

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.1
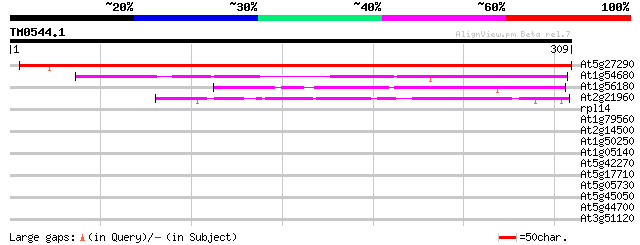
(309 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g27290 unknown protein 449 e-127
At1g54680 148 4e-36
At1g56180 unknown protein 88 7e-18
At2g21960 unknown protein 64 1e-10
rpl14 -chloroplast genome- ribosomal protein L14 31 0.97
At1g79560 FtsH like cell division protein 30 1.7
At2g14500 hypothetical protein 29 3.7
At1g50250 putative chloroplast FtsH protease 29 3.7
At1g05140 Unknown protein (YUP8H12.25) 29 3.7
At5g42270 cell division protein FtsH 28 4.8
At5g17710 chloroplast GrpE protein 28 6.3
At5g05730 anthranilate synthase component I-1 precursor (sp|P32068) 28 6.3
At5g45050 disease resistance protein-like 28 8.2
At5g44700 receptor-like protein kinase 28 8.2
At3g51120 putative protein 28 8.2
>At5g27290 unknown protein
Length = 341
Score = 449 bits (1155), Expect = e-127
Identities = 221/311 (71%), Positives = 264/311 (84%), Gaps = 7/311 (2%)
Query: 6 LMHCVGVTSCPGQLK------LVRVRIQCSTTNVGVS-RRQLLEKLDKELLKGDDRAALA 58
L+H VG+ C Q K R R ++ G+S RRQ LE++D +L GD+RAAL+
Sbjct: 4 LLHSVGLIPCSNQQKSFLFHSYYRYRCIVCSSETGLSIRRQALEQVDSKLSSGDERAALS 63
Query: 59 LVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAA 118
LVKDLQGKPDGLRCFGAARQVPQRLYTL+ELKLNGI A SLLSP D+TLGSIER LQIAA
Sbjct: 64 LVKDLQGKPDGLRCFGAARQVPQRLYTLEELKLNGINAASLLSPTDTTLGSIERNLQIAA 123
Query: 119 IAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRV 178
++GG+ AW AF +S QQLF+++LG +FLWTLD VS+ GG+G+LV+DT GH+FS++YHNRV
Sbjct: 124 VSGGIVAWKAFDLSSQQLFFLTLGFMFLWTLDLVSFNGGIGSLVLDTTGHTFSQRYHNRV 183
Query: 179 IQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAK 238
+QHEAGHFL+AYLVGILP+GYTLSSL+AL+K+GSLN+QAGSAFVD+EFLEEVN+GKVSA
Sbjct: 184 VQHEAGHFLVAYLVGILPRGYTLSSLEALQKEGSLNIQAGSAFVDYEFLEEVNSGKVSAT 243
Query: 239 TLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVL 298
LN+FSCIALAGV TEYL+YG++EGGLDDI KLD L+ LGFTQKK DSQVRWSVLNT+L
Sbjct: 244 MLNRFSCIALAGVATEYLLYGYAEGGLDDISKLDGLVKSLGFTQKKADSQVRWSVLNTIL 303
Query: 299 LLRRHEAARAK 309
LLRRHE AR+K
Sbjct: 304 LLRRHEIARSK 314
>At1g54680
Length = 289
Score = 148 bits (373), Expect = 4e-36
Identities = 96/286 (33%), Positives = 152/286 (52%), Gaps = 63/286 (22%)
Query: 37 RRQLLEKLDKELLKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEA 96
RR+ LE++DKELL+G+ AL+LVK L+ K L FG+A+ +P+ KL+
Sbjct: 23 RRKSLERVDKELLRGNYETALSLVKQLKSKHGCLSAFGSAKLLPK--------KLDMSSK 74
Query: 97 ESLLSPVDSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGG 156
L S +DS SIE ++ + + + SP++ ++
Sbjct: 75 TDLRSLIDSVSRSIE-SVYVQEDSVRTSKEMEIKTSPEEDWF------------------ 115
Query: 157 GLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQ 216
V+QHE+GHFL+ YL+G+LP+ Y + +L+A++++ S NV
Sbjct: 116 --------------------SVVQHESGHFLVGYLLGVLPRHYEIPTLEAVRQNVS-NVT 154
Query: 217 AGSAFVDFEFLEEV---------------NAGKVSAKTLNKFSCIALAGVCTEYLIYGFS 261
FV FEFL++V N G +S+KTLN FSC+ L G+ TE++++G+S
Sbjct: 155 GRVEFVGFEFLKQVGAANQLMKDDVDGQMNQGNISSKTLNNFSCVILGGMVTEHILFGYS 214
Query: 262 EGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAAR 307
EG DI KL+ +L LGFT+ + ++ ++W+V NTV LL H+ AR
Sbjct: 215 EGLYSDIVKLNDVLRWLGFTESEKEAHIKWAVSNTVSLLHSHKEAR 260
>At1g56180 unknown protein
