

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.4
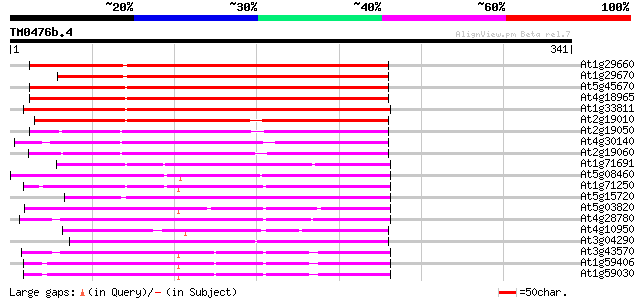
(341 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g29660 lipase/hydrolase like protein 197 5e-51
At1g29670 lipase/hydrolase-like protein 194 4e-50
At5g45670 GDSL-motif lipase/hydrolase-like protein 192 2e-49
At4g18965 unknown protein 192 2e-49
At1g33811 unknown protein 187 7e-48
At2g19010 putative GDSL-motif lipase/hydrolase 184 5e-47
At2g19050 putative GDSL-motif lipase/hydrolase 167 6e-42
At4g30140 unknown protein 167 1e-41
At2g19060 putative GDSL-motif lipase/hydrolase 159 3e-39
At1g71691 unknown protein 158 5e-39
At5g08460 GDSL-motif lipase/acylhydrolase-like protein 145 3e-35
At1g71250 putative GDSL-motif lipase/acylhydrolase 144 7e-35
At5g15720 unknown protein 139 2e-33
At5g03820 putative protein 129 2e-30
At4g28780 Proline-rich APG - like protein 127 1e-29
At4g10950 unknown protein 125 3e-29
At3g04290 GDSL-motif lipase/acylhydrolase like protein 119 2e-27
At3g43570 putative protein 118 5e-27
At1g59406 proline-rich protein, putative 116 2e-26
At1g59030 proline-rich protein, putative 116 2e-26
>At1g29660 lipase/hydrolase like protein
Length = 364
Score = 197 bits (502), Expect = 5e-51
Identities = 98/218 (44%), Positives = 143/218 (64%), Gaps = 2/218 (0%)
Query: 13 LVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFP 72
LV+V V + V E QVPC F+FGDSL D+GN+N L + A+A++ YGIDF P
Sbjct: 10 LVSVWVLLLGLGFKVKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGIDFGG--P 67
Query: 73 TERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
T R+SNGR +D + +LLGF +IP ++ ++G +IL+GVNYAS +AGIR+E+G+QLG +
Sbjct: 68 TGRFSNGRTTVDVLTELLGFDNYIPAYSTVSGQEILQGVNYASAAAGIREETGAQLGQRI 127
Query: 133 NLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMY 192
Q+ +++ V+ + LG A L +C+Y V + +ND+ NYF+P ++TSR Y
Sbjct: 128 TFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQFYSTSRQY 187
Query: 193 TPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
TP+QYA LI + L LY++GARK LVG+ +GC
Sbjct: 188 TPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGC 225
>At1g29670 lipase/hydrolase-like protein
Length = 363
Score = 194 bits (494), Expect = 4e-50
Identities = 93/201 (46%), Positives = 136/201 (67%), Gaps = 2/201 (0%)
Query: 30 ESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQL 89
++QVPC FVFGDSL D+GN+N L + A++N+ YGIDF PT R+SNG+ +D IA+L
Sbjct: 27 QAQVPCFFVFGDSLVDNGNNNGLISIARSNYFPYGIDFGG--PTGRFSNGKTTVDVIAEL 84
Query: 90 LGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIA 149
LGF +IP + ++G IL GVNYAS +AGIR+E+G QLG ++ Q+ +++ VS +
Sbjct: 85 LGFNGYIPAYNTVSGRQILSGVNYASAAAGIREETGRQLGQRISFSGQVRNYQTTVSQVV 144
Query: 150 HKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYL 209
LG +A L +C+Y V + +ND+ NYF+P +++SR +TP+QYA LI + S L
Sbjct: 145 QLLGDETRAADYLKRCIYSVGLGSNDYLNNYFMPTFYSSSRQFTPEQYANDLISRYSTQL 204
Query: 210 QTLYHFGARKSVLVGLDRLGC 230
LY++GARK L G+ +GC
Sbjct: 205 NALYNYGARKFALSGIGAVGC 225
>At5g45670 GDSL-motif lipase/hydrolase-like protein
Length = 362
Score = 192 bits (489), Expect = 2e-49
Identities = 92/218 (42%), Positives = 142/218 (64%), Gaps = 1/218 (0%)
