

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0487.5
(658 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
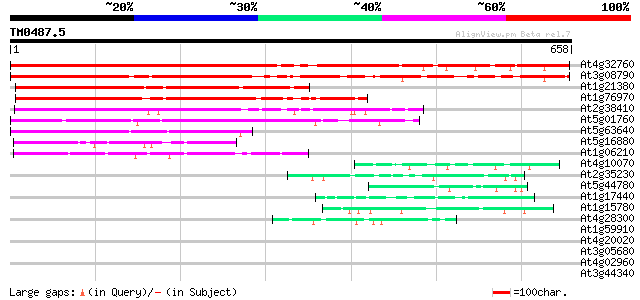
Score E
Sequences producing significant alignments: (bits) Value
At4g32760 unknown protein 692 0.0
At3g08790 hypothetical protein 536 e-152
At1g21380 unknown protein 327 2e-89
At1g76970 unknown protein 316 3e-86
At2g38410 unknown protein 290 2e-78
At5g01760 unknown protein 243 2e-64
At5g63640 unknown protein (At5g63640) 208 9e-54
At5g16880 TOM (target of myb1) -like protein 135 6e-32
At1g06210 unknown protein 101 2e-21
At4g10070 putative DNA-directed RNA polymerase 46 6e-05
At2g35230 unknown protein 45 1e-04
At5g44780 unknown protein 44 3e-04
At1g17440 unknown protein 44 4e-04
At1g15780 unknown protein 44 4e-04
At4g28300 predicted proline-rich protein 43 6e-04
At1g59910 hypothetical protein 41 0.002
At4g20020 DAG-like protein 40 0.003
At3g05680 unknown protein 40 0.003
At4g02960 putative polyprotein of LTR transposon 39 0.009
At3g44340 putative protein 39 0.012
>At4g32760 unknown protein
Length = 675
Score = 692 bits (1787), Expect = 0.0
Identities = 400/697 (57%), Positives = 466/697 (66%), Gaps = 70/697 (10%)
Query: 1 MVNPLVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALT 60
MVN +VERATS+MLIGPDWA+N+EICD+LN DPAQ+KDVVKG+KKRIGSR+ K QLLALT
Sbjct: 1 MVNAMVERATSEMLIGPDWAMNLEICDMLNSDPAQAKDVVKGIKKRIGSRNPKAQLLALT 60
Query: 61 LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARY 120
LLETI+KNCGD+VHMHVAE+ V+HEMV+IVKKKPD V+EKI++LIDTWQEAFGGPRARY
Sbjct: 61 LLETIVKNCGDMVHMHVAEKGVIHEMVRIVKKKPDFHVKEKILVLIDTWQEAFGGPRARY 120
Query: 121 PQYYAAYQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQNIRDSDAQPHTAESSAESE 180
PQYYA YQELL +GAVFPQRSE+SAPVFTP QTQPLTSYP N+R++ E SAE E
Sbjct: 121 PQYYAGYQELLRAGAVFPQRSERSAPVFTPPQTQPLTSYPPNLRNAGPGNDVPEPSAEPE 180
Query: 181 FPTLSLSEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVNST 240
FPTLSLSEIQNA+GIMDVLAEMLSA+EPGNKE L+QEV+VDLVEQCRTYKQRVVHLVNST
Sbjct: 181 FPTLSLSEIQNAKGIMDVLAEMLSALEPGNKEDLKQEVMVDLVEQCRTYKQRVVHLVNST 240
Query: 241 SDESLLCQGLALNDDLQRVLAKHESIASGTPG-QNHTEKLNPAPPGALIDVGGPLVDTGD 299
SDESLLCQGLALNDDLQRVL +E+IASG PG + EK +L+DV GPL+DTGD
Sbjct: 241 SDESLLCQGLALNDDLQRVLTNYEAIASGLPGTSSQIEKPKSETGKSLVDVDGPLIDTGD 300
Query: 300 TSKQSDERSPSGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDPKLDLLSGDDYNSPKADNS 359
+S Q++ + S G LNQL LPAPP +NGSA + K+DLLSGDD
Sbjct: 301 SSNQANGATSSSGNG---VLNQLALPAPPVTNGSA-----NSKIDLLSGDD--------- 343
Query: 360 LALVPVGQQQPASPM-SQQNALVLFDMFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQT 418
LALVPVG QPASP+ S QNAL L DMFS+ +N PS P+ N PL+PQ QQ
Sbjct: 344 LALVPVGPPQPASPVASDQNALALIDMFSDNTNNPSPATAPSGNPAQNIPLNPQGHQQPN 403
Query: 419 FIS--QGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFP 476
+ G+ NG P VG Q+EQ Y Q W+ Q A QQP P GAQ +FP
Sbjct: 404 SQAGEAGLQQSNGFAPQVGYSQFEQPSYGQGVSSPWSSQPA--QQPVQPSYGAQDSTAFP 461
Query: 477 PPPWEAQ------AAENGSPVAGNQYPQPLQVTQMVVTHVQ----GAPLPQGPQAMGYDQ 526
PPPWEAQ +AE+GSP + +P TQ TH Q P PQ PQ
Sbjct: 462 PPPWEAQLQDYSPSAESGSPFSPGMHP-----TQTAFTHAQPVNNNNPYPQIPQTGPPVN 516
Query: 527 AVGMYMQQPNASHMSAMNN---QVQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGPVAHP 583
Y Q P A + Q+ N M Q Q + + G P Q Q +
Sbjct: 517 NNSPYAQMPQTGQAVANISPYPQIPQNGVYMPNQPNQALGS----GYQPQQQQQQQMMMA 572
Query: 584 MY-----------AQQMYGNQYMGGYGYG--QQPQQGVQYVEQQMHGLSMSDDRN----- 625
Y QQ YGNQ MGGYGYG QQ Q Y++QQM+GLSM D +
Sbjct: 573 QYYAQQQQLQQQQQQQAYGNQ-MGGYGYGYNQQQQGSSPYLDQQMYGLSMRDQTSHQVAS 631
Query: 626 ----SSYVPPKKP-SKPEDKLFGDLVDMAKVKP-KPT 656
+SY+PP KP +KPEDKLFGDLVD++K KP KPT
Sbjct: 632 SSSTTSYLPPMKPKNKPEDKLFGDLVDISKFKPTKPT 668
>At3g08790 hypothetical protein
Length = 607
Score = 536 bits (1380), Expect = e-152
Identities = 341/692 (49%), Positives = 421/692 (60%), Gaps = 128/692 (18%)
Query: 1 MVNPLVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALT 60
MV+PLV+RATSDMLIGPDWA+N+EICD+LN +P Q+++VV G+KKR+ SR+SKVQLLALT
Sbjct: 1 MVHPLVDRATSDMLIGPDWAMNLEICDMLNHEPGQTREVVSGIKKRLTSRTSKVQLLALT 60
Query: 61 LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARY 120
LLETII NCG+++HM VAE+D+LH+MVK+ K+KP+ +V+EKI+ILIDTWQE+F GP+ R+
Sbjct: 61 LLETIITNCGELIHMQVAEKDILHKMVKMAKRKPNIQVKEKILILIDTWQESFSGPQGRH 120
