

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.9
(157 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
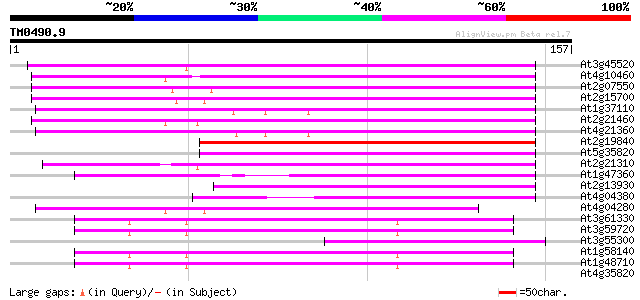
Score E
Sequences producing significant alignments: (bits) Value
At3g45520 copia-like polyprotein 74 2e-14
At4g10460 putative retrotransposon 69 1e-12
At2g07550 putative retroelement pol polyprotein 67 5e-12
At2g15700 copia-like retroelement pol polyprotein 65 2e-11
At1g37110 65 2e-11
At2g21460 putative retroelement pol polyprotein 63 7e-11
At4g21360 putative transposable element 62 1e-10
At2g19840 copia-like retroelement pol polyprotein 58 2e-09
At5g35820 copia-like retrotransposable element 57 5e-09
At2g21310 putative retroelement pol polyprotein 57 5e-09
At1g47360 polyprotein, putative 55 1e-08
At2g13930 putative retroelement pol polyprotein 54 3e-08
At4g04380 putative polyprotein 53 8e-08
At4g04280 putative transposon protein 52 2e-07
At3g61330 copia-type polyprotein 42 2e-04
At3g59720 copia-type reverse transcriptase-like protein 42 2e-04
At3g55300 putative protein 42 2e-04
At1g58140 hypothetical protein 42 2e-04
At1g48710 hypothetical protein 42 2e-04
At4g35820 putative protein 38 0.002
>At3g45520 copia-like polyprotein
Length = 1363
Score = 74.3 bits (181), Expect = 2e-14
Identities = 46/160 (28%), Positives = 81/160 (49%), Gaps = 18/160 (11%)
Query: 6 GAKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIA----------------- 48
GA+ EV + +G ++ +W+ ++ + GL LR+ +
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEERE 62
Query: 49 -IVDWNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRR 107
+ + E K+KA I L VSD V+ T+ + + L+ YMSK L +++ +KQ+
Sbjct: 63 KMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKL 122
Query: 108 YSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
YS KM E ++ +++ F +I+ADL L V V DE++AI+
Sbjct: 123 YSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAIL 162
>At4g10460 putative retrotransposon
Length = 1230
Score = 68.6 bits (166), Expect = 1e-12
Identities = 46/159 (28%), Positives = 82/159 (50%), Gaps = 20/159 (12%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRD------------------KSLDIA 48
A+ E+ + +G ++ LW+ ++ + GL ALR+ K A
Sbjct: 4 ARVEMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEA 63
Query: 49 IVDWNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRY 108
+++ E ++KA I L VSD V+ + T + + L+ YMSK L +++ +KQ+ Y
Sbjct: 64 LME--EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLY 121
Query: 109 SLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
S KMQE ++ +++ F ++ADL V V DE++AI+
Sbjct: 122 SYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAIL 160
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 66.6 bits (161), Expect = 5e-12
Identities = 45/159 (28%), Positives = 80/159 (50%), Gaps = 18/159 (11%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKS--------LDIAIVDWNEM--- 55
A+ EV + +G ++ +W+ ++ + GL+ AL++ LD + D+ E
Sbjct: 4 ARIEVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEK 63
Query: 56 -------KKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRY 108
KKKA I L V+D V+ +T + L+ YMSK L +++ KQ+ Y
Sbjct: 64 FEALEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLY 123
Query: 109 SLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
S KM E ++ +++ F I+ DL + V + DE++AI+
Sbjct: 124 SFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAIL 162
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 64.7 bits (156), Expect = 2e-11