Length = 389
Score = 87.8 bits (216), Expect = 7e-18
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 175 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGTCLAQV-----SCYWP 226
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 227 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 284
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 285 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 344
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 345 WSVLQSYNLLKWHKAA 360
>At2g21960 unknown protein
Length = 332
Score = 63.5 bits (153), Expect = 1e-10
Identities = 73/247 (29%), Positives = 115/247 (46%), Gaps = 44/247 (17%)
Query: 81 QRLYTLDELKLNGIE-AESLLSP--------VDSTLGSIERTLQIAAIAGGLTA-WNAFG 130
+R +L EL GI+ AE+L P + + +GS T IA +AG L W F
Sbjct: 92 RRTTSLRELTTLGIKNAETLAIPSVRNDAAFLFTVVGS---TGFIAVLAGQLPGDWGFF- 147
Query: 131 ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
+ Y+ +G + L L S GL + +F Y R+ HEA HFL+AY
Sbjct: 148 -----VPYL-VGSISLVVLAVGSVSPGLLQAAISGFS-TFFPDYQERIAAHEAAHFLVAY 200
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
L+G+ GY SLD K+ +L +D + + +GK+ +K L++ + +A+AG
Sbjct: 201 LIGLPILGY---SLDIGKEHVNL--------IDERLAKLIYSGKLDSKELDRLAAVAMAG 249
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQ-----VRWSVLNTVLLLRR--- 302
+ E L Y G D+ L ++ +Q K+ ++ RW+VL + LL+
Sbjct: 250 LAAEGLKYDKVIGQSADLFSLQRFINR---SQPKISNEQQQNLTRWAVLYSASLLKNNKT 306
Query: 303 -HEAARA 308
HEA A
Sbjct: 307 IHEALMA 313
>rpl14 -chloroplast genome- ribosomal protein L14
Length = 122
Score = 30.8 bits (68), Expect = 0.97
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 5/59 (8%)
Query: 41 LEKLDKELLKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESL 99
L++ + +++ DD AA+ + D +G P G R FGA +P+ L L+ K+ + E L
Sbjct: 69 LKRNNGTIIRYDDNAAVVI--DQEGNPKGTRVFGA---IPRELRQLNFTKIVSLAPEVL 122
>At1g79560 FtsH like cell division protein
Length = 619
Score = 30.0 bits (66), Expect = 1.7
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 10/96 (10%)
Query: 181 HEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTL 240
HEAGH ++A+L P+ + + L G A S F E + V+ G + +
Sbjct: 380 HEAGHIVLAHL---FPR-FDWHAFSQLLPGG--KETAVSVFYPRE--DMVDQGYTTFGYM 431
Query: 241 NKFSCIALAGVCTEYLIYG--FSEGGLDDIRKLDSL 274
+A G C E +++G ++GG DD+ K+ +
Sbjct: 432 KMQMVVAHGGRCAERVVFGDNVTDGGKDDLEKITKI 467
>At2g14500 hypothetical protein
Length = 347
Score = 28.9 bits (63), Expect = 3.7
Identities = 17/48 (35%), Positives = 28/48 (57%), Gaps = 1/48 (2%)
Query: 255 YLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRR 302
Y+I FSE +D + L+SLLS ++K D +++W N +L+ R
Sbjct: 102 YIIDVFSENRID-LPPLESLLSDNFTFEQKGDKELKWQASNDQILVFR 148
>At1g50250 putative chloroplast FtsH protease
Length = 716
Score = 28.9 bits (63), Expect = 3.7
Identities = 27/120 (22%), Positives = 51/120 (42%), Gaps = 15/120 (12%)
Query: 171 SKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEV 230
S++ V HEAGH L+ ++P+ ++ + + + QAG E +
Sbjct: 514 SEEKKRLVAYHEAGHALVG---ALMPEYDPVAKISIIPRG-----QAGGLTFFAPSEERL 565
Query: 231 NAGKVSAKTLNKFSCIALAGVCTEYLIYG---FSEGGLDDIRKLD----SLLSGLGFTQK 283
+G S L +AL G E +I+G + G +D ++ ++ GF++K
Sbjct: 566 ESGLYSRSYLENQMAVALGGRVAEEVIFGDENVTTGASNDFMQVSRVARQMIERFGFSKK 625