Query: 13 LVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFP 72
++ +I+ ++ + + PC F+FGDSL D+GN+N L + A+AN+ YGIDF A P
Sbjct: 7 MIMMIMVAVTMINIAKSDPIAPCYFIFGDSLVDNGNNNQLQSLARANYFPYGIDFAAG-P 65
Query: 73 TERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
T R+SNG +D IAQLLGF+ +I P+A+ G DIL+GVNYAS +AGIR E+G QLG +
Sbjct: 66 TGRFSNGLTTVDVIAQLLGFEDYITPYASARGQDILRGVNYASAAAGIRDETGRQLGGRI 125
Query: 133 NLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMY 192
Q+ +H VS + + LG ++A+ LS+C+Y + + +ND+ NYF+P ++T +
Sbjct: 126 AFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFMPTFYSTGNQF 185
Query: 193 TPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
+P+ YA L+ + + L+ LY GARK L+G+ +GC
Sbjct: 186 SPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGC 223
>At4g18965 unknown protein
Length = 361
Score = 192 bits (488), Expect = 2e-49
Identities = 93/218 (42%), Positives = 141/218 (64%), Gaps = 1/218 (0%)
Query: 13 LVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFP 72
++ + +A M + +G+ PC F+FGDSL DSGN+N L + A+AN+ YGIDF P
Sbjct: 6 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYG-P 64
Query: 73 TERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
T R+SNG+ +D I +LLGF +I P++ G DIL+GVNYAS +AGIR+E+G QLG +
Sbjct: 65 TGRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARI 124
Query: 133 NLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMY 192
Q+ +H VS + + LG ++A LS+C+Y + + +ND+ NYF+P ++T Y
Sbjct: 125 TFAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQY 184
Query: 193 TPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
+P YA LI++ + L+ +Y+ GARK LVG+ +GC
Sbjct: 185 SPDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGC 222
>At1g33811 unknown protein
Length = 370
Score = 187 bits (475), Expect = 7e-48
Identities = 91/224 (40%), Positives = 147/224 (65%), Gaps = 2/224 (0%)
Query: 9 LILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFP 68
L++SL V+ S ++QVPC+F+FGDSL D+GN+N L + A+AN+ YGIDFP
Sbjct: 8 LLISLNLVLFGFKTTVSQPQQQAQVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGIDFP 67
Query: 69 ATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQL 128
T R++NGR +D +AQ+LGF+ +IPP++ + G IL+G N+ASG+AGIR E+G L
Sbjct: 68 QG-TTGRFTNGRTYVDALAQILGFRNYIPPYSRIRGQAILRGANFASGAAGIRDETGDNL 126
Query: 129 GHNVNLGLQL-LHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFN 187
G + ++ Q+ L+ A+ + + G ++ + LS+C++Y + +ND+ NYF+P+ ++
Sbjct: 127 GAHTSMNQQVELYTTAVQQMLRYFRGDTNELQRYLSRCIFYSGMGSNDYLNNYFMPDFYS 186
Query: 188 TSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
TS Y K +A+ LI + L LY FGARK ++ G+ ++GC+
Sbjct: 187 TSTNYNDKTFAESLIKNYTQQLTRLYQFGARKVIVTGVGQIGCI 230
>At2g19010 putative GDSL-motif lipase/hydrolase
Length = 344
Score = 184 bits (468), Expect = 5e-47
Identities = 95/215 (44%), Positives = 132/215 (61%), Gaps = 8/215 (3%)
Query: 16 VIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTER 75
++ A I +TV+ Q PC FVFGDS+SD+GN+NNL + AK NF YG DFP PT R
Sbjct: 7 LVAAIIFTAATVVYGQQAPCFFVFGDSMSDNGNNNNLKSEAKVNFSPYGNDFPKG-PTGR 65
Query: 76 YSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLG 135
+SNGR D I +L GF+ FIPPFA + G+NYASG +G+R+E+ LG +++
