

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.11
(418 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
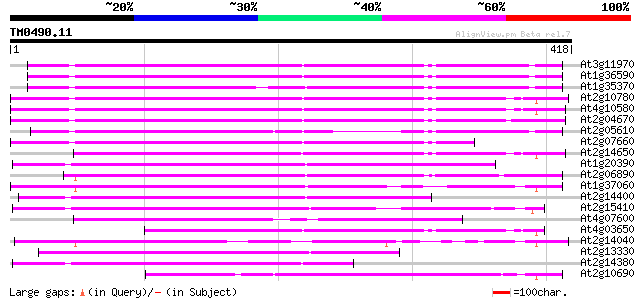
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 195 4e-50
At1g36590 hypothetical protein 195 4e-50
At1g35370 hypothetical protein 176 2e-44
At2g10780 pseudogene 171 5e-43
At4g10580 putative reverse-transcriptase -like protein 167 1e-41
At2g04670 putative retroelement pol polyprotein 166 2e-41
At2g05610 putative retroelement pol polyprotein 166 2e-41
At2g07660 putative retroelement pol polyprotein 157 1e-38
At2g14650 putative retroelement pol polyprotein 145 3e-35
At1g20390 hypothetical protein 137 1e-32
At2g06890 putative retroelement integrase 135 4e-32
At1g37060 Athila retroelment ORF 1, putative 123 2e-28
At2g14400 putative retroelement pol polyprotein 119 4e-27
At2g15410 putative retroelement pol polyprotein 103 2e-22
At4g07600 102 5e-22
At4g03650 putative reverse transcriptase 100 1e-21
At2g14040 putative retroelement pol polyprotein 98 1e-20
At2g13330 F14O4.9 96 4e-20
At2g14380 putative retroelement pol polyprotein 95 6e-20
At2g10690 putative retroelement pol polyprotein 94 1e-19
>At3g11970 hypothetical protein
Length = 1499
Score = 195 bits (495), Expect = 4e-50
Identities = 127/402 (31%), Positives = 209/402 (51%), Gaps = 16/402 (3%)
Query: 14 QQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPKIYSL 73
+ L+ G ++ + S +A V K+ R L VDYR LN D FP+P I L
Sbjct: 622 EDLLTNGTVQASSSPYASPVVLVKKKDGTWR----LCVDYRELNGMTVKDSFPIPLIEDL 677
Query: 74 YSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKA 133
+ A IFSK D+++G+ Q+ +DP D KTAF + H+++ V+PFGL AP+ FQ
Sbjct: 678 MDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGL 737
Query: 134 MTKIFEPILHT-SLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEV 192
M IF+P L LV+ DD+L++S E H++ L+Q +++ + + K A +V
Sbjct: 738 MNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKV 797
Query: 193 DFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLS 252
++LG + P ++ + ++P + T+KQ++ FLG+ Y R F+ L
Sbjct: 798 EYLGHFISAQGIETDPAKIKAVKEWP-QPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLH 856
Query: 253 KMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASDHYWAAVLIEEID 311
+ + +A W +A + LK A P L +P D + +++TDA AVL++E
Sbjct: 857 ALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQE-- 914
Query: 312 GQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLE 371
G L + S Q K +LH KE LAV + ++++ +L HF ++ D S +LE
Sbjct: 915 GHPL--AYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLE 972
Query: 372 FKNKTPPNPQIL-RLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
+ TP Q L +L E +D+ +++ QG +N++ D LSR
Sbjct: 973 QRLNTPIQQQWLPKLLE----FDYEIQYRQGKENVVADALSR 1010
>At1g36590 hypothetical protein
Length = 1499
Score = 195 bits (495), Expect = 4e-50
Identities = 127/402 (31%), Positives = 209/402 (51%), Gaps = 16/402 (3%)
Query: 14 QQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPKIYSL 73
+ L+ G ++ + S +A V K+ R L VDYR LN D FP+P I L
Sbjct: 622 EDLLTNGTVQASSSPYASPVVLVKKKDGTWR----LCVDYRELNGMTVKDSFPIPLIEDL 677
Query: 74 YSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKA 133
+ A IFSK D+++G+ Q+ +DP D KTAF + H+++ V+PFGL AP+ FQ
Sbjct: 678 MDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGL 737
Query: 134 MTKIFEPILHT-SLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEV 192
M IF+P L LV+ DD+L++S E H++ L+Q +++ + + K A +V
Sbjct: 738 MNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKV 797
Query: 193 DFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLS 252
++LG + P ++ + ++P + T+KQ++ FLG+ Y R F+ L
Sbjct: 798 EYLGHFISAQGIETDPAKIKAVKEWP-QPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLH 856