>At1g05140 Unknown protein (YUP8H12.25)
Length = 441
Score = 28.9 bits (63), Expect = 3.7
Identities = 15/39 (38%), Positives = 20/39 (50%)
Query: 178 VIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQ 216
++ HE GHFL A L GI + + L K S NV+
Sbjct: 93 IVVHETGHFLAASLQGIRVSKFAIGFGPILAKFNSNNVE 131
>At5g42270 cell division protein FtsH
Length = 704
Score = 28.5 bits (62), Expect = 4.8
Identities = 27/120 (22%), Positives = 51/120 (42%), Gaps = 15/120 (12%)
Query: 171 SKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEV 230
S++ V HEAGH L+ ++P+ ++ + + + QAG E +
Sbjct: 502 SEEKKRLVAYHEAGHALVG---ALMPEYDPVAKISIIPRG-----QAGGLTFFAPSEERL 553
Query: 231 NAGKVSAKTLNKFSCIALAGVCTEYLIYG---FSEGGLDDIRKLD----SLLSGLGFTQK 283
+G S L +AL G E +I+G + G +D ++ ++ GF++K
Sbjct: 554 ESGLYSRSYLENQMAVALGGRVAEEVIFGDENVTTGASNDFMQVSRVARQMVERFGFSKK 613
>At5g17710 chloroplast GrpE protein
Length = 326
Score = 28.1 bits (61), Expect = 6.3
Identities = 15/63 (23%), Positives = 30/63 (46%), Gaps = 2/63 (3%)
Query: 156 GGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNV 215
G LG + V+T+G F H +++ ++ + ++ KG+ L + L + + V
Sbjct: 239 GSLGVIHVETVGKQFDPMLHEAIMREDSAEYEEGIVLEEYRKGFLLG--ERLLRPSMVKV 296
Query: 216 QAG 218
AG
Sbjct: 297 SAG 299
>At5g05730 anthranilate synthase component I-1 precursor (sp|P32068)
Length = 595
Score = 28.1 bits (61), Expect = 6.3
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Query: 1 MGYLALMHCVGVTSCPGQLKLVRVRIQCSTTNVGVSRRQLLE----KLDKELLKGDDRAA 56
MGYL C+ V S P L V+ + G S+R E +L+KELL+ + + A
Sbjct: 358 MGYLQARGCILVASSPEILTKVKQNKIVNRPLAGTSKRGKNEVEDKRLEKELLENEKQCA 417
>At5g45050 disease resistance protein-like
Length = 1372
Score = 27.7 bits (60), Expect = 8.2
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 4/73 (5%)
Query: 25 RIQCSTTNVGVSRRQLLEKLDKELLKGDDRAALALVKDLQGKPDGLR----CFGAARQVP 80
++ C N R L ++ ELLK D + + ++ +QG P L+ A RQVP
Sbjct: 725 KLSCLELNDCSRLRSLPNMVNLELLKALDLSGCSELETIQGFPRNLKELYLVGTAVRQVP 784
Query: 81 QRLYTLDELKLNG 93
Q +L+ +G
Sbjct: 785 QLPQSLEFFNAHG 797
>At5g44700 receptor-like protein kinase
Length = 1236
Score = 27.7 bits (60), Expect = 8.2
Identities = 12/46 (26%), Positives = 21/46 (45%)
Query: 79 VPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAAIAGGLT 124
+PQ + L L +E L P+ ST+G + + ++ LT
Sbjct: 712 IPQEIGNLQALNALNLEENQLSGPLPSTIGKLSKLFELRLSRNALT 757
>At3g51120 putative protein
Length = 1247
Score = 27.7 bits (60), Expect = 8.2
Identities = 15/45 (33%), Positives = 24/45 (53%)
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSG 277
G+V + L+ ++ I + + YL F E LDDI K+D + G
Sbjct: 396 GRVQNENLDAYAAIDVHNINLIYLRRKFLESLLDDINKVDEKVVG 440
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,555,141
Number of Sequences: 26719
Number of extensions: 272802
Number of successful extensions: 632
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 618
Number of HSP's gapped (non-prelim): 16
length of query: 309
length of database: 11,318,596
effective HSP length: 99
effective length of query: 210
effective length of database: 8,673,415
effective search space: 1821417150
effective search space used: 1821417150
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0544.1


