

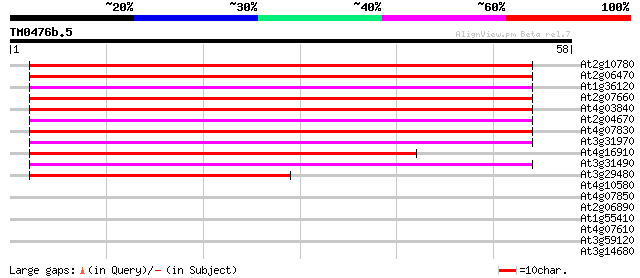
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.5
(58 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 56 4e-09
At2g06470 putative retroelement pol polyprotein 56 4e-09
At1g36120 putative reverse transcriptase gb|AAD22339.1 55 5e-09
At2g07660 putative retroelement pol polyprotein 55 9e-09
At4g03840 putative transposon protein 54 1e-08
At2g04670 putative retroelement pol polyprotein 53 3e-08
At4g07830 putative reverse transcriptase 52 4e-08
At3g31970 hypothetical protein 50 2e-07
At4g16910 retrotransposon like protein 49 6e-07
At3g31490 hypothetical protein 46 3e-06
At3g29480 hypothetical protein 40 3e-04
At4g10580 putative reverse-transcriptase -like protein 32 0.046
At4g07850 putative polyprotein 32 0.060
At2g06890 putative retroelement integrase 26 3.3
At1g55410 hypothetical protein 26 3.3
At4g07610 25 5.6
At3g59120 putative protein 25 7.3
At3g14680 putative cytochrome P450 25 9.6
>At2g10780 pseudogene
Length = 1611
Score = 55.8 bits (133), Expect = 4e-09
Identities = 27/52 (51%), Positives = 33/52 (62%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H+S+ SIHPG KMY+DLK ++ GMKK V +V C T Q K E
Sbjct: 1163 ILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAE 1214
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 55.8 bits (133), Expect = 4e-09
Identities = 27/52 (51%), Positives = 33/52 (62%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H+S+ SIHPG KMY+DLK ++ GMKK V +V C T Q K E
Sbjct: 613 ILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAE 664
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 55.5 bits (132), Expect = 5e-09
Identities = 27/52 (51%), Positives = 31/52 (58%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H S SIHPG TKMY+DLK H+ GMK+ V +V C Q K E
Sbjct: 879 ILSEAHASMFSIHPGATKMYRDLKRHYQWVGMKRDVANWVTECDVCQLVKAE 930
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 54.7 bits (130), Expect = 9e-09
Identities = 26/52 (50%), Positives = 33/52 (63%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H+S+ SIHPG KMY+DLK ++ GM+K V +V C T Q K E
Sbjct: 555 ILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAE 606
>At4g03840 putative transposon protein
Length = 973
Score = 54.3 bits (129), Expect = 1e-08
Identities = 26/52 (50%), Positives = 33/52 (63%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H+S+ SIHPG K+Y+DLK ++ GMKK V +V C T Q K E
Sbjct: 613 ILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAE 664
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 53.1 bits (126), Expect = 3e-08
Identities = 26/52 (50%), Positives = 31/52 (59%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H S SIHPG TKMY+DLK ++ GMK+ V +V C Q K E
Sbjct: 1019 ILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQLVKAE 1070
>At4g07830 putative reverse transcriptase
Length = 611
Score = 52.4 bits (124), Expect = 4e-08
Identities = 26/52 (50%), Positives = 32/52 (61%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H S SIHPG TKMY+DLK ++ GMK+ V +V C Q K+E
Sbjct: 173 ILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEECDVCQLVKIE 224
>At3g31970 hypothetical protein
Length = 1329
Score = 50.1 bits (118), Expect = 2e-07
Identities = 25/52 (48%), Positives = 30/52 (57%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H S SIHP TKMY+DLK ++ GMK+ V +V C Q K E
Sbjct: 906 ILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAE 957
>At4g16910 retrotransposon like protein