Identities = 45/167 (26%), Positives = 78/167 (45%), Gaps = 26/167 (15%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSL----------------DIAIV 50
A+ EV R +GT +F LW++R+ GL L D+ L D +
Sbjct: 5 ARVEVDRFDGTGDFSLWKVRMLAHYGVLGLKGILNDEQLLRDPPVIEEEAAVAGRDYHVG 64
Query: 51 DWN----------EMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDK 100
D+ E +KA LI L V + V+ + T +W TL+ YM +L ++
Sbjct: 65 DFELPSNVDLIKFEKSEKAKDLIVLNVGNQVLRKIKNCETAAAMWSTLKRLYMETSLPNR 124
Query: 101 LLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
+ ++ + Y+ KM + + +V+ F ++ DL +GV V +E +AI+
Sbjct: 125 IYLQLKFYTYKMTDSRSIDGNVDDFLKLVTDLNNIGVDVTEEVQAIL 171
>At1g37110
Length = 1356
Score = 64.7 bits (156), Expect = 2e-11
Identities = 45/164 (27%), Positives = 79/164 (47%), Gaps = 24/164 (14%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAG------ 61
+ E+ NG R+F LW++R++ L GL L D SL + K+ +G
Sbjct: 7 RVEIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSG 66
Query: 62 ------LIKLCVSDD----VMNHTLDLTTLN--------DVWDTLESQYMSKTLMDKLLV 103
+K+ S+ ++NH D+ L D+W TL +YM +L +++
Sbjct: 67 TKEVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYT 126
Query: 104 KQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
+ + YS KM + +V+ F I+A+L L ++VD+E +AI+
Sbjct: 127 QLKLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAIL 170
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 62.8 bits (151), Expect = 7e-11
Identities = 43/159 (27%), Positives = 78/159 (49%), Gaps = 18/159 (11%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRD------KSLDIAIVD--------- 51
A+ EV + +G ++ +W+ ++ L GL AL++ K ++ + +
Sbjct: 4 ARIEVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLR 63
Query: 52 ---WNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRY 108
E ++KA I L V+D V+ + + L+ YMSK L +++ KQ+ Y
Sbjct: 64 RELLEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLY 123
Query: 109 SLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
S KM E ++ +++ F I+ADL V V DE++AI+
Sbjct: 124 SFKMSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAIL 162
>At4g21360 putative transposable element
Length = 1308
Score = 62.4 bits (150), Expect = 1e-10
Identities = 46/167 (27%), Positives = 77/167 (45%), Gaps = 27/167 (16%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGL----- 62
K E+ NG R+F LW++R++ L GL AL D +L I+ KK++
Sbjct: 6 KVEIKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDDETD 65
Query: 63 ----------IKLCVSDD----VMNHTLDLTTLN--------DVWDTLESQYMSKTLMDK 100
IK SD ++NH D L ++W TL +M +L ++
Sbjct: 66 SKKTEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSLPNR 125
Query: 101 LLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
+ + R YS KM + + + + F I+A+L L ++V +E +AI+
Sbjct: 126 IYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAIL 172
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 58.2 bits (139), Expect = 2e-09
Identities = 27/94 (28%), Positives = 58/94 (60%)
Query: 54 EMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQ 113
+ +KA +I + V D V+ + + T + W TL+ Y+ K+L +++ ++ + Y+ +MQ
Sbjct: 39 DQDEKAMDMIFINVGDKVLRNIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYRMQ 98
Query: 114 EGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
+ L+++V+ F +++DL L ++V DE +AI+
Sbjct: 99 DSKTLEENVDEFQKMISDLNNLQIQVPDEVQAIL 132
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 56.6 bits (135), Expect = 5e-09
Identities = 29/94 (30%), Positives = 55/94 (57%)
Query: 54 EMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQ 113
E + KA I L + ++V+ + T + L+ +M+K+L +++ +KQR Y KM
