGKSIPNERGLVVSSMENGVLPEYQLAPDGVAQARLAGESFLQQLKESNIELDK 70
Query: 91 VYFYVSPYARTRSTLREIGRSFSKHRLLGVREECR----IREQDFG-NFQVQE-----RI 140
V SP++RT T R + + + L +C+ +RE+ FG F+++ I
Sbjct: 71 VRICYSPFSRTTHTARVVAKVLN---LPFDAPQCKMMEDLRERYFGPTFELKSHDKYPEI 127
Query: 141 NALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVS 200
AL E G GESA DV R+++ ++S+ + +++VS
Sbjct: 128 WALDEKDPFMGP-----EGGESADDVVSRLATAMKSMEAEYQR----------CAILVVS 172
Query: 201 HG 202
HG
Sbjct: 173 HG 174
>At1g22170 phosphoglycerate mutase like protein
Length = 334
Score = 33.5 bits (75), Expect = 0.15
Identities = 48/227 (21%), Positives = 96/227 (42%), Gaps = 44/227 (19%)
Query: 20 IPNQNNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRH 79
IP+ + K + +IL+RHGES N + + +T D + LT +G+ +A AG RI +
Sbjct: 67 IPDNSQKKSN--EAALILIRHGESLWN-EKNLFTGCVD--VPLTEKGVEEAIEAGKRISN 121
Query: 80 V---ISSTSS---------------TDWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVR 121
+ + TSS +V + + T ++ +K++ + V
Sbjct: 122 IPVDVIFTSSLIRAQMTAMLAMIQHRRKKVPIILHDESEQAKTWSQVFSDETKNQSIPVI 181
Query: 122 EECRIREQDFGNFQVQERINALKETRQRFGR---------FFFRFPDGESAADVFDRVSS 172
++ E+ +G Q + +ET +R+G+ + P GES +R +
Sbjct: 182 PAWQLNERMYGELQGLNK----QETAERYGKEQVHEWRRSYDIPPPKGESLEMCAERAVA 237
Query: 173 FLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQ 219
+ + + + + N++I +HG + R +M K T ++
Sbjct: 238 YFQ--------DNIEPKLAAGKNVMIAAHGNSLRSIIMYLDKLTCQE 276
>At5g41410 homeotic protein BEL1 homolog
Length = 611
Score = 32.3 bits (72), Expect = 0.33
Identities = 20/80 (25%), Positives = 35/80 (43%), Gaps = 18/80 (22%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPS-------------VLPKRIILVRH-----GES 43
H +NNN+N ++NHHQ QI + P+ + +++++H G+
Sbjct: 176 HQNNNNNNHQHHNHHQFQIGSSKYLSPAQELLSEFCSLGVKESDEEVMMMKHKKKQKGKQ 235
Query: 44 QGNLDPSAYTTTPDHKISLT 63
Q D S ++ H S T
Sbjct: 236 QEEWDTSHHSNNDQHDQSAT 255
>At5g22620 phosphoglycerate mutase like protein
Length = 482
Score = 32.0 bits (71), Expect = 0.43
Identities = 34/122 (27%), Positives = 56/122 (45%), Gaps = 18/122 (14%)
Query: 33 KRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRVY 92
KR++LVRHG+S N + + D + LT +G +QA I+ R ++ S
Sbjct: 48 KRVVLVRHGQSTWN-EEGRIQGSSDFSV-LTKKGESQAEIS----RQMLIDDSFD----V 97
Query: 93 FYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQRFGR 152
+ SP R++ T I S + + +RE D +FQ + KE +++FG
Sbjct: 98 CFTSPLKRSKKTAEIIWGSRESEMIF----DYDLREIDLYSFQGLLK----KEGKEKFGE 149
Query: 153 FF 154
F
Sbjct: 150 AF 151
>At1g05120 hypothetical protein, 3' partial
Length = 773
Score = 31.6 bits (70), Expect = 0.56
Identities = 35/136 (25%), Positives = 54/136 (38%), Gaps = 34/136 (25%)
Query: 113 SKHRLLGVREECRIREQDFGNFQVQERINALKETRQRFGRFFFRFPDGESAADVFDRVSS 172
SK L G R + +FQ +I AL+E + F DG + A VF + +S
Sbjct: 637 SKTTLKGFRASSILNRIKLDDFQTSTKIEALREEIR-----FMVERDGSAKAIVFSQFTS 691
Query: 173 FLESL---------------------WRDIDMNRLNHDPSNDLNLIIVSHG-------LA 204
FL+ + RD +N+ DP + L+ + G +A
Sbjct: 692 FLDLINYTLGKCGVSCVQLVGSMTMAARDTAINKFKEDPDCRVFLMSLKAGGVALNLTVA 751
Query: 205 SRVFLM-KWFKWTVEQ 219
S VF+M W+ VE+
Sbjct: 752 SHVFMMDPWWNPAVER 767
>At2g29410 putative zinc transporter
Length = 385
Score = 30.8 bits (68), Expect = 0.96