Query: 253 KMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASDHYWAAVLIEEID 311
+ + +A W +A + LK A P L +P D + +++TDA AVL++E
Sbjct: 857 ALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQE-- 914
Query: 312 GQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLE 371
G L + S Q K +LH KE LAV + ++++ +L HF ++ D S +LE
Sbjct: 915 GHPL--AYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLE 972
Query: 372 FKNKTPPNPQIL-RLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
+ TP Q L +L E +D+ +++ QG +N++ D LSR
Sbjct: 973 QRLNTPIQQQWLPKLLE----FDYEIQYRQGKENVVADALSR 1010
>At1g35370 hypothetical protein
Length = 1447
Score = 176 bits (447), Expect = 2e-44
Identities = 122/402 (30%), Positives = 198/402 (48%), Gaps = 24/402 (5%)
Query: 14 QQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPKIYSL 73
Q +IK G I+ + S +A V K+ R L VDY LN D+F +P I L
Sbjct: 601 QDMIKSGTIQVSSSPFASPVVLVKKKDGTWR----LCVDYTELNGMTVKDRFLIPLIEDL 656
Query: 74 YSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKA 133
+ + +FSK D+++G+ Q+ +DP D KTAF N H+++ V+ FGL AP+ FQ
Sbjct: 657 MDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSL 716
Query: 134 MTKIFEPILHT-SLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEV 192
M +F L LV+ DD+L++S E HK+ L+ +++ H + K
Sbjct: 717 MNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSK-------- 768
Query: 193 DFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLS 252
+ LG + + P ++ + ++P T+KQ++ FLG Y R F+ + L
Sbjct: 769 EHLGHFISAREIETDPAKIQAVKEWPTP-TTVKQVRGFLGFAGYYRRFVRNFGVIAGPLH 827
Query: 253 KMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASDHYWAAVLIEEID 311
+ + + W E A LK N P L +P D + +++TDA AVL+++
Sbjct: 828 ALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQK-- 885
Query: 312 GQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLE 371
G L + S Q K +LH KE LA + ++++ +L HF ++ D S +LE
Sbjct: 886 GHPL--AYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLE 943
Query: 372 FKNKTPPNPQIL-RLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
+ TP Q L +L E +D+ +++ QG +NL+ D LSR
Sbjct: 944 QRLNTPVQQQWLPKLLE----FDYEIQYRQGKENLVADALSR 981
>At2g10780 pseudogene
Length = 1611
Score = 171 bits (434), Expect = 5e-43
Identities = 119/422 (28%), Positives = 206/422 (48%), Gaps = 21/422 (4%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P+E+ +++ ++L+ +G I P+ S W +V K+ R L +DYR LN+
Sbjct: 667 MAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGSFR----LCIDYRGLNKVT 722
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+K+PLP+I L + A FSK D+ SG+ Q+ ++P D KTAF H+++ V+P
Sbjct: 723 VKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMP 782
Query: 121 FGLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVM 179
FGL AP+ F K M +F L +++I+D+L++S E+H++ L+ + +H +
Sbjct: 783 FGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELF 842
Query: 180 LSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRD 239
K + V FLG ++ P + + ++P +I+ FLG+ Y R
Sbjct: 843 AKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-RPRNATEIRSFLGLAGYYRR 901
Query: 240 FIPHVSKYTSKLSKMLRKN-APPWGKEQTEAIKFLKKAAQNPPPLKIPGDGK-RILQTDA 297
F+ + L+++ K+ A W E ++ LK N P L +P +G+ + TDA
Sbjct: 902 FVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDA 961
Query: 298 SDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHF 357
S VL+++ G + +AS Q + E +Y T E AV + +K + +L G
Sbjct: 962 SIVGLGCVLMQK--GSVI--AYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKV 1017
Query: 358 EVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDFSVKHVQGSKNLIPDYLSRPK 414
++ D+ S + T P LR + W L Y+ + + G N + D LSR +
Sbjct: 1018 QIYTDHKSLKYIF-----TQPELN-LRQRRWMELVADYNLDIAYHPGKANQVADALSRRR 1071
Query: 415 QQ 416
+
Sbjct: 1072 SE 1073
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 167 bits (422), Expect = 1e-41
