

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0541.9
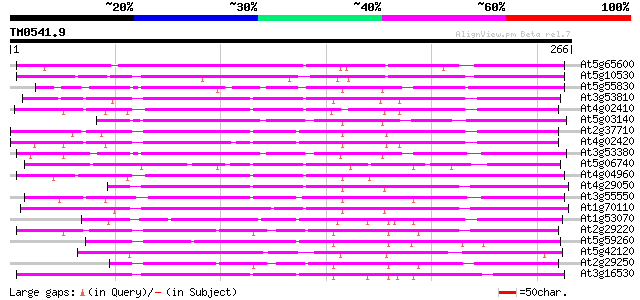
(266 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g65600 receptor protein kinase-like protein 138 3e-33
At5g10530 lectin-like protein kinase - like 130 9e-31
At5g55830 serine/threonine-specific kinase like protein 110 8e-25
At3g53810 serine/threonine-specific kinase like protein 99 2e-21
At4g02410 unknown protein 98 5e-21
At5g03140 receptor like protein kinase 96 3e-20
At2g37710 putative receptor-like protein kinase 96 3e-20
At4g02420 94 8e-20
At3g53380 receptor lectin kinase -like protein 93 1e-19
At5g06740 lectin-like protein kinase 91 8e-19
At4g04960 unknown protein 86 2e-17
At4g29050 serine/threonine-specific kinase like protein 86 3e-17
At3g55550 probable serine/threonine-specific protein kinase 85 3e-17
At1g70110 hypothetical protein 84 1e-16
At1g53070 protein kinase, putative 82 2e-16
At2g29220 putative protein kinase 77 1e-14
At5g59260 receptor-like protein kinase 75 4e-14
At5g42120 receptor lectin kinase-like protein 74 6e-14
At2g29250 putative protein kinase 74 8e-14
At3g16530 putative lectin 74 1e-13
>At5g65600 receptor protein kinase-like protein
Length = 675
Score = 138 bits (347), Expect = 3e-33
Identities = 92/269 (34%), Positives = 141/269 (52%), Gaps = 21/269 (7%)
Query: 4 MSLFVILISLFM-VAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVPL 62
MS ++ +SLF+ + V+S F F F+ + DI + GDA + T
Sbjct: 16 MSNSILFLSLFLFLPFVVDSLYFNFTSFRQGDPGDIFYHGDATPDE---DGTVNFNNAEQ 72
Query: 63 PSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKN 122
S G ++++ V +W KTG + F+TSF+ + + G GI FF+AP +++P
Sbjct: 73 TSQVGWITYSKKVPIWSHKTGKASDFSTSFSFKIDARNLSADGHGICFFLAPMGAQLPAY 132
Query: 123 STGGFLGLFSKDSAFDSYQNQIVAVEFDTYADT-WDP--FTSHIGIDINSIISSKTVPWR 179
S GGFL LF++ + + S +V VEFDT+ + WDP SH+GI+ NS++SS W
Sbjct: 133 SVGGFLNLFTRKNNYSS-SFPLVHVEFDTFNNPGWDPNDVGSHVGINNNSLVSSNYTSW- 190
Query: 180 TGNSLSTSAAFATVSYEPVTKTLSV-----LVKQIDQQKGLKFSTITTSLSFVVDLRTIL 234
+S S A +SY+ VTK LSV L D ++ ++SLS+++DL +L
Sbjct: 191 NASSHSQDICHAKISYDSVTKNLSVTWAYELTATSDPKE-------SSSLSYIIDLAKVL 243
Query: 235 PEWVRVGFSGATGQLVEQHRIHVWDFKSS 263
P V GF A G E+HR+ W+ SS
Sbjct: 244 PSDVMFGFIAAAGTNTEEHRLLSWELSSS 272
>At5g10530 lectin-like protein kinase - like
Length = 651
Score = 130 bits (326), Expect = 9e-31
Identities = 93/269 (34%), Positives = 140/269 (51%), Gaps = 25/269 (9%)
Query: 4 MSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVPLP 63
M+ ++L S +V V S F F + +I ++GDA A NG +ELT Y
Sbjct: 1 MANSILLFSFVLVLPFVCSVQFNISRFG-SDVSEIAYQGDARA-NGAVELTNIDY----T 54
Query: 64 SSSGRASFAEPVHLWDKKTGALAAFTT--SFTLDVQPTSSGFHGDGIAFFIAPFNSKIPK 121
+G A++ + V LW+ T + F+T SF +D + G +G G AFF+AP ++P
Sbjct: 55 CRAGWATYGKQVPLWNPGTSKPSDFSTRFSFRIDTRNVGYGNYGHGFAFFLAPARIQLPP 114
Query: 122 NSTGGFLGLF----SKDSAFDSYQNQIVAVEFDTYAD-TWDPF--TSHIGIDINSIISSK 174