Identities = 10/29 (34%), Positives = 21/29 (71%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSV 30
HNH+++ +D +++HH + +Q++HK V
Sbjct: 192 HNHSHHHHDHHHHHHNHKHQHQHHHKEVV 220
>At1g06250 lipase like protein
Length = 423
Score = 30.8 bits (68), Expect = 0.96
Identities = 19/51 (37%), Positives = 23/51 (44%), Gaps = 2/51 (3%)
Query: 258 LSPDMIADQKWRAHATKGAWH-DQGPLSLDTFF-DHLHDSDNEDNDDQTNS 306
L + + WR A KG D G LD DH D D +DNDD + S
Sbjct: 364 LKDEYLVPSTWRCLANKGMLQMDDGTWKLDVHRRDHDDDVDADDNDDSSTS 414
>At4g11610 phosphoribosylanthranilate transferase like protein
Length = 1011
Score = 30.4 bits (67), Expect = 1.3
Identities = 18/80 (22%), Positives = 32/80 (39%), Gaps = 6/80 (7%)
Query: 8 DNDDNNNHHQQQIPNQNNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGI 67
+ D++N HH +P + P R + H S + P+ D + T +
Sbjct: 197 EQDNHNEHHHHYVPKHQVDEMRSEPARPSKLVHAHSIASAQPA------DFALKETSPHL 250
Query: 68 TQARIAGARIRHVISSTSST 87
R+ G R+ H + +ST
Sbjct: 251 GGGRVVGGRVIHKDKTATST 270
>At3g60420 unknown protein
Length = 270
Score = 30.4 bits (67), Expect = 1.3
Identities = 59/240 (24%), Positives = 98/240 (40%), Gaps = 45/240 (18%)
Query: 33 KRIILVRHGESQGNLDPSAYTTTPDHKIS--LTPQGITQARIAGARIRHVISSTSSTDWR 90
+ +IL+RHG+ N +P +T+T L G +A G RIR S R
Sbjct: 12 QNVILMRHGDRLDNFEP-LWTSTAARPWDPPLAQDGKDRAFRTGQRIR---SQLGVPIHR 67
Query: 91 VYFYVSPYARTRSTLREIGR----------SFSKHRLLGVREECRIREQ-DFGNFQVQER 139
V +VSP+ R T E+ + S +L + + +I+ +FG ++
Sbjct: 68 V--FVSPFLRCIQTASEVVAALSAVDFDPIAMSSKDVLSI-DNTKIKVAIEFGLSEIPHP 124
Query: 140 INALKETRQRFGRFFFR-------FPDG--ESAAD-VFDRVSSFLESLW----RDIDMNR 185
I E + G+F F+ FP+G +S D V+ V + ES R +
Sbjct: 125 IFIKSEVAPKDGKFDFKISDLEAMFPEGTVDSNVDMVYKEVPEWGESAQAFEDRYYKTVK 184
Query: 186 LNHDPSNDLNLIIVSHGLASRVFLMKWFKWTVE-----------QFDLLNNFGNGEFRVM 234
+ + NL++V+H A V +FK + + +LN G G+F V+
Sbjct: 185 ILAEKYPSENLLLVTHWGAVSVAFYNYFKDATKYVVDYCGSVEMRRQILNGDGFGKFEVV 244
>At1g72790 unknown protein
Length = 561
Score = 30.4 bits (67), Expect = 1.3
Identities = 10/29 (34%), Positives = 21/29 (71%)
Query: 6 NNDNDDNNNHHQQQIPNQNNHKPSVLPKR 34
N +ND++N+H ++ I N+ + PS++ +R
Sbjct: 95 NTNNDESNHHKEEDIRNKFSTSPSIIDRR 123
>At5g49120 putative protein
Length = 150
Score = 30.0 bits (66), Expect = 1.6
Identities = 13/37 (35%), Positives = 24/37 (64%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSVLPKRIILV 38
+N+NNN+N++NNN+++ + P V+ K I+V
Sbjct: 12 NNNNNNNNNNNNNNNKNPLSEGVLISPKVVNKANIIV 48
>At3g24650 abscisic acid-insensitive protein 3
Length = 720
Score = 30.0 bits (66), Expect = 1.6
Identities = 10/17 (58%), Positives = 13/17 (75%)
Query: 2 HNHNNNDNDDNNNHHQQ 18
H HNNN+N++NNN Q
Sbjct: 474 HRHNNNNNNNNNNQQNQ 490
Score = 28.9 bits (63), Expect = 3.6
Identities = 9/17 (52%), Positives = 16/17 (93%)
Query: 2 HNHNNNDNDDNNNHHQQ 18
H+H +N+N++NNN++QQ
Sbjct: 472 HHHRHNNNNNNNNNNQQ 488
Score = 27.7 bits (60), Expect = 8.1
Identities = 9/18 (50%), Positives = 15/18 (83%)
Query: 2 HNHNNNDNDDNNNHHQQQ 19
H+ +NN+N++NNN+ Q Q
Sbjct: 473 HHRHNNNNNNNNNNQQNQ 490
>At2g46630 putative extensin
Length = 394
Score = 30.0 bits (66), Expect = 1.6
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 7/50 (14%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPS------VLPKRIILVRHGESQG 45
HN+N N + + N +HQ P + + +PS ++ R+I + GE++G