Query: 121 PQYYAAYQELLHSGAVFPQRSEQSAPVFTPL--QTQPLTSYPQNIRDSDAQPHTAESSAE 178
PQYYAAYQELL +G VFPQR P TP Q P T YPQN R +A+ ++S E
Sbjct: 121 PQYYAAYQELLRAGIVFPQR-----PQITPSSGQNGPSTRYPQNSR--NARQEAIDTSTE 173
Query: 179 SEFPTLSLSEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVN 238
SEFPTLSL+EIQNARGIMDVLAEM++AI+ NKEGL+QEV+VDLV QCRTYKQRVVHLVN
Sbjct: 174 SEFPTLSLTEIQNARGIMDVLAEMMNAIDGNNKEGLKQEVVVDLVSQCRTYKQRVVHLVN 233
Query: 239 STSDESLLCQGLALNDDLQRVLAKHESIASGTPGQNHTEKLNPAPPGALIDVGGPLVDTG 298
STSDES+LCQGLALNDDLQR+LAKHE+IASG EK P
Sbjct: 234 STSDESMLCQGLALNDDLQRLLAKHEAIASGNSMIKKEEKSKKEVP-------------K 280
Query: 299 DTSKQSDERSPSGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDPKLDLLSGDDYNSPKADN 358
DT++ D G S+T N ++ T+NG PK+DLLSGDD+ +P ADN
Sbjct: 281 DTTQIID-------VGSSETKNGSVVAY--TTNG--------PKIDLLSGDDFETPNADN 323
Query: 359 SLALVPVGQQQPASPMSQ-QNALVLFDMFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQ 417
SLALVP+G QP+SP+++ N++VL DM S+ NN ++ N + Q QQ
Sbjct: 324 SLALVPLGPPQPSSPVAKPDNSIVLIDMLSD-------NNCESSTPTSNPHANHQKVQQN 376
Query: 418 TFISQGVYYPNGSMPSVGSPQYEQSRYAQ-STGPAWNGQAAQQ---------------QQ 461
Y NG G EQS Y Q S+ P WN Q QQ
Sbjct: 377 --------YSNG----FGPGHQEQSYYGQGSSAPVWNLQITQQPSSPAYGNQPFSPNFSP 424
Query: 462 PPSPVNGAQSGG--SFPPPPWEAQAAENGSPVAGNQY--PQPLQVTQMVVTHVQGAPLPQ 517
P SP G Q+ + PPPPWEAQ SP + QY P+QVTQ+V+T PL
Sbjct: 425 PASPHYGGQNNNVLALPPPPWEAQ-----SPSSSPQYSPTHPMQVTQVVITTHTHQPLGY 479
Query: 518 GPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQN 577
PQ + H + NN +N FGM G P G N
Sbjct: 480 NPQG--------------GSPHATNNNN---NNMFGMFLPPMTGGHMPPPFG------HN 516
Query: 578 GPVAHPMYAQQMYGNQYMGGYGYGQQPQQGVQY-VEQQMHGLSMSDDRN--------SSY 628
G V + Y MYG GYG Q Q QY VEQQM+G+S+ D+ N SS+
Sbjct: 517 GHVTNNNYNPNMYG-------GYGGQAQPPQQYLVEQQMYGMSLQDNGNNNTNPYQVSSH 569
Query: 629 VPP--KKP--SKPEDKLFGDLVDMAKVKPKPT 656
PP KP KPEDKLFGDLV+++K K KPT
Sbjct: 570 QPPPMMKPMNKKPEDKLFGDLVELSKFK-KPT 600
>At1g21380 unknown protein
Length = 506
Score = 327 bits (837), Expect = 2e-89
Identities = 181/348 (52%), Positives = 236/348 (67%), Gaps = 13/348 (3%)
Query: 7 ERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLETII 66
ERAT+DMLIGPDWA+NIE+CDI+N +P+Q+K+ VK LKKR+GS++SKVQ+LAL LET+
Sbjct: 10 ERATNDMLIGPDWAINIELCDIINMEPSQAKEAVKVLKKRLGSKNSKVQILALYALETLS 69
Query: 67 KNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARYPQYYAA 126
KNCG+ V+ + +RD+L +MVKIVKKKPD VREKI+ L+DTWQEAFGG R+PQYY A
Sbjct: 70 KNCGESVYQLIVDRDILPDMVKIVKKKPDLTVREKILSLLDTWQEAFGGSGGRFPQYYNA 129
Query: 127 YQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQNIRDSDAQPHTAESSAESEFPTLSL 186
Y EL +G FP R+E S P FTP QTQP+ + D DA + S ++ LS+
Sbjct: 130 YNELRSAGIEFPPRTESSVPFFTPPQTQPIVA-QATASDEDAAIQASLQSDDAS--ALSM 186
Query: 187 SEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVNSTSDESLL 246
EIQ+A+G +DVL +ML A++P + EGL++E+IVDLVEQCRTY++RV+ LVN+TSDE L+
Sbjct: 187 EEIQSAQGSVDVLTDMLGALDPSHPEGLKEELIVDLVEQCRTYQRRVMALVNTTSDEELM 246
Query: 247 CQGLALNDDLQRVLAKHESIASGTPGQNHTEKLNPAP-PGALIDVGGPLVDTGDTSKQSD 305
CQGLALND+LQRVL H+ A G N P P P I+ ++ D Q
Sbjct: 247 CQGLALNDNLQRVLQHHDDKAKG----NSVPATAPTPIPLVSINHDDDDDESDDDFLQLA 302
Query: 306 ERSP-SGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDP-KLDLLSGDDY 351
RS A G Q +LP PP+ S P+ +D +D LSGD Y
Sbjct: 303 HRSKRESARGTGQGNFNPILPPPPS---SMRPVHVDSGAMDFLSGDVY 347
Score = 36.6 bits (83), Expect = 0.046
Identities = 19/40 (47%), Positives = 26/40 (64%), Gaps = 1/40 (2%)
Query: 614 QMHGLSMSDDRNSSYV-PPKKPSKPEDKLFGDLVDMAKVK 652
Q LS++ +++ V PPKK KPED LF DL+D AK +
Sbjct: 450 QSRNLSLNPTASAAPVTPPKKDDKPEDILFKDLMDFAKTR 489
>At1g76970 unknown protein
Length = 446
Score = 316 bits (809), Expect = 3e-86
Identities = 185/413 (44%), Positives = 256/413 (61%), Gaps = 35/413 (8%)
Query: 7 ERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLETII 66
ERAT+DMLIGPDWA+NIE+CD++N DP+Q+K+ VK LKKR+GS++SKVQ+LAL LET+
Sbjct: 10 ERATNDMLIGPDWAINIELCDLINMDPSQAKEAVKVLKKRLGSKNSKVQILALYALETLS 69
Query: 67 KNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARYPQYYAA 126
KNCG+ V+ + +R +L++MVKIVKKKP+ VREKI+ L+DTWQEAFGG RYPQYY A
Sbjct: 70 KNCGENVYQLIIDRGLLNDMVKIVKKKPELNVREKILTLLDTWQEAFGGRGGRYPQYYNA 129
Query: 127 YQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQNIRDSDAQPHTAESSAESEFPTLSL 186
