

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.9
(69 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
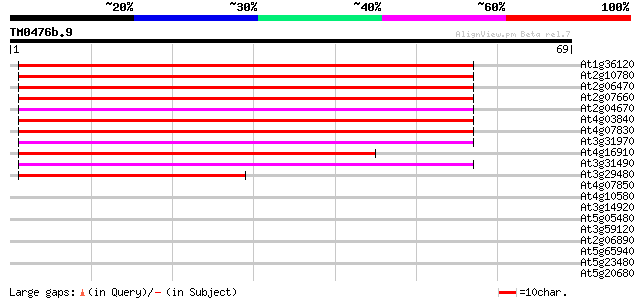
At1g36120 putative reverse transcriptase gb|AAD22339.1 59 3e-10
At2g10780 pseudogene 58 8e-10
At2g06470 putative retroelement pol polyprotein 58 8e-10
At2g07660 putative retroelement pol polyprotein 57 2e-09
At2g04670 putative retroelement pol polyprotein 57 2e-09
At4g03840 putative transposon protein 57 2e-09
At4g07830 putative reverse transcriptase 56 4e-09
At3g31970 hypothetical protein 54 1e-08
At4g16910 retrotransposon like protein 51 1e-07
At3g31490 hypothetical protein 49 6e-07
At3g29480 hypothetical protein 44 1e-05
At4g07850 putative polyprotein 32 0.058
At4g10580 putative reverse-transcriptase -like protein 31 0.13
At3g14920 hypothetical protein 27 1.4
At5g05480 unknown protein 25 5.4
At3g59120 putative protein 25 5.4
At2g06890 putative retroelement integrase 25 5.4
At5g65940 3-hydroxyisobutyryl-coenzyme A hydrolase 25 9.3
At5g23480 putative protein 25 9.3
At5g20680 unknown protein 25 9.3
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 59.3 bits (142), Expect = 3e-10
Identities = 30/56 (53%), Positives = 34/56 (60%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
LR+ IL E H S FSIHPG TKMY+DLK H GMK+ V +V C Q K E
Sbjct: 875 LRREILSEAHASMFSIHPGATKMYRDLKRHYQWVGMKRDVANWVTECDVCQLVKAE 930
>At2g10780 pseudogene
Length = 1611
Score = 58.2 bits (139), Expect = 8e-10
Identities = 29/56 (51%), Positives = 36/56 (63%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
L++ IL E H+S+FSIHPG KMY+DLK + GMKK V +V C T Q K E
Sbjct: 1159 LKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAE 1214
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 58.2 bits (139), Expect = 8e-10
Identities = 29/56 (51%), Positives = 36/56 (63%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
L++ IL E H+S+FSIHPG KMY+DLK + GMKK V +V C T Q K E
Sbjct: 609 LKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAE 664
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 57.0 bits (136), Expect = 2e-09
Identities = 28/56 (50%), Positives = 36/56 (64%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
L++ IL E H+S+FSIHPG KMY+DLK + GM+K V +V C T Q K E
Sbjct: 551 LKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAE 606
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 57.0 bits (136), Expect = 2e-09
Identities = 29/56 (51%), Positives = 34/56 (59%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
LR+ IL E H S FSIHPG TKMY+DLK + GMK+ V +V C Q K E
Sbjct: 1015 LRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQLVKAE 1070
>At4g03840 putative transposon protein
Length = 973
Score = 56.6 bits (135), Expect = 2e-09
Identities = 28/56 (50%), Positives = 36/56 (64%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
L++ IL E H+S+FSIHPG K+Y+DLK + GMKK V +V C T Q K E
Sbjct: 609 LKEEILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAE 664
>At4g07830 putative reverse transcriptase
Length = 611
Score = 55.8 bits (133), Expect = 4e-09
Identities = 29/56 (51%), Positives = 35/56 (61%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
LR+ IL E H S FSIHPG TKMY+DLK + GMK+ V +V C Q K+E
Sbjct: 169 LRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEECDVCQLVKIE 224
>At3g31970 hypothetical protein
Length = 1329
Score = 53.9 bits (128), Expect = 1e-08
Identities = 28/56 (50%), Positives = 33/56 (58%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
LR+ IL E H S FSIHP TKMY+DLK + GMK+ V +V C Q K E
Sbjct: 902 LRREILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAE 957
>At4g16910 retrotransposon like protein
Length = 687
Score = 50.8 bits (120), Expect = 1e-07
Identities = 24/44 (54%), Positives = 31/44 (69%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYV 45
L++ IL E H+S+FSIHPG KMY+DLK + GMKK V +V
Sbjct: 251 LKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV 294
>At3g31490 hypothetical protein
Length = 285