NS GGFLGLF ++ SAF +V VEFDT+ + WDP SH+GI+ NS++SS
Sbjct: 115 NSAGGFLGLFNGTNNQSSAF-----PLVYVEFDTFTNPEWDPLDVKSHVGINNNSLVSSN 169
Query: 175 TVPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTIL 234
W S + + Y+ + LSV L+ +SLS+++DL +L
Sbjct: 170 YTSW-NATSHNQDIGRVLIFYDSARRNLSVSWTYDLTSDPLE----NSSLSYIIDLSKVL 224
Query: 235 PEWVRVGFSGATGQLVEQHRIHVWDFKSS 263
P V +GFS +G + E +R+ W+F SS
Sbjct: 225 PSEVTIGFSATSGGVTEGNRLLSWEFSSS 253
>At5g55830 serine/threonine-specific kinase like protein
Length = 681
Score = 110 bits (275), Expect = 8e-25
Identities = 89/266 (33%), Positives = 136/266 (50%), Gaps = 37/266 (13%)
Query: 13 LFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVPLPSSSGRASFA 72
+F+ + N+N FTF F + N +TF GD+ NGV+ LT+ GVP +SSG +
Sbjct: 24 IFVSSDNMN---FTFKSFTIRN---LTFLGDSHLRNGVVGLTREL-GVP-DTSSGTVIYN 75
Query: 73 EPVHLWDKKTGALAAFTTSFTLDVQ-----PTSSGFHGDGIAFFIAPFNSKIPKNSTGGF 127
P+ +D + A+F+T F+ VQ PTS+G DG+AFF++ N + S GG+
Sbjct: 76 NPIRFYDPDSNTTASFSTHFSFTVQNLNPDPTSAG---DGLAFFLSHDNDTL--GSPGGY 130
Query: 128 LGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIGIDINSIISSKT--------VP 177
LGL + +N+ VA+EFDT D DP +HIG+D++S+ S T +
Sbjct: 131 LGLVNSSQPM---KNRFVAIEFDTKLDPHFNDPNGNHIGLDVDSLNSISTSDPLLSSQID 187
Query: 178 WRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEW 237
++G S+++ + Y+ + L+V + D K LS +DL L
Sbjct: 188 LKSGKSITS-----WIDYKNDLRLLNVFLSYTDPVTTTK-KPEKPLLSVNIDLSPFLNGE 241
Query: 238 VRVGFSGATGQLVEQHRIHVWDFKSS 263
+ VGFSG+T E H I W FK+S
Sbjct: 242 MYVGFSGSTEGSTEIHLIENWSFKTS 267
>At3g53810 serine/threonine-specific kinase like protein
Length = 677
Score = 99.0 bits (245), Expect = 2e-21
Identities = 81/267 (30%), Positives = 131/267 (48%), Gaps = 26/267 (9%)
Query: 7 FVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFAS-NGVLELTKTAYGVPLPSS 65
F L+ M++ + N FT+ GF P T DI+ +G A + NG+L+LT T+
Sbjct: 10 FFFLLCQIMISSSQNLN-FTYNGFHPPLT-DISLQGLATVTPNGLLKLTNTSV-----QK 62
Query: 66 SGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTG 125
+G A E + D + G +++F+T+F + G GIAF +AP +P
Sbjct: 63 TGHAFCTERIRFKDSQNGNVSSFSTTFVFAIHSQIPTLSGHGIAFVVAP-TLGLPFALPS 121
Query: 126 GFLGLFSKDSAFDSYQNQIVAVEFDTY--ADTWDPFTSHIGIDINSIISSK--TVPWRTG 181
++GLF+ + + N I AVEFDT ++ DP +H+GID+N + S+ T +R
Sbjct: 122 QYIGLFNISNNGND-TNHIFAVEFDTIQSSEFGDPNDNHVGIDLNGLRSANYSTAGYRDD 180
Query: 182 NS-------LSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTIL 234
+ +S + Y+ + + V V D K K +S+V DL +IL
Sbjct: 181 HDKFQNLSLISRKRIQVWIDYDNRSHRIDVTVAPFDSDKPRK-----PLVSYVRDLSSIL 235
Query: 235 PEWVRVGFSGATGQLVEQHRIHVWDFK 261
E + VGFS ATG ++ +H + W F+
Sbjct: 236 LEDMYVGFSSATGSVLSEHFLVGWSFR 262
>At4g02410 unknown protein
Length = 674
Score = 97.8 bits (242), Expect = 5e-21
Identities = 86/276 (31%), Positives = 134/276 (48%), Gaps = 35/276 (12%)
Query: 3 LMSLFVILISLFMVAHNVNSET--FTFPGFQLPNTKDITFEGDA-FASNGVLELT-KTAY 58