Sbjct: 74 EKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQRLYGYKMS 133
Query: 114 EGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
E ++++VN F +++DL + V V DE++AI+
Sbjct: 134 ENMTMEENVNDFFKLISDLENVKVVVPDEDQAIV 167
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 56.6 bits (135), Expect = 5e-09
Identities = 39/156 (25%), Positives = 77/156 (49%), Gaps = 21/156 (13%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVD------------------ 51
EV +L+G ++ LW+ ++ + GL + L + D AI +
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEE---DEAIEEEESTAETDSLLTKTEDKV 63
Query: 52 WNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLK 111
E + KA + L + + V+ + T + L+ +M+K+L +++ +KQR Y K
Sbjct: 64 LKEKRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYK 123
Query: 112 MQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
M + ++++VN F +++DL + V V DE++AI+
Sbjct: 124 MSDSMTIEENVNDFFKLISDLENVKVSVPDEDQAIV 159
>At1g47360 polyprotein, putative
Length = 1182
Score = 55.5 bits (132), Expect = 1e-08
Identities = 35/129 (27%), Positives = 65/129 (50%), Gaps = 15/129 (11%)
Query: 19 NFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSDDVMNHTLDL 78
+F LW+ R+ L+ GL + K++ + E +K I+LC
Sbjct: 14 DFSLWKTRIMAHLSVIGLKDVVIGKTITPLTAEEEEDPEKK---IELC------------ 58
Query: 79 TTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVK 138
T + W+TL+ +M ++L ++ + Y+ KMQE + ++++ F I+ADL L ++
Sbjct: 59 QTAQEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLKIVADLNHLQIE 118
Query: 139 VDDENKAII 147
V DE +AI+
Sbjct: 119 VTDEVQAIL 127
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 53.9 bits (128), Expect = 3e-08
Identities = 29/90 (32%), Positives = 51/90 (56%)
Query: 58 KAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGD 117
KA +I L V+D V+ T + W+TL+ +M ++L ++ + Y+ KMQE
Sbjct: 48 KAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKK 107
Query: 118 LQDHVNAFNNILADLTRLGVKVDDENKAII 147
+ ++++ F I+ADL L + V DE +AI+
Sbjct: 108 IDENIDDFLKIVADLNHLQIDVTDEVQAIL 137
>At4g04380 putative polyprotein
Length = 778
Score = 52.8 bits (125), Expect = 8e-08
Identities = 31/96 (32%), Positives = 51/96 (52%), Gaps = 13/96 (13%)
Query: 52 WNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLK 111
+ E K+K I L VSD V+ + + YMSK L +++ +KQ+ Y K
Sbjct: 49 FEEKKRKTRSTIVLSVSDRVL-------------EASDRLYMSKALPNQIYLKQKLYRFK 95
Query: 112 MQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII 147
M E ++ +++ F +I+ADL L V V DE++ I+
Sbjct: 96 MSENLSMEGNIDEFLHIVADLENLNVLVSDEDQTIL 131
>At4g04280 putative transposon protein
Length = 1104
Score = 51.6 bits (122), Expect = 2e-07
Identities = 36/151 (23%), Positives = 66/151 (42%), Gaps = 27/151 (17%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRD----KSLDIAIVDWN---------- 53
K E+ NG R+F W++R++ L GL +L D K++ +A +
Sbjct: 2 KVEIKTFNGDRDFSFWKIRIEAQLGVLGLKNSLTDFKLTKTVPVAKKEEKESEYEDDASD 61
Query: 54 -------------EMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDK 100
E ++A I ++D V+ T ++W TL +M +L+++
Sbjct: 62 IKQATEEPDPIKFEQSEQAKNFIINHITDTVLLKVQHCKTAAEIWATLNKLFMETSLLNR 121
Query: 101 LLVKQRRYSLKMQEGGDLQDHVNAFNNILAD 131
+ + + YS KM + + +V+ F ILAD
Sbjct: 122 IYTQLKLYSFKMVDTLSIDQNVDEFLRILAD 152
>At3g61330 copia-type polyprotein
Length = 1352
Score = 41.6 bits (96), Expect = 2e-04
Identities = 32/138 (23%), Positives = 63/138 (45%), Gaps = 15/138 (10%)
Query: 19 NFGLWQMRVKDLLA--------QQGLHKALRDKSLDIA----IVDWNEMKKKAAGLIKLC 66
N+ W +R+K +L ++G + + SL + D + KKA LI
Sbjct: 17 NYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQG 76