Identities = 118/420 (28%), Positives = 202/420 (48%), Gaps = 21/420 (5%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P+E+ +++ + L+ +G I P+ S W +V K+ R L +DYR LN+
Sbjct: 499 MAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKDGSFR----LCIDYRELNRVT 554
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+++PLP+I L ++ A FSK D+ SG+ Q+ + D KTAF H+++ V+P
Sbjct: 555 VKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMP 614
Query: 121 FGLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVM 179
FGL AP++F + M +F+ L +++IDD+L++S E + L++ + + +
Sbjct: 615 FGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLF 674
Query: 180 LSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRD 239
K +E+ FLG + P +E + +P +I+ FLG Y R
Sbjct: 675 AKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWP-RPTNATEIRSFLGWAGYYRR 733
Query: 240 FIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIPGDGK-RILQTDA 297
F+ + ++K+ K+ P W +E E LK+ + P L +P G+ ++ TDA
Sbjct: 734 FVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDA 793
Query: 298 SDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHF 357
S VL++ G+ + +AS Q E +Y T E AV + +K + +L G
Sbjct: 794 SRVGLGCVLMQH--GKVI--AYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKV 849
Query: 358 EVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDFSVKHVQGSKNLIPDYLSRPK 414
+V D+ S + T P LR + W L YD + + G N++ D LSR +
Sbjct: 850 QVFTDHKSLKYIF-----TQPELN-LRQRRWMELVADYDLEIAYHPGKANVVVDALSRKR 903
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 166 bits (421), Expect = 2e-41
Identities = 115/417 (27%), Positives = 200/417 (47%), Gaps = 15/417 (3%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P+E+ +++ + L+ +G I P+ S W +V K+ R L +DYR LN
Sbjct: 525 MAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDGSFR----LCIDYRGLNWVT 580
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+K+PLP+I L ++ A FSK D+ SG+ Q+ + D KTAF H+++ V+P
Sbjct: 581 VKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMP 640
Query: 121 FGLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVM 179
F L AP+ F + M +F+ L +++IDD+L++S E H+ L++ + + +
Sbjct: 641 FALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLF 700
Query: 180 LSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRD 239
K +E+ FLG + P +E + +P +I+ FL + Y R
Sbjct: 701 AKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWP-RPTNATEIRSFLRLTGYYRR 759
Query: 240 FIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIPGDGK-RILQTDA 297
F+ + ++K+ K+ P W E E LK+ + P L +P G+ ++ TDA
Sbjct: 760 FVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDA 819
Query: 298 SDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHF 357
S VL++ G+ + +AS Q + E +Y T E V + +K + +L G
Sbjct: 820 SRVGLGCVLMQR--GKVI--AYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKV 875
Query: 358 EVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSRPK 414
+V D+ S + N+ N + +R E + YD + + G N++ D LSR +
Sbjct: 876 QVFTDHKSLKYIF---NQPELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKR 929
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 166 bits (420), Expect = 2e-41
Identities = 119/400 (29%), Positives = 187/400 (46%), Gaps = 67/400 (16%)
Query: 16 LIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPKIYSLYS 75
++ G I+ + S +A V K+ R L VDYR LN D+FP+P I L
Sbjct: 66 MLASGTIQASSSPYASPVVLVKKKDGTWR----LCVDYRELNGMTVKDRFPIPLIEDLMD 121
Query: 76 YIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKAMT 135
+ ++++SK D+++G+ Q+ +DP D KTAF N HY++ V+PFGL AP+ FQ M
Sbjct: 122 ELGGSNVYSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMN 181
Query: 136 KIFEPILHT-SLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDF 194
F+P L LV+ DD+L++S E HKK L+ +++ H + K A V++