L ++F I L N +S++ FT+ F P T +I+ +G A SNG+L+LT KT
Sbjct: 5 LFTIFFFFIILLSKPLNSSSQSLNFTYNSFHRPPT-NISIQGIATVTSNGILKLTDKTVI 63
Query: 59 GVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSK 118
S+G A + EP+ D +++F+T+F + + G G+AFFIAP N
Sbjct: 64 ------STGHAFYTEPIRFKDSPNDTVSSFSTTFVIGIYSGIPTISGHGMAFFIAP-NPV 116
Query: 119 IPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDT-----YADTWDPFTSHIGIDINSIISS 173
+ +LGLFS + + N I+AVEFDT + DT D +H+GI+INS+ S
Sbjct: 117 LSSAMASQYLGLFSSTNNGND-TNHILAVEFDTIMNPEFDDTND---NHVGININSLTSV 172
Query: 174 KTV---PWRTGNS------LSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSL 224
K+ W N +S V Y+ T + V + + K K +
Sbjct: 173 KSSLVGYWDEINQFNNLTLISRKRMQVWVDYDDRTNQIDVTMAPFGEVKPRK-----ALV 227
Query: 225 SFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDF 260
S V DL ++ + + +GFS ATG ++ +H + W F
Sbjct: 228 SVVRDLSSVFLQDMYLGFSAATGYVLSEHFVFGWSF 263
>At5g03140 receptor like protein kinase
Length = 711
Score = 95.5 bits (236), Expect = 3e-20
Identities = 72/232 (31%), Positives = 120/232 (51%), Gaps = 28/232 (12%)
Query: 42 GDAFASNGVLELTKTAYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSS 101
GDA +NG ++LT+ VP S++G+A + +PV +T + A+FTT F+ V +
Sbjct: 51 GDAHLNNGTIKLTREL-SVPT-STAGKALYGKPVKFRHPETKSPASFTTYFSFSVTNLNP 108
Query: 102 GFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPF 159
G G+AF I+P + STGGFLGL + + + VAVEFDT D D
Sbjct: 109 SSIGGGLAFVISPDEDYL--GSTGGFLGLTEETGSGSGF----VAVEFDTLMDVQFKDVN 162
Query: 160 TSHIGIDINSIISSKT-------VPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQ 212
+H+G+D+N+++S+ + ++GN+++ + ++Y+ + L+V V +
Sbjct: 163 GNHVGLDLNAVVSAAVADLGNVDIDLKSGNAVN-----SWITYDGSGRVLTVYVSYSN-- 215
Query: 213 KGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDFKSSF 264
+ LS +DL + + + VGFSG+T E H + W F SSF
Sbjct: 216 ----LKPKSPILSVPLDLDRYVSDSMFVGFSGSTQGSTEIHSVDWWSFSSSF 263
>At2g37710 putative receptor-like protein kinase
Length = 675
Score = 95.5 bits (236), Expect = 3e-20
Identities = 76/275 (27%), Positives = 129/275 (46%), Gaps = 29/275 (10%)
Query: 1 MGLMSLFVILISLFMVAHNVNSETFTFP---GFQLPNTKDITFEG-DAFASNGVLELTKT 56
M L L + F + +S++ F GF P D++ +G NG+L+LT T
Sbjct: 1 MFLKLLTIFFFFFFNLIFQSSSQSLNFAYNNGFNPPT--DLSIQGITTVTPNGLLKLTNT 58
Query: 57 AYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFN 116
+G A + +P+ D G +++F+TSF + + G GIAF +AP N
Sbjct: 59 TV-----QKTGHAFYTKPIRFKDSPNGTVSSFSTSFVFAIHSQIAILSGHGIAFVVAP-N 112
Query: 117 SKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIGIDINSIISSK 174
+ +P + ++GLF+ + + N + AVE DT T D +H+GIDINS+ S +
Sbjct: 113 ASLPYGNPSQYIGLFNLANN-GNETNHVFAVELDTILSTEFNDTNDNHVGIDINSLKSVQ 171
Query: 175 TVP---------WRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLS 225
+ P ++ +S V Y+ T + V + ++ K + ++
Sbjct: 172 SSPAGYWDEKGQFKNLTLISRKPMQVWVDYDGRTNKIDVTMAPFNEDKPTR-----PLVT 226
Query: 226 FVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDF 260