Sbjct: 225 HNYNQNHSYNQNQNHQGNNPKKMHRQPSSSDSENIMSTRVITIA-GENKG 273
Score = 28.1 bits (61), Expect = 6.2
Identities = 13/35 (37%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Query: 2 HNHNNN-DNDDNNNHHQQQIPNQNNHKPSVLPKRI 35
HN N+N D + N+N++Q NQN + PK++
Sbjct: 213 HNQNHNHDYNQNHNYNQNHSYNQNQNHQGNNPKKM 247
>At4g24060 unknown protein
Length = 342
Score = 29.3 bits (64), Expect = 2.8
Identities = 17/58 (29%), Positives = 28/58 (47%), Gaps = 4/58 (6%)
Query: 1 MHNHNNNDNDDNNNHHQQQIPNQNNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDH 58
+HN NN N NNN++ + + + S L L+R+ + GN S++ P H
Sbjct: 179 IHNIGNNTNKSNNNNNPLIVSSCSAMATSSLD----LIRNNSNNGNSSNSSFMGFPVH 232
>At3g18810 protein kinase, putative
Length = 700
Score = 29.3 bits (64), Expect = 2.8
Identities = 17/73 (23%), Positives = 29/73 (39%), Gaps = 11/73 (15%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKIS 61
+N NNN+ +DNN ++ N NN++ +G N P + D
Sbjct: 111 NNGNNNNGNDNNGNNNNGNNNDNNNQ-----------NNGGGSNNRSPPPPSRNSDRNSP 159
Query: 62 LTPQGITQARIAG 74
P+ + R +G
Sbjct: 160 SPPRALAPPRSSG 172
Score = 29.3 bits (64), Expect = 2.8
Identities = 11/26 (42%), Positives = 17/26 (65%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHK 27
+ +NNNDN++NNN + N N+K
Sbjct: 77 NGNNNNDNNNNNNGNNNNDNNNGNNK 102
Score = 28.1 bits (61), Expect = 6.2
Identities = 10/25 (40%), Positives = 18/25 (72%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNH 26
+N+ NN+ND+NN +++ N NN+
Sbjct: 87 NNNGNNNNDNNNGNNKDNNNNGNNN 111
Score = 27.7 bits (60), Expect = 8.1
Identities = 10/24 (41%), Positives = 17/24 (70%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNN 25
+N+NNN N++N+N++ N NN
Sbjct: 84 NNNNNNGNNNNDNNNGNNKDNNNN 107
Score = 27.7 bits (60), Expect = 8.1
Identities = 10/25 (40%), Positives = 17/25 (68%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNH 26
+N+N N+N+DNNN + + N N+
Sbjct: 86 NNNNGNNNNDNNNGNNKDNNNNGNN 110
>At1g79580 unknown protein
Length = 371
Score = 29.3 bits (64), Expect = 2.8
Identities = 10/30 (33%), Positives = 17/30 (56%)
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSVL 31
H+H +++++HH N NNH P +L
Sbjct: 183 HHHQYISTNNDHDHHHHIDSNSNNHSPLIL 212
>At2g43970 unknown protein
Length = 545
Score = 28.9 bits (63), Expect = 3.6
Identities = 9/26 (34%), Positives = 17/26 (64%)
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHKP 28
NHN+N ++++HH Q+ Q ++ P
Sbjct: 481 NHNHNGRGNHHHHHHHQVGTQPSNNP 506
>At5g13680 putative protein
Length = 1319
Score = 28.5 bits (62), Expect = 4.8
Identities = 14/49 (28%), Positives = 24/49 (48%)
Query: 59 KISLTPQGITQARIAGARIRHVISSTSSTDWRVYFYVSPYARTRSTLRE 107
K+ T + +++A + H S+ S D VY + +TRST R+
Sbjct: 1257 KLQQTAENFQVSQVAAVELAHDTVSSESVDEEVYCFERYAQKTRSTARD 1305
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,676,059
Number of Sequences: 26719
Number of extensions: 337422
Number of successful extensions: 1534
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1386
Number of HSP's gapped (non-prelim): 88
length of query: 306
length of database: 11,318,596
effective HSP length: 99
effective length of query: 207
effective length of database: 8,673,415
effective search space: 1795396905
effective search space used: 1795396905
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0544.10