Y +L +G FP R+E S FTP QTQP D DA + ++ +LSL
Sbjct: 130 YNDLRSAGIEFPPRTESSLSFFTPPQTQP---------DEDAAIQASLQGDDAS--SLSL 178
Query: 187 SEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVNSTSDESLL 246
EIQ+A G +DVL +ML A +PGN E L++EVIVDLVEQCRTY++RV+ LVN+T+DE LL
Sbjct: 179 EEIQSAEGSVDVLMDMLGAHDPGNPESLKEEVIVDLVEQCRTYQRRVMTLVNTTTDEELL 238
Query: 247 CQGLALNDDLQRVLAKHESIASGTPGQNHTEKLNPAPPGALIDVGGPLVDTGDTSKQSDE 306
CQGLALND+LQ VL +H+ IA+ ++ PP ++D ++ D +SD+
Sbjct: 239 CQGLALNDNLQHVLQRHDDIANVGSVPSNGRNTRAPPPVQIVD-----INHDDEDDESDD 293
Query: 307 RSPSGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDPKLDLLSGDDYNSPKADNSLALVPVG 366
A S + P + +G +D+LSGD Y P+ ++S G
Sbjct: 294 EFARLAHRSSTPTRR---PVHGSDSG---------MVDILSGDVY-KPQGNSS----SQG 336
Query: 367 QQQPASPMSQQNALVLFDMFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQTF 419
++P P ++ +F + S S +++ P P S Q+QQ F
Sbjct: 337 VKKPPPPPPHTSSSSSSPVFDDASPQQSKSSEVIRNLPP--PPSRHNQRQQFF 387
Score = 32.3 bits (72), Expect = 0.86
Identities = 25/98 (25%), Positives = 39/98 (39%), Gaps = 7/98 (7%)
Query: 556 PQYAQGVAASP-YMGMVPHQMQNGPVAHPMYAQQMYGNQYMGGYGYGQQPQQGVQYVEQQ 614
P + ++SP + P Q ++ V + NQ + + E Q
Sbjct: 344 PPHTSSSSSSPVFDDASPQQSKSSEVIRNLPPPPSRHNQRQQFFEHHHSSSGSDSSYEGQ 403
Query: 615 MHGLSMSDDRNSSYVPPKKPSKPEDKLFGDLVDMAKVK 652
LS++ P+K KPED LF DLV+ AK +
Sbjct: 404 TRNLSLTSSE------PQKEEKPEDLLFKDLVEFAKTR 435
>At2g38410 unknown protein
Length = 671
Score = 290 bits (741), Expect = 2e-78
Identities = 202/578 (34%), Positives = 293/578 (49%), Gaps = 121/578 (20%)
Query: 6 VERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLETI 65
V++ATSD+L+GPDW N+EICD +N Q+KDVVK +KKR+ +SS+VQLLALTLLET+
Sbjct: 12 VDKATSDLLLGPDWTTNMEICDSVNSLHWQAKDVVKAVKKRLQHKSSRVQLLALTLLETL 71
Query: 66 IKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARYPQYYA 125
+KNCGD +H VAE+++L EMVKIVKKK D +VR+KI++++D+WQ+AFGGP +YPQYY
Sbjct: 72 VKNCGDYLHHQVAEKNILGEMVKIVKKKADMQVRDKILVMVDSWQQAFGGPEGKYPQYYW 131
Query: 126 AYQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQ---NIRDSDAQPHTA--------- 173
AY EL SG FP+RS ++P+ TP + P PQ + + H A
Sbjct: 132 AYDELRRSGVEFPRRSPDASPIITPPVSHPPLRQPQGGYGVPPAGYGVHQAGYGVPQAGY 191
Query: 174 -------------------------------ESSAESEFPTLSLSEIQNARGIMDVLAEM 202
+ + +E LSLS I++ R +MD+L +M
Sbjct: 192 GIPQAGYGVPQAGYGIPQVGYGMPSGSSRRLDEAMATEVEGLSLSSIESMRDVMDLLGDM 251
Query: 203 LSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVNSTSDESLLCQGLALNDDLQRVLAK 262
L A++P ++E ++ EVIVDLVE+CR+ +++++ ++ ST D+ LL +GL LND LQ +LAK
Sbjct: 252 LQAVDPSDREAVKDEVIVDLVERCRSNQKKLMQMLTSTGDDELLGRGLDLNDSLQILLAK 311
Query: 263 HESIASGTPGQNHTEKLNPAPPGALIDVGGPLVDTGDTSKQSDERSPSGAEGGSQTLNQL 322
H++IASG+P L G+ + V D+S +S E S + GS +
Sbjct: 312 HDAIASGSP-------LPVQASGSPLSVQAS--KPADSSPKSSEAKDSSSIAGSSS---- 358
Query: 323 LLPAPPT-SNG----------------------SAPPIKIDPKLDLLSGDDYNSPKADNS 359
P P T S G S PP + D S + +N+ A N+
Sbjct: 359 --PIPATVSTGKSPIDEEYEEEEDEFAQLARRHSKPPASV--TTDPTSLESHNA--ASNA 412
Query: 360 LALVPVGQQQPASPMSQQNALVLFDM-FSNGSNAPSVNNQP-----------TNV----- 402
LAL P + +Q+ + L + S P+ ++QP T++
Sbjct: 413 LALALPDPPPPVNTTKEQDMIDLLSITLCTPSTPPAPSSQPSPPPPAGSDQNTHIYPQPQ 472
Query: 403 ----------AVPNSPLSPQFQQ-----QQTFISQGVYYPNGSMPSVGSPQYEQSRYAQS 447
A P PQ QQ QQ QG P S G Q +Q + Q
Sbjct: 473 PRFDSYVAPWAQQQQPQQPQAQQGYSQHQQHQQQQGYSQPQHSQQQQGYSQLQQPQPQQ- 531
Query: 448 TGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAA 485
G + + AQ Q PS Q+ +PPPPW + +A
Sbjct: 532 -GYSQSQPQAQVQMQPS--TRPQNPYEYPPPPWASTSA 566
>At5g01760 unknown protein
Length = 539
Score = 243 bits (621), Expect = 2e-64
Identities = 164/492 (33%), Positives = 257/492 (51%), Gaps = 40/492 (8%)
Query: 2 VNPLVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTL 61
V V++ATS++L PDW + I ICD LN + Q KD +K +K+R+ +SS+VQLL LT
Sbjct: 22 VTVAVDKATSELLRTPDWTIIIAICDSLNSNRWQCKDAIKAVKRRLQHKSSRVQLLTLTA 81
Query: 62 LETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARYP 121
+ +KNCGD VH H+AE+ +L +MVK+V+KK D VR K++IL+DTW EAF G ++P
Sbjct: 82 M---LKNCGDFVHSHIAEKHLLEDMVKLVRKKGDFEVRNKLLILLDTWNEAFSGVACKHP 138
Query: 122 QYYAAYQELLHSGAVFPQRSEQSAPVF---TPLQTQPLTSYPQNIRDSDAQPHTAESSAE 178
Y AYQEL G FPQRS++ AP+ P TQ +S N+ + E+ A
Sbjct: 139 HYNWAYQELKRCGVKFPQRSKE-APLMLEPPPPVTQSSSSSSMNLMSIGSFRRLDETMA- 196