V DL ++L + + VGFS ATG ++ +H I W F
Sbjct: 227 AVRDLSSVLLQDMYVGFSSATGSVLSEHYILGWSF 261
>At4g02420
Length = 669
Score = 94.0 bits (232), Expect = 8e-20
Identities = 85/275 (30%), Positives = 135/275 (48%), Gaps = 30/275 (10%)
Query: 1 MGLMSLFVIL-ISLFMVAHNVNSET--FTFPGFQLPNTKDITFEGDA-FASNGVLELTKT 56
M + LF I +S F + +S+ FT+ GF+ P T DI+ G A NG+L+LT T
Sbjct: 1 MFFIKLFTIFFLSFFWQSLKSSSQIIDFTYNGFRPPPT-DISILGIATITPNGLLKLTNT 59
Query: 57 AYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFN 116
S+G A + +P+ D G +++F+T+F + HG +AF IAP N
Sbjct: 60 TM-----QSTGHAFYTKPIRFKDSPNGTVSSFSTTFVFAIHSQIPIAHG--MAFVIAP-N 111
Query: 117 SKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIGIDINSIISSK 174
++P S +LGLF+ + + +N + AVE DT + D +H+GIDINS+ S K
Sbjct: 112 PRLPFGSPLQYLGLFNVTNN-GNVRNHVFAVELDTIMNIEFNDTNNNHVGIDINSLNSVK 170
Query: 175 TVP---WRTGNS------LSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLS 225
+ P W + +S+ V ++ T + V + + K K +S
Sbjct: 171 SSPAGYWDENDQFHNLTLISSKRMQVWVDFDGPTHLIDVTMAPFGEVKPRK-----PLVS 225
Query: 226 FVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDF 260
V DL ++L + + VGFS ATG +V + + W F
Sbjct: 226 IVRDLSSVLLQDMFVGFSSATGNIVSEIFVLGWSF 260
>At3g53380 receptor lectin kinase -like protein
Length = 715
Score = 93.2 bits (230), Expect = 1e-19
Identities = 80/272 (29%), Positives = 134/272 (48%), Gaps = 33/272 (12%)
Query: 4 MSLFV-ILISLFMVAHNVNSET-FTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVP 61
MSLF+ IS+ + N + T F F + N K + GDA SNG++ LT+ VP
Sbjct: 1 MSLFLSFFISILLCFFNGATTTQFDFSTLAISNLKLL---GDARLSNGIVGLTRDL-SVP 56
Query: 62 LPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPK 121
S +G+ ++ P+ T +F++ F+ + + G G+AF I+P + I
Sbjct: 57 -NSGAGKVLYSNPIRFRQPGTHFPTSFSSFFSFSITNVNPSSIGGGLAFVISPDANSI-- 113
Query: 122 NSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADT--WDPFTSHIGIDINSIISSKT---- 175
GG LGL + + + VAVEFDT D D ++H+G D+N ++SS +
Sbjct: 114 GIAGGSLGLTGPNGSGSKF----VAVEFDTLMDVDFKDINSNHVGFDVNGVVSSVSGDLG 169
Query: 176 ---VPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRT 232
+ ++GN++++ + Y+ +T+ +V V + + + LSF +DL
Sbjct: 170 TVNIDLKSGNTINS-----WIEYDGLTRVFNVSVSYSNLKPKVPI------LSFPLDLDR 218
Query: 233 ILPEWVRVGFSGATGQLVEQHRIHVWDFKSSF 264
+ +++ VGFSG+T E H I W F SSF
Sbjct: 219 YVNDFMFVGFSGSTQGSTEIHSIEWWSFSSSF 250
>At5g06740 lectin-like protein kinase
Length = 652
Score = 90.5 bits (223), Expect = 8e-19
Identities = 75/265 (28%), Positives = 132/265 (49%), Gaps = 28/265 (10%)
Query: 8 VILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVP---LPS 64
+ LI + V F FPGF + N ++ +++ G +++T G P + +
Sbjct: 10 LFLILTCKIETQVKCLKFDFPGFNVSNELEL-IRDNSYIVFGAIQVTPDVTGGPGGTIAN 68
Query: 65 SSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQ-PTSSGFHGDGIAFFIAPFNSKIPKNS 123
+GRA + +P LW K A F T+F +++ T G G+G+AF + P P+NS
Sbjct: 69 QAGRALYKKPFRLWSKHKSA--TFNTTFVINISNKTDPG--GEGLAFVLTP-EETAPQNS 123