Query: 67 VSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRR---YSLKMQEGGDLQDHVN 123
+ +D ++ T+ + W+ L + Y + K+ ++ R +L+M+EG + D+ +
Sbjct: 77 LDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFS 136
Query: 124 AFNNILADLTRLGVKVDD 141
+ +L R G K+DD
Sbjct: 137 RVLTVTNNLKRNGEKLDD 154
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 41.6 bits (96), Expect = 2e-04
Identities = 32/138 (23%), Positives = 63/138 (45%), Gaps = 15/138 (10%)
Query: 19 NFGLWQMRVKDLLA--------QQGLHKALRDKSLDIA----IVDWNEMKKKAAGLIKLC 66
N+ W +R+K +L ++G + + SL + D + KKA LI
Sbjct: 17 NYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQG 76
Query: 67 VSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRR---YSLKMQEGGDLQDHVN 123
+ +D ++ T+ + W+ L + Y + K+ ++ R +L+M+EG + D+ +
Sbjct: 77 LDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFS 136
Query: 124 AFNNILADLTRLGVKVDD 141
+ +L R G K+DD
Sbjct: 137 RVLTVTNNLKRNGEKLDD 154
>At3g55300 putative protein
Length = 262
Score = 41.6 bits (96), Expect = 2e-04
Identities = 19/62 (30%), Positives = 36/62 (57%)
Query: 89 ESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAIIF 148
E Y +KTL + + +K + S KM E ++++++ ++ADL L + DE++A+ F
Sbjct: 37 EGNYQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQALQF 96
Query: 149 CA 150
A
Sbjct: 97 MA 98
>At1g58140 hypothetical protein
Length = 1320
Score = 41.6 bits (96), Expect = 2e-04
Identities = 32/138 (23%), Positives = 63/138 (45%), Gaps = 15/138 (10%)
Query: 19 NFGLWQMRVKDLLA--------QQGLHKALRDKSLDIA----IVDWNEMKKKAAGLIKLC 66
N+ W +R+K +L ++G + + SL + D + KKA LI
Sbjct: 17 NYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQG 76
Query: 67 VSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRR---YSLKMQEGGDLQDHVN 123
+ +D ++ T+ + W+ L + Y + K+ ++ R +L+M+EG + D+ +
Sbjct: 77 LDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFS 136
Query: 124 AFNNILADLTRLGVKVDD 141
+ +L R G K+DD
Sbjct: 137 RVLTVTNNLKRNGEKLDD 154
>At1g48710 hypothetical protein
Length = 1352
Score = 41.6 bits (96), Expect = 2e-04
Identities = 32/138 (23%), Positives = 63/138 (45%), Gaps = 15/138 (10%)
Query: 19 NFGLWQMRVKDLLA--------QQGLHKALRDKSLDIA----IVDWNEMKKKAAGLIKLC 66
N+ W +R+K +L ++G + + SL + D + KKA LI
Sbjct: 17 NYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQG 76
Query: 67 VSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRR---YSLKMQEGGDLQDHVN 123
+ +D ++ T+ + W+ L + Y + K+ ++ R +L+M+EG + D+ +
Sbjct: 77 LDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFS 136
Query: 124 AFNNILADLTRLGVKVDD 141
+ +L R G K+DD
Sbjct: 137 RVLTVTNNLKRNGEKLDD 154
>At4g35820 putative protein
Length = 272
Score = 38.1 bits (87), Expect = 0.002
Identities = 17/52 (32%), Positives = 32/52 (60%), Gaps = 1/52 (1%)
Query: 101 LLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENK-AIIFCAL 151
L ++QR LK+ E DL H+N F+ ++ + + VK++++ K I+ C+L
Sbjct: 11 LYLRQRLQGLKIYETSDLIQHINTFDELVGEQVSVDVKIEEKTKDMILLCSL 62
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,388,380
Number of Sequences: 26719
Number of extensions: 125472
Number of successful extensions: 447
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 400
Number of HSP's gapped (non-prelim): 56
length of query: 157
length of database: 11,318,596
effective HSP length: 91
effective length of query: 66
effective length of database: 8,887,167
effective search space: 586553022
effective search space used: 586553022
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0490.9