Sbjct: 182 SFFKPFLRKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEY 241
Query: 195 LGMHFTEGK-FVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSK 253
LG HF G+ P ++ + +P + +KQ+ FLG+ Y R F
Sbjct: 242 LG-HFISGEGIATDPAKIKAVQDWP-VPVNLKQLCGFLGLTGYYRRFF------------ 287
Query: 254 MLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIPGDGKRILQTDASDHYWAAVLIEEIDGQ 313
+++ DA H AVL++E G
Sbjct: 288 --------------------------------------VVEMDACGHGIGAVLMQE--GH 307
Query: 314 KLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFK 373
L S Q K +LH KE LAV + ++++ +L HF ++ D S +LE +
Sbjct: 308 PL--AFISRQLKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQR 365
Query: 374 NKTPPNPQIL-RLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
TP Q L +L E +D+ +++ QG +NL+ D LSR
Sbjct: 366 LNTPIQQQWLPKLLE----FDYEIQYKQGKENLVADALSR 401
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 157 bits (397), Expect = 1e-38
Identities = 103/349 (29%), Positives = 175/349 (49%), Gaps = 12/349 (3%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P+E+ +++ ++L+ +G I P+ S W +V K+ R L +DYR LN+
Sbjct: 162 MAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGSFR----LCIDYRGLNKVT 217
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+K+PLP+I L + A FSK D+ SG+ Q+ ++P D KTAF H+++ V+P
Sbjct: 218 VKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMP 277
Query: 121 FGLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVM 179
FGL AP+ F K M +F L +++IDD+L+ S E+H++ L+ + +H +
Sbjct: 278 FGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELF 337
Query: 180 LSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRD 239
K + V FLG ++ P + + ++P +I+ FLG+ Y R
Sbjct: 338 AKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-RPRNATEIRSFLGLAGYYRR 396
Query: 240 FIPHVSKYTSKLSKMLRKN-APPWGKEQTEAIKFLKKAAQNPPPLKIPGDGK-RILQTDA 297
F+ + L+++ K+ A W E ++ LK N P L +P +G+ + TDA
Sbjct: 397 FVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEPYTVYTDA 456
Query: 298 SDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIK 346
S VL+++ G + +AS Q + E +Y T E AV + +K
Sbjct: 457 SIVGLGCVLMQK--GSVI--AYASRQLRKHEKNYPTHDLEMAAVVFALK 501
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 145 bits (367), Expect = 3e-35
Identities = 106/373 (28%), Positives = 180/373 (47%), Gaps = 17/373 (4%)
Query: 48 RLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAF 107
RL +DYR LN +K+PLP+I L ++ A FSK D+ SG+ + + D KTAF
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 108 CIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFL 166
H+++ V+PFGL AP+ F + M +F+ +L +++IDD+L++S E H+ L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 167 QQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQ 226
++ + + + K +E+ FLG + P +E + + +
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW-HTPTNATE 695
Query: 227 IQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKI 285
I+ FLG+ Y R F+ + ++K+ K+ P W E E LK+ + P L +
Sbjct: 696 IRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLAL 755
Query: 286 PGDGK-RILQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYG 344
P G+ ++ TDAS VL++ G+ + +AS Q + E +Y T E AV +
Sbjct: 756 PEHGEPYMVYTDASGVGLGCVLMQR--GKVI--AYASRQLRKHEGNYPTHDLEMAAVIFA 811
Query: 345 IKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDFSVKHVQG 401
+K + +L G + +V D+ S + T P LR ++W L YD + + G
Sbjct: 812 LKIWRSYLYGGNVQVFTDHKSLKYIF-----TQPELN-LRQRQWMELVADYDLEIAYHPG 865
Query: 402 SKNLIPDYLSRPK 414
N++ D LS +
Sbjct: 866 KANVVADALSHKR 878
>At1g20390 hypothetical protein
Length = 1791
Score = 137 bits (344), Expect = 1e-32
Identities = 103/364 (28%), Positives = 166/364 (45%), Gaps = 9/364 (2%)
Query: 3 PSELRAAQEECQQLIKQG-LIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
P RA EE ++L+K G +IE EW V K++ K R+ VDY LN+