Query: 124 TGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTWDPFT-SHIGIDINSIISSKTVPWRTGN 182
+G +LG+ ++ + ++ +++IV+VEFDT D +H+ +++N+I +S +G
Sbjct: 124 SGMWLGMVNERTNRNN-ESRIVSVEFDTRKSHSDDLDGNHVALNVNNI-NSVVQESLSGR 181
Query: 183 SLSTSAAF---ATVSYEPVTKTLSVLVKQ---IDQQKGLKFSTITTSLSFVVDLRTILPE 236
+ + A V Y+ K LSV V + + +Q+ L FS +DL LPE
Sbjct: 182 GIKIDSGLDLTAHVRYDG--KNLSVYVSRNLDVFEQRNLVFSR-------AIDLSAYLPE 232
Query: 237 WVRVGFSGATGQLVEQHRIHVWDFK 261
V VGF+ +T E + + W F+
Sbjct: 233 TVYVGFTASTSNFTELNCVRSWSFE 257
>At4g04960 unknown protein
Length = 686
Score = 86.3 bits (212), Expect = 2e-17
Identities = 73/267 (27%), Positives = 128/267 (47%), Gaps = 16/267 (5%)
Query: 4 MSLFVILISLFMVAHN-VNSETFTFPGFQLPNTKDITFEGDAFASNGVLELT-KTAYGVP 61
M + L++LF++ N +++ F F GF ++ +++ G A + +L LT +T++
Sbjct: 1 MKALLFLLTLFLILPNPISAIDFIFNGFN-DSSSNVSLFGIATIESKILTLTNQTSF--- 56
Query: 62 LPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPK 121
++GRA + + D T ++ F+TSF + P + G GI F AP ++ I
Sbjct: 57 ---ATGRALYNRTIRTKDPITSSVLPFSTSFIFTMAPYKNTLPGHGIVFLFAP-STGING 112
Query: 122 NSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIGIDINS---IISSKTV 176
+S+ LGLF+ + + N I VEFD +A+ D +H+GID+NS + S+ +
Sbjct: 113 SSSAQHLGLFNLTNNGNP-SNHIFGVEFDVFANQEFSDIDANHVGIDVNSLHSVYSNTSG 171
Query: 177 PWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPE 236
W + V V + Q K LS ++L ++ +
Sbjct: 172 YWSDDGVVFKPLKLNDGRNYQVWIDYRDFVVNVTMQVAGKIRPKIPLLSTSLNLSDVVED 231
Query: 237 WVRVGFSGATGQLVEQHRIHVWDFKSS 263
+ VGF+ ATG+LV+ H+I W F +S
Sbjct: 232 EMFVGFTAATGRLVQSHKILAWSFSNS 258
>At4g29050 serine/threonine-specific kinase like protein
Length = 669
Score = 85.5 bits (210), Expect = 3e-17
Identities = 68/231 (29%), Positives = 109/231 (46%), Gaps = 24/231 (10%)
Query: 47 SNGVLELTKTAYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGD 106
S G+++LT ++ S G + PV + G +++F+T+F + + G
Sbjct: 45 SKGLMKLTNSS-----EFSYGHVFYNSPVRFKNSPNGTVSSFSTTFVFAIVSNVNALDGH 99
Query: 107 GIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIG 164
G+AF I+P +P +S+ +LGLF+ + D N IVAVEFDT+ + D +H+G
Sbjct: 100 GLAFVISP-TKGLPYSSSSQYLGLFNLTNNGDP-SNHIVAVEFDTFQNQEFDDMDNNHVG 157
Query: 165 IDINSIISSKTV----------PWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKG 214
IDINS+ S K ++ ++ A + Y+ + L+V + I K
Sbjct: 158 IDINSLSSEKASTAGYYEDDDGTFKNIRLINQKPIQAWIEYDSSRRQLNVTIHPIHLPK- 216
Query: 215 LKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDFKSSFT 265
LS DL L + + VGF+ ATG+L H I W FK + T
Sbjct: 217 ----PKIPLLSLTKDLSPYLFDSMYVGFTSATGRLRSSHYILGWTFKLNGT 263
>At3g55550 probable serine/threonine-specific protein kinase
Length = 684
Score = 85.1 bits (209), Expect = 3e-17
Identities = 80/270 (29%), Positives = 122/270 (44%), Gaps = 29/270 (10%)
Query: 8 VILISLFMVAHNVNS--ETFTFPGFQLPNTKDITFEGDA-FASNGVLELTKTAYGVPLPS 64
VIL+ L + H V+S + F+F GF+ + ++T G A A G + LT V