Score = 48.5 bits (114), Expect = 6e-07
Identities = 26/56 (46%), Positives = 32/56 (56%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
LR IL E H S FSI+P TKMY+DLK + GMK+ V ++ C Q K E
Sbjct: 213 LRWKILSEVHASMFSIYPRATKMYRDLKRYYQWVGMKRDVANWIAECDVCQLVKAE 268
>At3g29480 hypothetical protein
Length = 718
Score = 44.3 bits (103), Expect = 1e-05
Identities = 20/28 (71%), Positives = 22/28 (78%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLK 29
LR+ IL E H S FSIHPG TKMY+DLK
Sbjct: 688 LRREILSEAHASMFSIHPGATKMYRDLK 715
>At4g07850 putative polyprotein
Length = 1138
Score = 32.0 bits (71), Expect = 0.058
Identities = 21/76 (27%), Positives = 34/76 (44%), Gaps = 10/76 (13%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVEAKDE 61
LR+L + E H H G++K + ++ H P MK+ V C T ++AK +++
Sbjct: 758 LRELFIREAHGGGLMGHFGVSKTLKVMQDHFHWPHMKRDVERMCERCTTCKQAKAKSQPH 817
Query: 62 S----------*WNDI 67
WNDI
Sbjct: 818 GLCTPLPIPLHPWNDI 833
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 30.8 bits (68), Expect = 0.13
Identities = 16/36 (44%), Positives = 19/36 (52%)
Query: 22 TKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAKVE 57
TKMY+DLK + GMK V +V C Q K E
Sbjct: 959 TKMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAE 994
>At3g14920 hypothetical protein
Length = 609
Score = 27.3 bits (59), Expect = 1.4
Identities = 15/42 (35%), Positives = 21/42 (49%), Gaps = 1/42 (2%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVE 43
L L+ + HK FS+ + Y D LHLW +K +VE
Sbjct: 345 LGSLLDGKTHKVGFSVTNALNVWYIDANLHLWL-DQEKEIVE 385
>At5g05480 unknown protein
Length = 608
Score = 25.4 bits (54), Expect = 5.4
Identities = 11/30 (36%), Positives = 18/30 (59%), Gaps = 1/30 (3%)
Query: 5 LILD-EGHKSRFSIHPGMTKMYQDLKLHLW 33
L+LD + H+ F ++ G++ D LHLW
Sbjct: 346 LLLDGKSHEFAFGVNDGISYWLVDANLHLW 375
>At3g59120 putative protein
Length = 602
Score = 25.4 bits (54), Expect = 5.4
Identities = 15/38 (39%), Positives = 18/38 (46%), Gaps = 1/38 (2%)
Query: 11 HKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTC 48
HKS H ++ Y + L C G KKRV Y L C
Sbjct: 431 HKSHAEHHVFISPSYSEENEAL-CQGCKKRVYNYHLHC 467
>At2g06890 putative retroelement integrase
Length = 1215
Score = 25.4 bits (54), Expect = 5.4
Identities = 17/54 (31%), Positives = 23/54 (42%)
Query: 2 LRKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCLTYQKAK 55
LR L + E H H G+ K + + H P MK V C T ++AK
Sbjct: 892 LRDLFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQAK 945
>At5g65940 3-hydroxyisobutyryl-coenzyme A hydrolase
Length = 378
Score = 24.6 bits (52), Expect = 9.3
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query: 4 KLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYVLTCL--TYQKAKV 56
KL++ +GH F + + +D+ W G EY+L + TY KA+V
Sbjct: 55 KLVILKGHGRAFCAGGDVAAVVRDINQGNWRLGANYFSSEYMLNYVMATYSKAQV 109
>At5g23480 putative protein
Length = 581
Score = 24.6 bits (52), Expect = 9.3
Identities = 15/43 (34%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Query: 3 RKLILDEGHKSRFSIHPGMTKMYQDLKLHLWCPGMKKRVVEYV 45
R ++D K+R S+H +TK Y + + L P +KR+V+ V
Sbjct: 254 RLTVVDMEEKAR-SLHEDVTKHYLEKRELLQNPDEQKRLVDEV 295
>At5g20680 unknown protein
Length = 551
Score = 24.6 bits (52), Expect = 9.3
Identities = 18/33 (54%), Positives = 21/33 (63%), Gaps = 5/33 (15%)
Query: 6 ILDEGHKSRFSIHPGMTKMYQDLKLHLWC-PGM 37
I DEGH SRFSI ++ QD LH WC PG+
Sbjct: 511 IRDEGHISRFSI--SASRGVQDC-LH-WCLPGV 539
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.338 0.149 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,461,427
Number of Sequences: 26719
Number of extensions: 44375
Number of successful extensions: 176
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 160
Number of HSP's gapped (non-prelim): 21
length of query: 69
length of database: 11,318,596
effective HSP length: 45
effective length of query: 24
effective length of database: 10,116,241
effective search space: 242789784
effective search space used: 242789784
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0476b.9