Sbjct: 787 PERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNG----KWRVCVDYTDLNKACP 842
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D +PLP I L + S D SG+ Q+ + D+ KT+F Y + V+ F
Sbjct: 843 KDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSF 902
Query: 122 GLKVAPSLFQKAMTKIF-EPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
GLK A + +Q+ + K+ + I T VYIDD+L+ S E H + L + +D+ + +G+ L
Sbjct: 903 GLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKL 962
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
+ K +FLG T+ P + + + P +++Q+ G + + F
Sbjct: 963 NPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSP-RNAREVQRLTGRIAALNRF 1021
Query: 241 IPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIPGDGKRI-LQTDAS 298
I + +L++ A W K+ EA + LK PP L P G+ + L S
Sbjct: 1022 ISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVS 1081
Query: 299 DHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFE 358
DH ++VL+ E G++ + S +AE Y K LAV ++ + + H
Sbjct: 1082 DHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIA 1141
Query: 359 VQID 362
V D
Sbjct: 1142 VLTD 1145
>At2g06890 putative retroelement integrase
Length = 1215
Score = 135 bits (340), Expect = 4e-32
Identities = 103/378 (27%), Positives = 178/378 (46%), Gaps = 14/378 (3%)
Query: 41 EQVRQKK---RLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGL 97
E RQK R+ D R +N P+P++ + + + IFSK D+KSG+ Q+ +
Sbjct: 460 ELQRQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRM 519
Query: 98 DPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHT-SLVYIDDVLLFS 156
+ D KTAF + Y+W V+PFGL APS F + M + + +VY DD+L++S
Sbjct: 520 NEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYS 579
Query: 157 HDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPK 216
H + L ++ K + + +K T + FLG + V+ +
Sbjct: 580 ESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVKAIRD 639
Query: 217 FPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKK 275
+P T+ +++ F G+ + R F S + L+++++K+ W K Q EA + LK
Sbjct: 640 WPSP-KTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQSLKD 698
Query: 276 AAQNPPPLKIPGDGKRI-LQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTT 334
N P L + K ++ DAS AVL+++ QKL S + A L+Y T
Sbjct: 699 KLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQD---QKL-IAFFSEKLGGATLNYPTY 754
Query: 335 FKETLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSLYDF 394
KE A+ ++R+ +L F + D+ S + K + N + R E+ + +
Sbjct: 755 DKELYALVRALQRWQHYLWPKVFVIHTDHES---LKHLKGQQKLNKRHARWVEFIETFAY 811
Query: 395 SVKHVQGSKNLIPDYLSR 412
+K+ +G N++ D LS+
Sbjct: 812 VIKYKKGKDNVVADALSQ 829
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 123 bits (308), Expect = 2e-28
Identities = 111/433 (25%), Positives = 186/433 (42%), Gaps = 56/433 (12%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEP-TKSEWACTAFYVNKRSEQVRQKK------------ 47
+ P+ ++E +L+ G+I P + S W V K+ K
Sbjct: 907 LNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTIT 966
Query: 48 --RLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKT 105
R+ ++YR LN + + FPLP I + + N + D SGF+Q+ + P D+ KT
Sbjct: 967 GHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKT 1026
Query: 106 AFCIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKK 164
F P + + +PFGL AP+ FQ+ MT IF ++ + V++DD ++ S
Sbjct: 1027 TFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLL 1086
Query: 165 FLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFT-EGKFVPQPHIVEELPKFPDEGLT 223
L + C + ++L+ K +E LG + EG V + I + P + T
Sbjct: 1087 NLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPK--T 1144
Query: 224 IKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPP 282
+K I+ FLG + R FI SK L+++L K + E A K +K+A P
Sbjct: 1145 VKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEALITAP- 1203