Sbjct: 7 VILLLLIFLTHLVSSLIQDFSFIGFKKASP-NLTLNGVAEIAPTGAIRLTTETQRV---- 61
Query: 65 SSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNST 124
G A ++ P+ +F+TSF + + P G G+AF I P + +
Sbjct: 62 -IGHAFYSLPIRFKPIGVNRALSFSTSFAIAMVPEFVTLGGHGLAFAITP-TPDLRGSLP 119
Query: 125 GGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIGIDINSIISSKTVPWRTGN 182
+LGL + ++ + AVEFDT D D +H+GIDINS+ SS + P
Sbjct: 120 SQYLGLLNSSRV--NFSSHFFAVEFDTVRDLEFEDINDNHVGIDINSMESSISTPAGYFL 177
Query: 183 SLSTSAAF---------ATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTI 233
+ ST A + Y+ K L V + ++ L LS+ VDL ++
Sbjct: 178 ANSTKKELFLDGGRVIQAWIDYDSNKKRLDVKLSPFSEKPKLSL------LSYDVDLSSV 231
Query: 234 LPEWVRVGFSGATGQLVEQHRIHVWDFKSS 263
L + + VGFS +TG L H I W+F S
Sbjct: 232 LGDEMYVGFSASTGLLASSHYILGWNFNMS 261
>At1g70110 hypothetical protein
Length = 666
Score = 83.6 bits (205), Expect = 1e-16
Identities = 72/272 (26%), Positives = 124/272 (45%), Gaps = 24/272 (8%)
Query: 6 LFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASN-GVLELTKTAYGVPLPS 64
+ ++ + LF V +V + G + + ++ G A+ +N G++ LT + P
Sbjct: 2 VLLLFLVLFFVPESVVCQRPNPNGVEFNTSGNMYTSGSAYINNNGLIRLTNST-----PQ 56
Query: 65 SSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNST 124
++G+ + + + + G +++F+T+F ++ + + G GIAF I P P T
Sbjct: 57 TTGQVFYNDQLRFKNSVNGTVSSFSTTFVFSIEFHNGIYGGYGIAFVICPTRDLSPTFPT 116
Query: 125 GGFLGLFSKDSAFDSYQNQIVAVEFDTYADTW--DPFTSHIGIDINSIISSKTV------ 176
+LGLF++ + D +N IVAVE DT D D +H+GIDIN+++S
Sbjct: 117 -TYLGLFNRSNMGDP-KNHIVAVELDTKVDQQFEDKDANHVGIDINTLVSDTVALAGYYM 174
Query: 177 ---PWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTI 233
+R+ S + Y+ K ++V + + K LS DL
Sbjct: 175 DNGTFRSLLLNSGQPMQIWIEYDSKQKQINVTLHPLYVPK-----PKIPLLSLEKDLSPY 229
Query: 234 LPEWVRVGFSGATGQLVEQHRIHVWDFKSSFT 265
L E + VGF+ TG L H I W FK + T
Sbjct: 230 LLELMYVGFTSTTGDLTASHYILGWTFKMNGT 261
>At1g53070 protein kinase, putative
Length = 272
Score = 82.4 bits (202), Expect = 2e-16
Identities = 75/250 (30%), Positives = 118/250 (47%), Gaps = 34/250 (13%)
Query: 35 TKDITFEGDAF--------ASNGVLELTKTAYGVPLPSSSGRASFAEPVHLWDKKTGALA 86
T I+F GDA + +G + LT+ +P S GRA F P+ + AL
Sbjct: 35 TDPISFHGDAEYGPDTDGKSRSGAIALTRDN----IPFSHGRAIFTTPI-TFKPNASALY 89
Query: 87 AFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVA 146
F TSFT + P ++ G G+AF + P N S G+L L ++ + + N + A
Sbjct: 90 PFKTSFTFSITPKTNPNQGHGLAFIVVPSNQN-DAGSGLGYLSLLNRTNN-GNPNNHLFA 147
Query: 147 VEFDTYAD--TWDPFTSHIGIDIN---SIISSKTVPW---RTG-----NSLSTSAAF-AT 192
VEFD + D D +H+GIDIN S++S K+ W R+G LS+ + A
Sbjct: 148 VEFDVFQDKSLGDMNDNHVGIDINSVDSVVSVKSGYWVMTRSGWLFKDLKLSSGDRYKAW 207
Query: 193 VSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQ 252
+ Y K +SV + +K + + DL ++ E + GF+G+ G+ VE+
Sbjct: 208 IEYNNNYKVVSVTIGLAHLKKPNR-----PLIEAKFDLSKVIHEVMYTGFAGSMGRGVER 262
Query: 253 HRIHVWDFKS 262
H I W F++