Length = 687
Score = 48.5 bits (114), Expect = 6e-07
Identities = 22/40 (55%), Positives = 28/40 (70%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYV 42
IL E H+S+ SIHPG KMY+DLK ++ GMKK V +V
Sbjct: 255 ILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV 294
>At3g31490 hypothetical protein
Length = 285
Score = 46.2 bits (108), Expect = 3e-06
Identities = 23/52 (44%), Positives = 30/52 (57%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
IL E H S SI+P TKMY+DLK ++ GMK+ V ++ C Q K E
Sbjct: 217 ILSEVHASMFSIYPRATKMYRDLKRYYQWVGMKRDVANWIAECDVCQLVKAE 268
>At3g29480 hypothetical protein
Length = 718
Score = 39.7 bits (91), Expect = 3e-04
Identities = 17/27 (62%), Positives = 20/27 (73%)
Query: 3 ILDEGHKSRLSIHPGMTKMYQDLKLHF 29
IL E H S SIHPG TKMY+DLK ++
Sbjct: 692 ILSEAHASMFSIHPGATKMYRDLKEYY 718
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 32.3 bits (72), Expect = 0.046
Identities = 16/36 (44%), Positives = 20/36 (55%)
Query: 19 TKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVE 54
TKMY+DLK ++ GMK V +V C Q K E
Sbjct: 959 TKMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAE 994
>At4g07850 putative polyprotein
Length = 1138
Score = 32.0 bits (71), Expect = 0.060
Identities = 17/56 (30%), Positives = 30/56 (53%)
Query: 1 KLILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKVEAK 56
+L + E H L H G++K + ++ HF P MK+ V C T ++AK +++
Sbjct: 760 ELFIREAHGGGLMGHFGVSKTLKVMQDHFHWPHMKRDVERMCERCTTCKQAKAKSQ 815
>At2g06890 putative retroelement integrase
Length = 1215
Score = 26.2 bits (56), Expect = 3.3
Identities = 17/51 (33%), Positives = 23/51 (44%)
Query: 2 LILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAK 52
L + E H L H G+ K + + HF P MK V C T ++AK
Sbjct: 895 LFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQAK 945
>At1g55410 hypothetical protein
Length = 672
Score = 26.2 bits (56), Expect = 3.3
Identities = 14/46 (30%), Positives = 22/46 (47%), Gaps = 8/46 (17%)
Query: 8 HKSRLSIHPGMTK--------MYQDLKLHFWCPGMKKRVVEYVLTC 45
HK L++ P T +YQD+ F C G ++R V ++ C
Sbjct: 437 HKHPLTLQPFATGQMPLSPYLLYQDINNMFRCDGCRRRGVGFLYKC 482
>At4g07610
Length = 276
Score = 25.4 bits (54), Expect = 5.6
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 1/35 (2%)
Query: 25 LKLHFWCPGMKKRVVEYVLTC-LTYQKAKVEAKDE 58
L+ FW P M K E+VL C ++K + ++E
Sbjct: 107 LQAGFWWPTMFKEAQEFVLKCDSCHRKGNISKRNE 141
>At3g59120 putative protein
Length = 602
Score = 25.0 bits (53), Expect = 7.3
Identities = 14/38 (36%), Positives = 18/38 (46%), Gaps = 1/38 (2%)
Query: 8 HKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTC 45
HKS H ++ Y + + C G KKRV Y L C
Sbjct: 431 HKSHAEHHVFISPSYSE-ENEALCQGCKKRVYNYHLHC 467
>At3g14680 putative cytochrome P450
Length = 512
Score = 24.6 bits (52), Expect = 9.6
Identities = 9/25 (36%), Positives = 15/25 (60%)
Query: 1 KLILDEGHKSRLSIHPGMTKMYQDL 25
KL+ D+G + + PG+T M D+
Sbjct: 184 KLVSDKGSSCEVDVWPGLTSMTADV 208
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,238,891
Number of Sequences: 26719
Number of extensions: 34256
Number of successful extensions: 98
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 82
Number of HSP's gapped (non-prelim): 18
length of query: 58
length of database: 11,318,596
effective HSP length: 34
effective length of query: 24
effective length of database: 10,410,150
effective search space: 249843600
effective search space used: 249843600
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0476b.5