Query: 179 SEFPTLSLSEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVN 238
+E +LSLS +++ R +MD++ +M+ A+ P +K L+ E+IVDLVEQCR+ +++++ ++
Sbjct: 197 TEIESLSLSSLESMRNVMDLVNDMVQAVNPSDKSALKDELIVDLVEQCRSNQKKLIQMLT 256
Query: 239 STSDESLLCQGLALNDDLQRVLAKHESIASGTPGQNHTEKLNP-APPGALIDVGGPLVDT 297
+T+DE +L +GL LND LQ VLA+H++IASG + P +L G +++
Sbjct: 257 TTADEDVLARGLELNDSLQVVLARHDAIASGVSLPLLLQAPEPRETSSSLKTCGAAALES 316
Query: 298 GDT-SKQSDERSPSGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDPKLDLLSGDDYNSPKA 356
D+ S S S S + + + A + +A + + LL D+ + +A
Sbjct: 317 ADSESSSSSSSSESETDEVEDVKDDFIQLAKRHALLNALHSDEEEETLLLGNDNEKTAEA 376
Query: 357 D-----NSLALVPVGQQQPASPMSQQNALVLFDMFSNGSNAPSVNNQPTNVAVPNSPLSP 411
+ LAL + S+Q+ + L + + A+P+ P
Sbjct: 377 EAKTQCKDLALFDT--TTTTTTKSEQDIIELLSL------------TLSTTALPSPQTQP 422
Query: 412 QFQQQQTFISQGVYYPNGSMP---SVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNG 468
Q Q F + + +P S PQ + +GP + QQQQP S
Sbjct: 423 QTQHPSFFADDNILMNSYVVPWAQSQEEPQVPKMTQFAPSGPQFQPWPLQQQQPYS---- 478
Query: 469 AQSGGSFPPPPW 480
+P P W
Sbjct: 479 ----YGYPQPQW 486
>At5g63640 unknown protein (At5g63640)
Length = 447
Score = 208 bits (529), Expect = 9e-54
Identities = 114/292 (39%), Positives = 168/292 (57%), Gaps = 10/292 (3%)
Query: 1 MVNPLVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALT 60
M LV ATS+ L DWA NIEIC++ RD Q+KDV+K +KKR+GS++ QL A+
Sbjct: 1 MAAELVSSATSEKLADVDWAKNIEICELAARDERQAKDVIKAIKKRLGSKNPNTQLYAVQ 60
Query: 61 LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARY 120
LLE ++ N G+ +H V + VL +VKIVKKK D VRE+I +L+D Q + GG ++
Sbjct: 61 LLEMLMNNIGENIHKQVIDTGVLPTLVKIVKKKSDLPVRERIFLLLDATQTSLGGASGKF 120
Query: 121 PQYYAAYQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQNIRDSDAQPHTAESSAESE 180
PQYY AY EL+++G F QR + PV Q P + + + + + ES+
Sbjct: 121 PQYYTAYYELVNAGVKFTQR-PNATPVVVTAQAVPRNTLNEQLASARNEGPATTQQRESQ 179
Query: 181 FPTLSLSEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVNST 240
+ S S +Q A +++L E+L A++ N EG + E +DLVEQC K+RV+HLV ++
Sbjct: 180 SVSPS-SILQKASTALEILKEVLDAVDSQNPEGAKDEFTLDLVEQCSFQKERVMHLVMTS 238
Query: 241 SDESLLCQGLALNDDLQRVLAKHESIASG--------TPGQNHTEKLNPAPP 284
DE + + + LN+ LQR+L +HE + SG T + L P P
Sbjct: 239 RDEKAVSKAIELNEQLQRILNRHEDLLSGRITVPSRSTTSNGYHSNLEPVRP 290
>At5g16880 TOM (target of myb1) -like protein
Length = 407
Score = 135 bits (341), Expect = 6e-32
Identities = 93/280 (33%), Positives = 153/280 (54%), Gaps = 35/280 (12%)
Query: 5 LVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLET 64
+VE AT++ L PDW +N+EICD++N++ S ++++G+KKRI + ++Q LAL LLET
Sbjct: 51 IVEDATTENLEEPDWDMNLEICDMINQETINSVELIRGIKKRIMMKQPRIQYLALVLLET 110
Query: 65 IIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRV----REKIMILIDTWQEAFGGPRARY 120
+KNC AER VL EMVK++ DP+ R K ++LI+ W E+ RY
Sbjct: 111 CVKNCEKAFSEVAAER-VLDEMVKLI---DDPQTVVNNRNKALMLIEAWGESTS--ELRY 164
Query: 121 -PQYYAAYQELLHSGAVFPQRSEQS-APVFTPLQTQPL----TSYPQNIR-------DSD 167
P + Y+ L G FP R +S AP+FTP ++ P PQ++ D
Sbjct: 165 LPVFEETYKSLKARGIRFPGRDNESLAPIFTPARSTPAPELNADLPQHVHEPAHIQYDVP 224
Query: 168 AQPHTAESSAESEFPTLSLSEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCR 227
+ TAE + E+ AR +++L+ +LS+ + L+ ++ LV+QCR
Sbjct: 225 VRSFTAEQTKEA---------FDIARNSIELLSTVLSS--SPQHDALQDDLTTTLVQQCR 273
Query: 228 TYKQRVVHLVNSTSD-ESLLCQGLALNDDLQRVLAKHESI 266
+ V ++ + + E+LL + L +ND+L + L+K+E +
Sbjct: 274 QSQTTVQRIIETAGENEALLFEALNVNDELVKTLSKYEEM 313
>At1g06210 unknown protein
Length = 383
Score = 101 bits (251), Expect = 2e-21
Identities = 93/361 (25%), Positives = 162/361 (44%), Gaps = 36/361 (9%)
Query: 5 LVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLET 64
+V+ AT + L P+W +N+ IC +N D ++V+ +K++I +S Q L+L LLE
Sbjct: 41 MVDEATLETLEEPNWGMNMRICAQINNDEFNGTEIVRAIKRKISGKSPVSQRLSLELLEA 100
Query: 65 IIKNCGDIVHMHVAERDVLHEMVKIVKK-KPDPRVREKIMILIDTWQEAFGGPRARYPQY 123
NC + V VA VL EMV ++K + D R++ LI W ++ P +
Sbjct: 101 CAMNC-EKVFSEVASEKVLDEMVWLIKNGEADSENRKRAFQLIRAWGQS--QDLTYLPVF 157
Query: 124 YAAYQELLHSGAVFPQRSEQSAP--------VFTPLQTQPLTSYPQNIRDSDAQPHTAES 175
+ Y L + + E S P + P+ P SYP + Q +
Sbjct: 158 HQTYMSLEGENGLHARGEENSMPGQSSLESLMQRPVPVPPPGSYPV---PNQEQALGDDD 214
Query: 176 SAESEFPTLSL----SEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQ 231
+ F LS+ +I+ R +++L+ ML+ G ++ V L+E+C+ +