Query: 283 LKIPGDGKRILQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVK 342
I+Q D + + ++ DA++ Y TT KE LAV
Sbjct: 1204 ---------IVQAPNWDFPFEIITMD-----------------DAQVRYATTEKELLAVV 1237
Query: 343 YGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDFSVKHV 399
+ ++F +L G + D+++ + K+ P RL W L +D +
Sbjct: 1238 FAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKP------RLLRWILLLQEFDMEIVDK 1291
Query: 400 QGSKNLIPDYLSR 412
+G +N + D+LSR
Sbjct: 1292 KGIENGVADHLSR 1304
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 119 bits (297), Expect = 4e-27
Identities = 88/312 (28%), Positives = 148/312 (47%), Gaps = 9/312 (2%)
Query: 7 RAAQEECQQLIKQGLIEPTK-SEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKF 65
+A +E +L+K G I + +W V K++ K R+ +D+ LN+ D F
Sbjct: 478 KAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNG----KDRVCIDFTDLNKACPKDSF 533
Query: 66 PLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKV 125
PLP I L + S D SG+ Q+ ++P+D+ KT F Y + V+PFGL+
Sbjct: 534 PLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRN 593
Query: 126 APSLFQKAMTKIF-EPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRK 184
A + + + + K+F E + T VYIDD+L+ S +E H K L++ + I +++ + L+ K
Sbjct: 594 AGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAK 653
Query: 185 SQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHV 244
+FLG T+ P+ + P K++Q+ G + + FI
Sbjct: 654 CTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSP-KNFKEVQRLTGRIAALNRFISRS 712
Query: 245 SKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIPG-DGKRILQTDASDHYW 302
+ + ++L+ N W ++ EA LK +PP L P D K L S+H
Sbjct: 713 TDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAV 772
Query: 303 AAVLIEEIDGQK 314
+ VLI E G++
Sbjct: 773 SGVLIREDRGEQ 784
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 103 bits (257), Expect = 2e-22
Identities = 97/401 (24%), Positives = 168/401 (41%), Gaps = 35/401 (8%)
Query: 3 PSELRAAQEECQQLIKQG-LIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
P + EE ++L+ G ++E +W V K++ K R+ +D+ LN+
Sbjct: 792 PDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNG----KWRVCIDFTDLNKACP 847
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D FPLP I L + S D SG+ Q+ + DR KT F Y + V+PF
Sbjct: 848 KDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPF 907
Query: 122 GLKVAPSLFQKAMTKIF-EPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
GLK A + + + + ++F + + H+ VYIDD+L+ S E H L+Q + + +++ + L
Sbjct: 908 GLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKL 967
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
+ K +FLG T P + + P T +++Q+ +G + + F
Sbjct: 968 NPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSPRNT-REVQRLIGRIAALNRF 1026
Query: 241 IPHVSKYTSKLSKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIPGDGKRILQTDASDH 300
I + ++LR N E+ E + L L S
Sbjct: 1027 ISRSTDKCLPFYQLLRANKRFEWDEKCEEGETL------------------YLYIAVSTS 1068
Query: 301 YWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQ 360
+ VL+ E G++ + S AEL Y T K AV ++ + + + EV
Sbjct: 1069 AVSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKLAFAVVISARKLRPYFKSYTVEV- 1127
Query: 361 IDNSSFPKMLEFKNKTPPNPQILRLKEW---FSLYDFSVKH 398
+ N +L +++ RL +W S YD + K+
Sbjct: 1128 LTNQPLRTILHSPSQSG------RLAKWAVELSEYDIAYKN 1162
>At4g07600
Length = 630
Score = 102 bits (253), Expect = 5e-22
Identities = 83/293 (28%), Positives = 133/293 (45%), Gaps = 22/293 (7%)
Query: 48 RLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAF 107
R+ +DYR LN ++D FPLP + + N + D SGF+Q+ + P D KT F
Sbjct: 356 RMCIDYRNLNAASRNDHFPLPFTDQMLERLANHPYYCFLDGYSGFFQIPIHPNDHEKTTF 415
Query: 108 CIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKKFL 166
P + + +PFGL AP+ FQ+ MT IF I+ + V++DD ++ S L