Sbjct: 66 FSNGRTIPDIIGELSGFKDFIPPFAEASPEQAHTGMNYASGGSGLREETSEHLGDRISIR 125
Query: 136 LQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPK 195
QL +H+ ++ + + L QCLY +NI +ND+ NYF+ +NT R YTPK
Sbjct: 126 KQLQNHKTSIT-------KANVPAERLQQCLYMINIGSNDYINNYFMSKPYNTKRRYTPK 178
Query: 196 QYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
QYA LI +L+ L+ GARK + GL ++GC
Sbjct: 179 QYAYSLIIIYRSHLKNLHRLGARKVAVFGLSQIGC 213
>At2g19050 putative GDSL-motif lipase/hydrolase
Length = 349
Score = 167 bits (424), Expect = 6e-42
Identities = 89/218 (40%), Positives = 131/218 (59%), Gaps = 9/218 (4%)
Query: 13 LVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFP 72
L+ + + LG+ +VPC FVFGDS+ D+GN+N L TSAK N+ YGIDF A P
Sbjct: 10 LLVIATTAFATTEAALGQ-RVPCYFVFGDSVFDNGNNNVLNTSAKVNYSPYGIDF-ARGP 67
Query: 73 TERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
T R+SNGRN D IA+L+ F +IPPF + G+NYASG GIR+E+ LG +
Sbjct: 68 TGRFSNGRNIPDIIAELMRFSDYIPPFTGASPEQAHIGINYASGGGGIREETSQHLGEII 127
Query: 133 NLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMY 192
+ Q+ +HR+++ + L++CLY +NI +ND+ NYF+P + T++ +
Sbjct: 128 SFKKQIKNHRSMIMT-------AKVPEEKLNKCLYTINIGSNDYLNNYFMPAPYMTNKKF 180
Query: 193 TPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
+ +YA LI YL++LY GARK + G+ +LGC
Sbjct: 181 SFDEYADSLIRSYRSYLKSLYVLGARKVAVFGVSKLGC 218
>At4g30140 unknown protein
Length = 348
Score = 167 bits (422), Expect = 1e-41
Identities = 97/228 (42%), Positives = 131/228 (56%), Gaps = 13/228 (5%)
Query: 4 ESKT-WLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLS 62
ESK W+IL+ V + A + + Q PC FVFGDS+ D+GN+N L T AK N+L
Sbjct: 5 ESKALWIILATVFAVAAV----APAVHGQQTPCYFVFGDSVFDNGNNNALNTKAKVNYLP 60
Query: 63 YGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRK 122
YGID+ PT R+SNGRN D IA+L GF IPPFA + + G+NYASG+ GIR+
Sbjct: 61 YGIDY-FQGPTGRFSNGRNIPDVIAELAGFNNPIPPFAGASQAQANIGLNYASGAGGIRE 119
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFL 182
E+ +G ++L Q+ +H + + A L L QCLY +NI +ND+ NYFL
Sbjct: 120 ETSENMGERISLRQQVNNHFSAIITAAVPLSR-------LRQCLYTINIGSNDYLNNYFL 172
Query: 183 PNVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
R++ P QYA+ LI YL LY GAR L G+ ++GC
Sbjct: 173 SPPTLARRLFNPDQYARSLISLYRIYLTQLYVLGARNVALFGIGKIGC 220
>At2g19060 putative GDSL-motif lipase/hydrolase
Length = 349
Score = 159 bits (401), Expect = 3e-39
Identities = 86/219 (39%), Positives = 125/219 (56%), Gaps = 9/219 (4%)
Query: 12 SLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATF 71
+L+ +++ V G+ VPC FVFGDS+ D+GN+N L T AK N+ YGIDF A
Sbjct: 8 ALLWAFATAVVMAEAVRGQL-VPCYFVFGDSVFDNGNNNELDTLAKVNYSPYGIDF-ARG 65
Query: 72 PTERYSNGRNPIDKIAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHN 131
PT R+SNGRN D IA+ L IPPF + G+NYASG AG+ +E+ LG
Sbjct: 66 PTGRFSNGRNIPDFIAEELRISYDIPPFTRASTEQAHTGINYASGGAGLLEETSQHLGER 125
Query: 132 VNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRM 191
++ Q+ +HR ++ + L +CLY +NI +ND+ NYF+P + T+
Sbjct: 126 ISFEKQITNHRKMIMTAG-------VPPEKLKKCLYTINIGSNDYLNNYFMPAPYTTNEN 178
Query: 192 YTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGC 230
++ +YA LI YL++LY GARK + G+ +LGC