Sbjct: 263 HEIWDWTFQN 272
>At2g29220 putative protein kinase
Length = 627
Score = 77.0 bits (188), Expect = 1e-14
Identities = 77/273 (28%), Positives = 124/273 (45%), Gaps = 34/273 (12%)
Query: 4 MSLFVILISLFMVAHNVNSET-FTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVPL 62
++L +I +S F+ + ET F GF N + F +G+LELT T+
Sbjct: 7 IALTIIFLSYFVSCVSSQRETKFLNHGFLGANL--LNFGSSKVYPSGLLELTNTSM---- 60
Query: 63 PSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAP---FNSKI 119
G+A P+ L + + +F+TSF + +G G G+AF I+P F+
Sbjct: 61 -RQIGQAFHGFPIPLSNPNSTNSVSFSTSFIFAITQ-GTGAPGHGLAFVISPSMDFSGAF 118
Query: 120 PKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTY--ADTWDPFTSHIGIDINSIISSKTVP 177
P N +LGLF+ + +S N+I+A+EFDT + D +H+GID+N +IS + P
Sbjct: 119 PSN----YLGLFNTSNNGNSL-NRILAIEFDTVQAVELNDIDDNHVGIDLNGVISIASAP 173
Query: 178 W-----RTGNSLSTSAAFAT-----VSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFV 227
R ++S A + Y L+V + +D+ K LS
Sbjct: 174 AAYFDDREAKNISLRLASGKPVRVWIEYNATETMLNVTLAPLDRPK-----PSIPLLSRK 228
Query: 228 VDLRTILPEWVRVGFSGATGQLVEQHRIHVWDF 260
++L I + VGFS +TG + H + W F
Sbjct: 229 MNLSGIFSQEHHVGFSASTGTVASSHFVLGWSF 261
>At5g59260 receptor-like protein kinase
Length = 674
Score = 75.1 bits (183), Expect = 4e-14
Identities = 65/243 (26%), Positives = 109/243 (44%), Gaps = 25/243 (10%)
Query: 37 DITFEGDAFASNGVLELTKTAYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDV 96
D+ +G A ++G L LT + +G A + P+ + + F+T F +
Sbjct: 39 DLELDGMANTNHGPLHLTNNTN-----TGTGHAFYNIPIKFTASSLSSFS-FSTEFVFAI 92
Query: 97 QPTSSGFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTY--AD 154
P +G G+AF ++P S LG+F++ + + I AVE DT ++
Sbjct: 93 FPLQKSTYGHGMAFVVSPTKDLRSNGSANSNLGIFNRAND-NKTATHIFAVELDTNQNSE 151
Query: 155 TWDPFTSHIGIDINSIISSKTVPW-----RTGNSLSTSAA-----FATVSYEPVTKTLSV 204
++D + +GIDINSI+S ++ R G ++S A + Y+ + K L+V
Sbjct: 152 SFDKGGNDVGIDINSIVSVESADASYFNARKGKNISLPLASGKSILVWIDYDGIEKVLNV 211
Query: 205 LVKQIDQQK--GLKFSTITTS----LSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVW 258
+ + K FS+ LS ++L I E + VGFSG+TG + I W
Sbjct: 212 TLAPVQTPKPDSPYFSSFIKPKVPLLSRSINLSEIFTETMYVGFSGSTGSIKSNQYILGW 271
Query: 259 DFK 261
FK
Sbjct: 272 SFK 274
>At5g42120 receptor lectin kinase-like protein
Length = 691
Score = 74.3 bits (181), Expect = 6e-14
Identities = 68/244 (27%), Positives = 106/244 (42%), Gaps = 29/244 (11%)
Query: 33 PNTKDITFEGDAFASNGVLELTK---------TAYGVPLPSSSGRASFAEPVHLWDKKTG 83
P +++T GDAF + + LT+ T P S GRA + P+ + T
Sbjct: 30 PPLQNLTLYGDAFFRDRTISLTQQQPCFPSVTTPPSKPSSSGIGRALYVYPIKFLEPSTN 89
Query: 84 ALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQ 143
A+F+ F+ + + S GDG AF I + GFLGL + D +F
Sbjct: 90 TTASFSCRFSFSIIASPSCPFGDGFAFLITSNADSFV--FSNGFLGLPNPDDSF------ 141
Query: 144 IVAVEFDTYADT--WDPFTSHIGIDINSIISSKTVP--WRTGNSLSTSAAFATVSYEPVT 199
+AVEFDT D D +H+GID++SI S +V + + S A + Y V