Sbjct: 416 TCPYGTFAYERMPFGLCNAPATFQRCMTSIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNL 475
Query: 167 QQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQ 226
+ C + ++L+ K KE LG H + E D+G
Sbjct: 476 GRVLTRCEETNLVLNWEKCHFMVKEGIMLG------------HKISEKGIEVDKGC---- 519
Query: 227 IQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKI 285
FLG + R FI SK L+++L K + ++ ++ +K+A + P ++
Sbjct: 520 ---FLGHAGFYRRFIKDFSKIARPLTRLLCKETEFEFDEDCPKSFHTIKEALVSAPVVRA 576
Query: 286 PG-DGKRILQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKE 337
D + DA D+ AVL ++ID + +AS +A+ Y TT KE
Sbjct: 577 TNWDYPFEIMCDALDYAVGAVLGQKIDKKLHVIYYASRTLDEAQGRYATTEKE 629
>At4g03650 putative reverse transcriptase
Length = 839
Score = 100 bits (249), Expect = 1e-21
Identities = 82/304 (26%), Positives = 139/304 (44%), Gaps = 17/304 (5%)
Query: 101 DRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDE 159
D KTAF H+++ V+PFGL AP+ F + M +F+ L +++IDD+L++S
Sbjct: 520 DVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSP 579
Query: 160 ESHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPD 219
E H+ L++ + + + K +E+ FLG + P +E + +P
Sbjct: 580 EEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWP- 638
Query: 220 EGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQ 278
+I+ FLG+ Y R FI + ++K+ K+ P W E E LK+
Sbjct: 639 RPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLT 698
Query: 279 NPPPLKIPGDGK-RILQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKE 337
+ P L +P G+ ++ TDAS L++ G+ + +AS Q + E +Y T E
Sbjct: 699 STPVLALPEHGEPYMVYTDASGVGLGCALMQR--GKVI--AYASRQLRKHEGNYPTHDLE 754
Query: 338 TLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDF 394
AV + +K + +L G +V D+ S + T P LR + W L YD
Sbjct: 755 MAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIF-----TQPELN-LRQRRWMELVADYDL 808
Query: 395 SVKH 398
+ +
Sbjct: 809 EIAY 812
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 97.8 bits (242), Expect = 1e-20
Identities = 111/440 (25%), Positives = 179/440 (40%), Gaps = 95/440 (21%)
Query: 4 SELR-AAQEECQQLIKQGLIEP-TKSEWACTAFYVNKRSEQVRQKK-------------- 47
S LR ++E +L+ G+I P + S W V K+ + K
Sbjct: 384 SNLRDVVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGH 443
Query: 48 RLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAF 107
R+ +DYR LN + D FPL I + + N + D GF+Q+ + P D+ KT F
Sbjct: 444 RMCIDYRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTF 503
Query: 108 CIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKKFL 166
P + + +PFGL AP+ FQ M IF ++ + V+IDD L+ E+ H
Sbjct: 504 TCPYGTFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDFLVLQRCEDKH---- 559
Query: 167 QQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTE-GKFVPQPHIVEELPKFPDEGLTIK 225
++L+ KS ++ LG +E G V + I
Sbjct: 560 -----------LVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKI--------------- 593
Query: 226 QIQQFLGILNYIRDFIPHVSKYTSKLSKML-RKNAPPWGKEQTEAIKFLKKAAQN----- 279
+I +FLG + R FI SK +++L ++ + +A +K++ +
Sbjct: 594 EIMRFLGHAGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQ 653
Query: 280 PPPLKIPGDGKRILQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETL 339
PP ++P + + DASD+ AVL GQ KD +LH
Sbjct: 654 PPEWELPFE----VICDASDYVVGAVL---------------GQRKDKKLH--------- 685
Query: 340 AVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDFSV 396
A+ Y + D G V +++ +L K+ P RL W L +D +
Sbjct: 686 AIYYASRTLD----GAQVIVHTVHAALRYLLSKKDAKP------RLLRWILLLQDFDLEI 735
Query: 397 KHVQGSKNLIPDYLSRPKQQ 416
K +G +N + D LSR + Q
Sbjct: 736 KDKKGIENEVADNLSRLRVQ 755
>At2g13330 F14O4.9
Length = 889
Score = 95.9 bits (237), Expect = 4e-20
Identities = 73/271 (26%), Positives = 122/271 (44%), Gaps = 3/271 (1%)
Query: 22 IEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAH 81