Sbjct: 215 GLDYNFGNLSIKDKKEQIEITRNSLELLSSMLNT--EGKPNHTEDDLTVSLMEKCKQSQP 272
Query: 232 RVVHLVNSTS-DESLLCQGLALNDDLQRVLAKHESIASGTPGQNHTEKLNPAPPGALIDV 290
+ ++ ST+ DE +L + L LND+LQ+VL+ ++ + TEK +
Sbjct: 273 LIQMIIESTTDDEGVLFEALHLNDELQQVLSSYKK-------PDETEK----KASIVEQE 321
Query: 291 GGPLVDTGDTSKQSDERSPSGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDPKLDL-LSGD 349
DTG + +E+ P G ++ A +SN + K K++L LS D
Sbjct: 322 SSGSKDTGPKPTEQEEQEPVKKTGADD--DKKHSEASGSSNKTVKEEKQAVKIELGLSSD 379
Query: 350 D 350
+
Sbjct: 380 E 380
>At4g10070 putative DNA-directed RNA polymerase
Length = 748
Score = 46.2 bits (108), Expect = 6e-05
Identities = 71/270 (26%), Positives = 92/270 (33%), Gaps = 63/270 (23%)
Query: 405 PNSPLSPQFQQQQTFISQGVYYPNGSMPSVGS--PQYEQSRYAQSTGPAWNGQAAQQQQP 462
P++P + + + SQG YY + P G PQ+ R T W+ Q+P
Sbjct: 399 PHAPHPYDYHPRGPYSSQGSYY---NSPGFGGYPPQHMPPRGGYGTD--WD------QRP 447
Query: 463 P--SPVN-----GAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQG--- 512
P P N GAQS G PPP + G P PL G
Sbjct: 448 PYSGPYNYYGRQGAQSAGPVPPPSGPVPSPAFGGP--------PLSQVSYGYGQSHGPEY 499
Query: 513 ---APLPQGPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQYAQGVAASPYM- 568
AP Q GY Q G +QP N +Q G +P G + M
Sbjct: 500 GHAAPYSQ----TGYQQTYGQTYEQPKYD----SNPPMQPPYGGSYPPAGGGQSGYYQMQ 551
Query: 569 --GMVPHQMQNGPVAHPMYAQQMYG-NQYMGGYGYGQQPQQGV-----------QYVEQQ 614
G+ P+ MQ GPV QQ YG Q G P QG +QQ
Sbjct: 552 QPGVRPYGMQQGPV------QQGYGPPQPAAAASSGDVPYQGATPAAPSYGSTNMAPQQQ 605
Query: 615 MHGLSMSDDRNSSYVPPKKPSKPEDKLFGD 644
+G + SD P S P + +
Sbjct: 606 QYGYTSSDGPVQQQTYPSYSSAPPSDAYNN 635
Score = 36.2 bits (82), Expect = 0.060
Identities = 55/242 (22%), Positives = 81/242 (32%), Gaps = 31/242 (12%)
Query: 325 PAPPTSNGSAPPIKIDPKLDLLSGDDYNSPKADNSLALVPVGQQQPASPMSQQNALVLFD 384
P P GS PP +G + + P G QQ P+ Q
Sbjct: 528 PMQPPYGGSYPP----------AGGGQSGYYQMQQPGVRPYGMQQ--GPVQQGYGPPQPA 575
Query: 385 MFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQTFISQGVY----YPNGSMPSVGSPQYE 440
++ + P P + ++ ++PQ QQ S G YP+ S + S Y
Sbjct: 576 AAASSGDVPYQGATPAAPSYGSTNMAPQQQQYGYTSSDGPVQQQTYPSYSS-APPSDAYN 634
Query: 441 QSRYAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAENG---SPVAGNQYP 497
+TGPA+ Q+ Q GAQ +A AA G +P G YP
Sbjct: 635 NGTQTPATGPAYQQQSVQPASSTYDQTGAQ----------QAAAAGYGGQVAPTGGYTYP 684
Query: 498 QPLQVTQMVVTHVQGAPLPQGPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQ 557
+ Q AP G + QA +Y P + + + Q Q+
Sbjct: 685 TSQPAYGSQAAYSQAAPTQTGYEQQPATQA-AVYATAPGTAPVKTQSPQSAYAQYDASQV 743
Query: 558 YA 559
YA
Sbjct: 744 YA 745
>At2g35230 unknown protein
Length = 402
Score = 45.1 bits (105), Expect = 1e-04
Identities = 79/311 (25%), Positives = 117/311 (37%), Gaps = 59/311 (18%)
Query: 326 APPTSNGSAPPIKIDPKLDLLSGDDYNS--------------PKADNSLALVPVG---QQ 368
A TSNG+AP ++ P++ +S +D+ S P+ + +L P Q+
Sbjct: 30 AASTSNGAAPRLQTQPQVYNISKNDFRSIVQQLTGSPSRESLPRPPQNNSLRPQNTRLQR 89
Query: 369 QPASPMSQQN-ALVLFDMFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQTFISQGVYYP 427
SP++Q N V + + P QP P+ P PQ QQ + ++
Sbjct: 90 IRPSPLTQLNRPAVPLPSMAPPQSHPQFARQP-----PHQPPFPQTTQQPMMGHRDQFWS 144
Query: 428 NGSMPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAEN 487
N + SP E RY QS+ +G A Q QP G + P E
Sbjct: 145 N----TAESPVSEYMRYLQSS-LGDSGPNANQMQP-----GHEQRPYIP-------GHEQ 187
Query: 488 GSPVAGNQ---YPQPLQVTQMVVTHVQGAPLPQGPQAMGYDQAVGMYMQQPNASHMSAMN 544
V GN+ Y + + H Q + +P Q+ Q Q MN
Sbjct: 188 RPYVPGNEQQPYMPGNEQRPYIPGHEQRSYMPAQSQSQSQPQPQPQPQQHMMPGPQPRMN 247
Query: 545 NQ--VQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGPV------AHPMYAQQMYG---NQ 593
Q +Q NQ+ P G+ SP +P N PV PM++ QMYG +
Sbjct: 248 MQGPLQPNQYLPPP----GLVPSPVPHNLPSPRFNAPVPVTPTQPSPMFS-QMYGGFPSP 302
Query: 594 YMGGYGYGQQP 604
G+G Q P
Sbjct: 303 RYNGFGPLQSP 313
>At5g44780 unknown protein
Length = 723
Score = 43.9 bits (102), Expect = 3e-04
Identities = 60/210 (28%), Positives = 79/210 (37%), Gaps = 32/210 (15%)
Query: 422 QGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWE 481
QG MP G Q ++S+ S G GQA Q+P + V Q G+ PP +
Sbjct: 312 QGNVKQRQEMPIHGQGQAQRSQMPSSQGTLRQGQAQGSQRPSNQVGYNQGQGAQTPPYHQ 371
Query: 482 AQAAENGS-PVAGNQYPQPLQVTQMVVTHVQGAP---LPQGPQAMGYDQAVGMYMQQPNA 537
Q A+ + N Y Q V + QG P + Q Q G Y Q
Sbjct: 372 GQGAQTPPYQESPNNYGQ-----GAFVQYNQGPPQGNVVQTTQEKYNQMGQGNYAPQSGG 426