Sbjct: 142 -IAVEFDTRFDPVHGDINDNHVGIDVSSIFSVSSVDAISKGFDLKSGKKMMAWIEYSDVL 200
Query: 200 KTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQ-HRIHVW 258
K + V V + + LS +DL + E++ VGFS + + H + W
Sbjct: 201 KLIRVWVGY------SRVKPTSPVLSTQIDLSGKVKEYMHVGFSASNAGIGSALHIVERW 254
Query: 259 DFKS 262
F++
Sbjct: 255 KFRT 258
>At2g29250 putative protein kinase
Length = 623
Score = 73.9 bits (180), Expect = 8e-14
Identities = 69/228 (30%), Positives = 105/228 (45%), Gaps = 31/228 (13%)
Query: 48 NGVLELTKTAYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDG 107
+G LELT T+ G+A P+ + + L +F TSF + P G G G
Sbjct: 50 SGHLELTNTSM-----RQIGQAFHGFPIPFLNPNSSNLVSFPTSFVFAITP-GPGAPGHG 103
Query: 108 IAFFIAP---FNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTY--ADTWDPFTSH 162
+AF I+P F+ +P N +LGLF+ + +S N I+AVEFDT + D +H
Sbjct: 104 LAFVISPSLDFSGALPSN----YLGLFNTSNNGNSL-NCILAVEFDTVQAVELNDIDDNH 158
Query: 163 IGIDINSIISSKTVPW-----RTGNSLSTSAAFAT-----VSYEPVTKTLSVLVKQIDQQ 212
+GID+N +IS ++ R ++S A + Y L+V + +D+
Sbjct: 159 VGIDLNGVISIESTSAEYFDDREAKNISLRLASGKPIRVWIEYNATETMLNVTLAPLDRP 218
Query: 213 KGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDF 260
K LS ++L I+ E VGFS ATG + H + W F
Sbjct: 219 KPK-----LPLLSRKLNLSGIISEENYVGFSAATGTVTSSHFVLGWSF 261
>At3g16530 putative lectin
Length = 276
Score = 73.6 bits (179), Expect = 1e-13
Identities = 67/278 (24%), Positives = 123/278 (44%), Gaps = 29/278 (10%)
Query: 4 MSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVPLP 63
+ V+ ++ A N ++F ++ D + +G L +T+ P
Sbjct: 6 LCFLVLFLANAAFAVKFNFDSFDGSNLLFLGDAELGPSSDGVSRSGALSMTRDEN----P 61
Query: 64 SSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNS 123
S G+ + + T + +F TSFT + P + G G AF I P + S
Sbjct: 62 FSHGQGLYINQIPFKPSNTSSPFSFETSFTFSITPRTKPNSGQGFAFIITP-EADNSGAS 120
Query: 124 TGGFLGLFSKDSAFDSYQNQIVAVEFDTY--ADTWDPFTSHIGIDIN---SIISSKTVPW 178
GG+LG+ +K + +N I+A+EFDT+ + D +H+G++IN S+++ K W
Sbjct: 121 DGGYLGILNKTND-GKPENHILAIEFDTFQNKEFLDISGNHVGVNINSMTSLVAEKAGYW 179
Query: 179 ---RTGN---------SLSTSAAF-ATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLS 225
R G +LS+ F A V + T++V + + +K + +
Sbjct: 180 VQTRVGKRKVWSFKDVNLSSGERFKAWVEFRNKDSTITVTLAPENVKKPKRALIEAPRV- 238
Query: 226 FVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDFKSS 263
L +L + + GF+G+ G+ VE+H I W F+++
Sbjct: 239 ----LNEVLLQNMYAGFAGSMGRAVERHDIWSWSFENA 272
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,748,605
Number of Sequences: 26719
Number of extensions: 240330
Number of successful extensions: 713
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 593
Number of HSP's gapped (non-prelim): 55
length of query: 266
length of database: 11,318,596
effective HSP length: 98
effective length of query: 168
effective length of database: 8,700,134
effective search space: 1461622512
effective search space used: 1461622512
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0541.9