++ K +A ++ + S + K R+ +D+R LN+ D FPLP I L
Sbjct: 269 LKENKDSFAWSSANLQGISLEKNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHE 328
Query: 82 IFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKAMTKIF-EP 140
S D G+ Q+ + + KTAF Y + V+PFGLK A + +Q+ + ++F +
Sbjct: 329 KLSFMDAFYGYNQILMRRDGQEKTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQ 388
Query: 141 ILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFT 200
+ T VYIDD+L+ S E+ H L++ + I K + L+ K + +FL T
Sbjct: 389 LGKTIEVYIDDMLVKSAHEKDHVPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVT 448
Query: 201 EGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP 260
E P + + P + +++Q+ G + + FI + + LRK
Sbjct: 449 ERGIEANPKQIAAFIEMPSPKMA-REVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKE 507
Query: 261 -PWGKEQTEAIKFLKKAAQNPPPLKIPGDGK 290
W K+ +A K LK PP L P G+
Sbjct: 508 FDWNKDCEQAFKQLKAYLTEPPILAKPEKGE 538
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 95.1 bits (235), Expect = 6e-20
Identities = 67/256 (26%), Positives = 117/256 (45%), Gaps = 7/256 (2%)
Query: 3 PSELRAAQEECQQLIKQG-LIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
P +A +E +L+ G ++E EW V K+++ K R+ +D+ LN+
Sbjct: 507 PERSKAVNDEVDKLLDAGSIVEVKYPEWLANPVVVKKKND----KWRVCIDFTDLNKACP 562
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D FPLP I + + S D SG+ Q+ + D+ KT+F I Y + V+PF
Sbjct: 563 KDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQIPMHKDDQEKTSFIIDRGTYCYKVMPF 622
Query: 122 GLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
GLK + +Q+ + ++F P L T VYIDD+L+ S H L+ ++ +K+ + L
Sbjct: 623 GLKNVGARYQRLVNQMFAPQLGKTMEVYIDDMLVKSTRSADHIDHLKACFETLNKYNMKL 682
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
+ K +FLG T+ P + + K++Q+ G + + F
Sbjct: 683 NPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRAILDL-QSPRNKKEVQRLTGRIAGLNRF 741
Query: 241 IPHVSKYTSKLSKMLR 256
I + + ++LR
Sbjct: 742 IARSTDKSLPFYQLLR 757
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 94.0 bits (232), Expect = 1e-19
Identities = 86/317 (27%), Positives = 146/317 (45%), Gaps = 17/317 (5%)
Query: 102 RPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEE 160
+ KT F PN + + + FGL AP FQ++MT IF + + V++DD FS
Sbjct: 163 KKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDDFSGFSSCLL 222
Query: 161 SHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDE 220
+ + L++ C + ++L+ K E LG H GK + ++
Sbjct: 223 NLCRVLER----CEETNLVLNWEKCHFMVHEGIVLG-HKISGKGIEVDKAKIDVMIQLQP 277
Query: 221 GLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQN 279
T+K I+ FLG + R FI SK L+++L K + ++ +A +K+ +
Sbjct: 278 PKTVKDIRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAFHLIKETLVS 337
Query: 280 PPPLKIPG-DGKRILQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKET 338
P + P D + DA D+ AVL ++ID + +AS +A+ Y T KE
Sbjct: 338 APIFQAPNWDHPFEIMCDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQTRYATIEKEL 397
Query: 339 LAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWFSL---YDFS 395
LAV + ++ +L G +V ID+++ + + K +T RL W L +D
Sbjct: 398 LAVVFAFEKIRSYLVGFKVKVYIDHAAL-RHIYAKKETKS-----RLLRWILLLQEFDME 451
Query: 396 VKHVQGSKNLIPDYLSR 412
+ +G +N + D+LSR
Sbjct: 452 IIDKKGVENGVADHLSR 468
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,136,089
Number of Sequences: 26719
Number of extensions: 456474
Number of successful extensions: 1183
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1059
Number of HSP's gapped (non-prelim): 68
length of query: 418
length of database: 11,318,596
effective HSP length: 102
effective length of query: 316
effective length of database: 8,593,258
effective search space: 2715469528
effective search space used: 2715469528
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0490.11