Query: 538 SHMSAMNNQVQSNQFGMHPQYAQGVAASPYMG--------MVPHQMQNGPVAHPMYAQQM 589
++ A S +FG + Q G SPY G +P Q Q G ++ M Q
Sbjct: 427 NYSPAQG--AGSPRFG-YGQGQGGQLLSPYRGNYNQGQGTPLPGQGQEGQPSYQMGFSQG 483
Query: 590 YG-----NQYM-GGYG------YGQQPQQG 607
G NQ + G YG Y Q P QG
Sbjct: 484 LGAPVPPNQVIPGNYGQWAFVNYNQGPPQG 513
>At1g17440 unknown protein
Length = 683
Score = 43.5 bits (101), Expect = 4e-04
Identities = 63/259 (24%), Positives = 92/259 (35%), Gaps = 38/259 (14%)
Query: 359 SLALVPVGQQQPASPMSQQNALVLFDMFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQT 418
S +L P Q P +PM A + S+ S + P + +V NS SP T
Sbjct: 7 SSSLSPKSLQSP-NPMEPSPASST-PLPSSSSQQQQLMTAPISNSV-NSAASPAMTVTTT 63
Query: 419 FISQGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPP 478
+G+ N S P++ SP S N Q PSP+ S P
Sbjct: 64 ---EGIVIQNNSQPNISSPNPTSS----------NPPIGAQIPSPSPL-------SHPSS 103
Query: 479 PWEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQ-GPQAMGYDQAVGMYMQQPNA 537
+ Q Q PQ Q ++ + +P+PQ PQ +QQ
Sbjct: 104 SLDQQTQTQQLVQQTQQLPQQ---QQQIMQQISSSPIPQLSPQQQ-------QILQQ--- 150
Query: 538 SHMSAMNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGPVAHPMYAQQ-MYGNQYMG 596
HM++ + S Q Q + ++ + Q G + + Q +YG G
Sbjct: 151 QHMTSQQIPMSSYQIAQSLQRSPSLSRLSQIQQQQQQQHQGQYGNVLRQQAGLYGTMNFG 210
Query: 597 GYGYGQQPQQGVQYVEQQM 615
G G QQ QQ Q V M
Sbjct: 211 GSGSVQQSQQNQQMVNPNM 229
>At1g15780 unknown protein
Length = 1335
Score = 43.5 bits (101), Expect = 4e-04
Identities = 78/309 (25%), Positives = 105/309 (33%), Gaps = 51/309 (16%)
Query: 367 QQQPASPMSQQNALVLFDMFSNGSNAPSVNN--------QPTNVAVPNS------PLSPQ 412
QQQP Q +L F +G N P+ N+ Q NV PN P P
Sbjct: 222 QQQPYLYQQQLQQQLLKQNFQSG-NVPNPNSLLPSHIQQQQQNVLQPNQLHSSQQPGVPT 280
Query: 413 FQQQQTFIS----QGVYYPNGSMPSVGSPQYEQS--RYAQSTGPAWNGQAAQ-----QQQ 461
Q + ++ QG++ S P + S Q QS R QS+ + Q+ Q QQQ
Sbjct: 281 SATQPSTVNSAPLQGLHTNQQSSPQLSSQQTTQSMLRQHQSSMLRQHPQSQQASGIHQQQ 340
Query: 462 PPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQGPQA 521
P P QAA + +Q QM+ HV G Q Q
Sbjct: 341 SSLPQQSISPLQQQPTQLMRQQAANSSG----------IQQKQMMGQHVVGDMQQQHQQR 390
Query: 522 MGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGP-- 579
+ Q M +QQ + Q Q Q P Q + + HQ G
Sbjct: 391 LLNQQNNVMNIQQQQSQQQPLQQPQQQQKQ---QPPAQQQLMSQQNSLQATHQNPLGTQS 447
Query: 580 -VAHPMYAQQMYGNQYMGGYGYGQ---------QPQQGVQYVEQQMHGLSMSDDRNSSYV 629
VA QQ N +G QP G+Q Q HGL S + S
Sbjct: 448 NVAGLQQPQQQMLNSQVGNSSLQNNQHSVHMLSQPTVGLQRTHQAGHGLYSSQGQQSQNQ 507
Query: 630 PPKKPSKPE 638
P ++ P+
Sbjct: 508 PSQQQMMPQ 516
>At4g28300 predicted proline-rich protein
Length = 496
Score = 42.7 bits (99), Expect = 6e-04
Identities = 69/243 (28%), Positives = 84/243 (34%), Gaps = 46/243 (18%)
Query: 309 PSGAEGGSQTLNQLLLPAPPTSNGSAPPIKIDPKLDLLSGDDYNSP---KADNSLALVPV 365
P +E S NQ L A P + AP ++ P+ Y P + N+ A VPV
Sbjct: 199 PKKSENTSDAHNQQLALALP--HQIAPQPQVQPQPQPQQHQYYMPPPPTQLQNTPAPVPV 256
Query: 366 GQQQPASPMSQQNALVLFDMFSNGSNAPSVNNQPTNVAVP----NSPLSPQFQQQQTFIS 421
++P SQ A F APS + + P N P PQ + Q S
Sbjct: 257 -----STPPSQLQAPPAQSQFMPPPPAPSHPSSAQTQSFPQYQQNWPPQPQARPQ----S 307
Query: 422 QGVY-----YPNGSMPSV--------------GSPQYEQSRYAQSTGPAWNGQAAQQQQP 462
G Y P G+ P V G PQ Y P Q +
Sbjct: 308 SGGYPTYSPAPPGNQPPVESLPSSMQMQSPYSGPPQQSMQAYGYGAAPPPQAPPQQTKMS 367
Query: 463 PSPVNGAQSGGSFPPPP-WEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQGPQA 521
SP G S PPPP A A G + QYP P Q Q A QGPQ
Sbjct: 368 YSPQTGDGYLPSGPPPPSGYANAMYEGGRM---QYPPPQPQQQQ-----QQAHYLQGPQG 419
Query: 522 MGY 524
GY
Sbjct: 420 GGY 422
Score = 36.2 bits (82), Expect = 0.060
Identities = 47/204 (23%), Positives = 73/204 (35%), Gaps = 35/204 (17%)
Query: 320 NQLLLPAPPTSNGSAP-PIKIDPKLDLLSGDDYNS-----------PKADNSLALVPVGQ 367
+Q +P PPT + P P+ + L S P + + + Q
Sbjct: 236 HQYYMPPPPTQLQNTPAPVPVSTPPSQLQAPPAQSQFMPPPPAPSHPSSAQTQSFPQYQQ 295
Query: 368 QQPASPMSQQNALVLFDMFSNG--SNAPSVNNQPTNVAVPNSPLS--------------- 410
P P ++ + + +S N P V + P+++ + SP S
Sbjct: 296 NWPPQPQARPQSSGGYPTYSPAPPGNQPPVESLPSSMQM-QSPYSGPPQQSMQAYGYGAA 354
Query: 411 --PQFQQQQTFISQGVYYPNGSMPSVGSPQ--YEQSRYAQSTGPAWNGQAAQQQQPPSPV 466
PQ QQT +S +G +PS P Y + Y Q QQQQ +
Sbjct: 355 PPPQAPPQQTKMSYSPQTGDGYLPSGPPPPSGYANAMYEGGRMQYPPPQPQQQQQQAHYL 414
Query: 467 NGAQSGGSFPPPPWEAQAAENGSP 490
G Q GG + P P +A G+P
Sbjct: 415 QGPQGGG-YSPQPHQAGGGNIGAP 437
>At1g59910 hypothetical protein
Length = 929
Score = 40.8 bits (94), Expect = 0.002
Identities = 37/129 (28%), Positives = 48/129 (36%), Gaps = 6/129 (4%)
Query: 390 SNAPSVNNQPTNVAVPNSPLSPQFQQQQTFISQGVYYPNGSMPSVGSPQYEQSRYAQSTG 449
++APSV P N+P S Q + G Y P + S +P +
Sbjct: 315 TSAPSVPLPPGQYTAVNAPFSTS--TQPVSLPPGQYMPGNAALSASTPLTPGQFTTANAP 372
Query: 450 PAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVTH 509
PA G A Q PP P A + PPPP + AA P G + P M
Sbjct: 373 PAPPGPANQTSPPPPPPPSAAAPPP-PPPPKKGPAAPPPPPPPGKKGAGPPPPPPM---S 428
Query: 510 VQGAPLPQG 518
+G P P G
Sbjct: 429 KKGPPKPPG 437
Score = 31.2 bits (69), Expect = 1.9
Identities = 58/243 (23%), Positives = 80/243 (32%), Gaps = 36/243 (14%)
Query: 427 PNGSMPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAE 486
P+ S S SP ++ YA S G + G +Q A GGSFP P + +
Sbjct: 184 PSSSSASAASPS--RTSYATSAGSDYGGGGGGKQSQSK--FQAPGGGSFPSSPSQIHSGG 239
Query: 487 NGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQGP--QAMGYDQAVGMYMQQPNASHMSAMN 544
SP P PL Q + Q P Q M + + P +M+
Sbjct: 240 GRSP------PLPLPPGQFTAGNASFPSSTQPPPGQYMAGNASFPSSTPPPPGQYMA--G 291
Query: 545 NQVQSNQFGMHP-QY----AQGVAASPYMGMVPHQMQ--NGPVAHPMYAQQMYGNQYMGG 597
N S+ + P QY AQ ++P + + P Q N P + + QYM G
Sbjct: 292 NAPFSSSTPLPPGQYPAVNAQLSTSAPSVPLPPGQYTAVNAPFSTSTQPVSLPPGQYMPG 351
Query: 598 YG--YGQQPQQGVQYVEQQMHGLSMSDDRNSSYVPPKKPSKPEDKLFGDLVDMAKVKPKP 655
P Q+ +S PP PS A P P
Sbjct: 352 NAALSASTPLTPGQFTTANAPPAPPGPANQTSPPPPPPPS-------------AAAPPPP 398
Query: 656 TPP 658
PP
Sbjct: 399 PPP 401
>At4g20020 DAG-like protein
Length = 406
Score = 40.4 bits (93), Expect = 0.003
Identities = 33/99 (33%), Positives = 40/99 (40%), Gaps = 12/99 (12%)
Query: 444 YAQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAE-NGSPVAGNQYPQPLQV 502
Y G A GQ Q PP G G PPPP++A + GSPV P Q
Sbjct: 236 YGPQQGYATPGQGQGTQAPPPFQGGYNQGPRSPPPPYQAGYNQGQGSPV------PPYQA 289
Query: 503 TQMVVTHVQGAPLP--QGPQAMGYDQAVGMYMQQPNASH 539
VQG+P+P QG Q+ G Y Q P +
Sbjct: 290 G---YNQVQGSPVPPYQGTQSSYGQGGSGNYSQGPQGGY 325
Score = 35.4 bits (80), Expect = 0.10
Identities = 71/282 (25%), Positives = 91/282 (32%), Gaps = 46/282 (16%)
Query: 335 PPIKIDPKLDLLSGDDYNSPKADNSLALVPVGQQQPASPMSQQNALVLFDMFSNGSNAPS 394
P IDP+ GD Y + + + G+ +P FD GS P
Sbjct: 165 PDSYIDPQNKEYGGDKYENGVITHRPPPIQSGRARPRPR---------FDRSGGGSGGPQ 215
Query: 395 VNNQPTNVAVPNSPLSPQFQQQQTFISQGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNG 454
N + Q+ QQ G Y P G Q + Q+ P G
Sbjct: 216 -----------NFQRNTQYGQQPPMQGGGGSYG----PQQGYATPGQGQGTQAPPPFQGG 260
Query: 455 QAAQQQQPPSPVN-GAQSGGSFPPPPWEAQAAE-NGSPVAGNQYPQPLQVTQMVVTHVQG 512
+ PP P G G P PP++A + GSPV Q TQ
Sbjct: 261 YNQGPRSPPPPYQAGYNQGQGSPVPPYQAGYNQVQGSPVPP------YQGTQSSYGQGGS 314
Query: 513 APLPQGPQAMGYDQAVGMYMQQP----NASHMSAMNNQVQSNQFGMHPQYAQGVAASPYM 568
QGPQ GY+Q G P N S N + G +P Y QG Y
Sbjct: 315 GNYSQGPQG-GYNQG-GPRNYNPQGAGNFGPASGAGNLGPAPGAG-NPGYGQG-----YS 366
Query: 569 GMVPHQMQNGPVAHPMYAQQMYGNQYMGGYGYGQQPQQGVQY 610
G P Q QN + + N G Q QG +Y
Sbjct: 367 G--PGQEQNQTFPQADQRNRDWNNNNPAGQPGSDQFPQGRRY 406
>At3g05680 unknown protein
Length = 2070
Score = 40.4 bits (93), Expect = 0.003
Identities = 79/304 (25%), Positives = 105/304 (33%), Gaps = 57/304 (18%)
Query: 351 YNSPKADNSLALVP-VGQQQPASPMSQQNALVLFDMFSNGS--NAPSVNNQPTNVAVPNS 407
Y+ P + ++ P VG Q S QQ+ + FS + NQP VP
Sbjct: 1709 YHMPGSSGQNSIDPRVGPQGFYSKSGQQHTGHIHGGFSGRGVYEQKVMPNQPPLPLVPPP 1768
Query: 408 PLSPQF--------QQQQTFISQGVYYPNGS---MPSVGS--PQYEQSRYA--------- 445
+SP Q FIS G G MP + S PQY + YA
Sbjct: 1769 SVSPVIPHSSDSLSNQSSPFISHGTQSSGGPTRLMPPLPSAIPQYSSNPYASLPPRTSTV 1828
Query: 446 QSTGPAWNGQAAQQQQPPSPVNGAQSGG-------SFPPPPWEAQAAENGSPVAGNQYPQ 498
QS G G +QQ P QSG S+PPP N P P
Sbjct: 1829 QSFGYNHAGVGTTEQQQSGPTIDHQSGNLSVTGMTSYPPP--------NLMPSHNFSRPS 1880
Query: 499 PLQVTQMVVTHVQGAPLPQG---PQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMH 555
L V G P QG PQ M ++ + + + +M Q +Q
Sbjct: 1881 SLPVP------FYGNPSHQGGDKPQTMLLVPSIPQSLNTQSIPQLPSM----QLSQLQRP 1930
Query: 556 PQYAQGVAASPYMGMVPHQ---MQNGPVAHPMYAQQMYGNQYMGGYGYGQQPQQGVQYVE 612
Q Q V + Q MQN P PM+ Q+ + Y + Q Q+ Q +
Sbjct: 1931 MQPPQHVRPPIQISQPSEQGVSMQN-PFQIPMHQMQLMQQTQVQPYYHPPQQQEISQVQQ 1989
Query: 613 QQMH 616
QQ H
Sbjct: 1990 QQQH 1993
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 38.9 bits (89), Expect = 0.009
Identities = 41/151 (27%), Positives = 52/151 (34%), Gaps = 23/151 (15%)
Query: 301 SKQSDERSPSGAEGGSQTL---NQLLLPAPPT-----SNGSAPPIKIDPKLDL------L 346
S ++RS S S T L+LPAPP PP P L
Sbjct: 738 STSQEQRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSSNL 797
Query: 347 SGDDYNSPKADNSLALVPVGQQQPASPMSQQNALVLFDMFSNGSNAPSVNNQPTNVAVPN 406
+SP + A G Q A P QN+ SN+P +NN N PN
Sbjct: 798 PSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNS---------NSNSPILNNPNPNSPSPN 848
Query: 407 SPLSPQFQQQQTFISQGVYYPNGSMPSVGSP 437
SP Q S + P+ S+ SP
Sbjct: 849 SPNQNSPLPQSPISSPHIPTPSTSISEPNSP 879
Score = 29.3 bits (64), Expect = 7.3
Identities = 32/165 (19%), Positives = 54/165 (32%), Gaps = 30/165 (18%)
Query: 387 SNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQTFISQGVYYPNGSMPSVGSPQYEQSRYAQ 446
S + S N P++ +P +PL + P +G R
Sbjct: 740 SQEQRSDSAPNWPSHTTLPTTPLVLP-----------------APPCLGPHLDTSPRPPS 782
Query: 447 STGPAWNGQAAQQQQPPSPVNGAQSGGSFPPP---------PWEAQAAENGSPVAGNQYP 497
S P Q + P S ++ S P P + Q + + SP+ N P
Sbjct: 783 SPSPLCTTQVSSSNLPSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNP 842
Query: 498 QPLQVTQMVVTHVQGAPLPQGPQAMGYDQAVGMYMQQPNASHMSA 542
+ Q +PLPQ P + + + +PN+ S+
Sbjct: 843 NSPSPN----SPNQNSPLPQSPISSPHIPTPSTSISEPNSPSSSS 883
>At3g44340 putative protein
Length = 1122
Score = 38.5 bits (88), Expect = 0.012
Identities = 93/414 (22%), Positives = 123/414 (29%), Gaps = 91/414 (21%)
Query: 281 PAPPGALI------DVGGPLVDTGDTSKQSDERSPSGAEGGSQTLNQLLLPAPPTSNGSA 334
P PPGA + GGP + +P+ Q LN P P +G
Sbjct: 4 PVPPGAYRPNNNQQNSGGP-----PNFVPGSQGNPNSLAANMQNLNINRPPPPMPGSGPR 58
Query: 335 PPIKIDPKLDLLSGDDYNSPKADNSLALVPVGQQQPA--SPMSQQNALVLFDMFSNGSNA 392
P SP S P QQQ SPM++ M G
Sbjct: 59 P-----------------SPPFGQSPQSFPQQQQQQPRPSPMARPGPPPPAAMARPGG-- 99
Query: 393 PSVNNQPTNVAVPNSPLSPQFQQQQ--------TFISQGVYYPN-GSMPSVGSPQYEQSR 443
P +QP P++P Q + G +P G P+ G P S
Sbjct: 100 PPQVSQPGGFPPVGRPVAPPSNQPPFGGRPSTGPLVGGGSSFPQPGGFPASGPPGGVPSG 159
Query: 444 YAQSTGPAWNGQA-----AQQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQ 498
P G PPS + G PP NG P +G
Sbjct: 160 PPSGARPIGFGSPPPMGPGMSMPPPSGMPGGPLSNGPPPSGMHGGHLSNGPPPSG----- 214
Query: 499 PLQVTQMVVTHVQGAPLPQGPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQY 558
+ G PL GP M P A + P Y
Sbjct: 215 -----------MPGGPLSNGPPPP---------MMGPGA--FPRGSQFTSGPMMAPPPPY 252
Query: 559 AQGVAASPYMGMVPHQMQNGPVAHPM-----YAQQMYGNQYM-GGYGYGQQPQQ------ 606
Q A P+ G P + P AH + + YG M GG+ YG PQQ
Sbjct: 253 GQPPNAGPFTGNSP---LSSPPAHSIPPPTNFPGVPYGRPPMPGGFPYGAPPQQLPSAPG 309
Query: 607 --GVQYVEQQMHGLSMSDDRNSSYVPPKKPSKPEDKLFGDLVDMAKVKPKPTPP 658
G Y M SM+ + S + + +P +V +V+ K PP
Sbjct: 310 TPGSIYGMGPMQNQSMTSVSSPSKIDLNQIPRPGSS-SSPIVYETRVENKANPP 362
Score = 33.9 bits (76), Expect = 0.30
Identities = 59/235 (25%), Positives = 75/235 (31%), Gaps = 45/235 (19%)
Query: 361 ALVPVGQQQPASPMSQQNALVLFDMFSNGS-NAPSVNNQPTNVAVPNSPL-------SPQ 412
A VP G +P + F S G+ N+ + N Q N+ P P+ SP
Sbjct: 3 APVPPGAYRPNNNQQNSGGPPNFVPGSQGNPNSLAANMQNLNINRPPPPMPGSGPRPSPP 62
Query: 413 F-QQQQTFISQGVYYPNGS-MPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPPSPVNGAQ 470
F Q Q+F Q P S M G P A +P P +Q
Sbjct: 63 FGQSPQSFPQQQQQQPRPSPMARPGPPP-----------------PAAMARPGGPPQVSQ 105
Query: 471 SGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQGPQAMGYDQAVGM 530
GG FPP N P G PL V P P G A G
Sbjct: 106 PGG-FPPVGRPVAPPSNQPPFGGRPSTGPL------VGGGSSFPQPGGFPASG------- 151
Query: 531 YMQQPNASHMSAMNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGPVAHPMY 585
P S + + FG P G++ P GM + NGP M+
Sbjct: 152 ----PPGGVPSGPPSGARPIGFGSPPPMGPGMSMPPPSGMPGGPLSNGPPPSGMH 202
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.130 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,930,401
Number of Sequences: 26719
Number of extensions: 759987
Number of successful extensions: 2260
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 2003
Number of HSP's gapped (non-prelim): 259
length of query: 658
length of database: 11,318,596
effective HSP length: 106
effective length of query: 552
effective length of database: 8,486,382
effective search space: 4684482864
effective search space used: 4684482864
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0487.5