Sbjct: 179 FSFDEYADFLIQSYRSYLKSLYVLGARKVAVFGVSKLGC 217
>At1g71691 unknown protein
Length = 384
Score = 158 bits (399), Expect = 5e-39
Identities = 80/203 (39%), Positives = 120/203 (58%), Gaps = 3/203 (1%)
Query: 29 GESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQ 88
G+ VP +FVFGDSL D+GN+NN+P+ AKAN+ YGIDF PT R+ NG +D IAQ
Sbjct: 49 GDGIVPALFVFGDSLIDNGNNNNIPSFAKANYFPYGIDFNGG-PTGRFCNGLTMVDGIAQ 107
Query: 89 LLGFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCI 148
LLG IP ++ G +L+GVNYAS +AGI ++G + Q+ + + +
Sbjct: 108 LLGLP-LIPAYSEATGDQVLRGVNYASAAAGILPDTGGNFVGRIPFDQQIHNFETTLDQV 166
Query: 149 AHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHY 208
A K GG +++ L+++ + +ND+ NY +PN F T Y +Q+ +L+ +
Sbjct: 167 ASKSGGAVAIADSVTRSLFFIGMGSNDYLNNYLMPN-FPTRNQYNSQQFGDLLVQHYTDQ 225
Query: 209 LQTLYHFGARKSVLVGLDRLGCV 231
L LY+ G RK V+ GL R+GC+
Sbjct: 226 LTRLYNLGGRKFVVAGLGRMGCI 248
>At5g08460 GDSL-motif lipase/acylhydrolase-like protein
Length = 385
Score = 145 bits (366), Expect = 3e-35
Identities = 82/233 (35%), Positives = 131/233 (56%), Gaps = 4/233 (1%)
Query: 1 MGCESKTWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANF 60
M C +T +++ V+ + + P +FVFGDSL D+GN+N+L + A++N+
Sbjct: 15 MSCTVQTLVLVPWFLVVFVLAGGEDSSETTAMFPAMFVFGDSLVDNGNNNHLNSLARSNY 74
Query: 61 LSYGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFANL--NGSDILKGVNYASGSA 118
L YGIDF PT R+SNG+ +D I +LLG IP F + G DIL GVNYAS +
Sbjct: 75 LPYGIDFAGNQPTGRFSNGKTIVDFIGELLGLPE-IPAFMDTVDGGVDILHGVNYASAAG 133
Query: 119 GIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQ 178
GI +E+G LG ++G Q+ + + I+ + + + +++ L V++ ND+
Sbjct: 134 GILEETGRHLGERFSMGRQVENFEKTLMEISRSM-RKESVKEYMAKSLVVVSLGNNDYIN 192
Query: 179 NYFLPNVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
NY P +F +S +Y P +A +L+ + +L LY G RK V+ G+ LGC+
Sbjct: 193 NYLKPRLFLSSSIYDPTSFADLLLSNFTTHLLELYGKGFRKFVIAGVGPLGCI 245
>At1g71250 putative GDSL-motif lipase/acylhydrolase
Length = 374
Score = 144 bits (363), Expect = 7e-35
Identities = 88/225 (39%), Positives = 133/225 (59%), Gaps = 7/225 (3%)
Query: 9 LILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFP 68
LIL+L ++ + Q V G+++VP +FV GDSL D+GN+N L T A+ANFL YGID
Sbjct: 17 LILALTVSVI--LQQPELVTGQARVPAMFVLGDSLVDAGNNNFLQTVARANFLPYGIDMN 74
Query: 69 ATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGS 126
PT R+SNG ID +A+LL + PPFA+ +G+ IL+GVNYAS +AGI SG
Sbjct: 75 YQ-PTGRFSNGLTFIDLLARLLEIPS-PPPFADPTTSGNRILQGVNYASAAAGILDVSGY 132
Query: 127 QLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVF 186
G +L Q+++ +S + + T L++ L + +ND+ NY +PN++
Sbjct: 133 NYGGRFSLNQQMVNLETTLSQLRTMMSP-QNFTDYLARSLVVLVFGSNDYINNYLMPNLY 191
Query: 187 NTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
++S + P +A +L+ Q + L TLY G RK + G+ LGC+
Sbjct: 192 DSSIRFRPPDFANLLLSQYARQLLTLYSLGLRKIFIPGVAPLGCI 236
>At5g15720 unknown protein
Length = 364
Score = 139 bits (351), Expect = 2e-33
Identities = 80/200 (40%), Positives = 112/200 (56%), Gaps = 4/200 (2%)
Query: 34 PCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGFQ 93
P FVFGDSL DSGN+N +PT A+AN+ YGIDF FPT R+ NGR +D A LG
Sbjct: 29 PAFFVFGDSLVDSGNNNYIPTLARANYFPYGIDFG--FPTGRFCNGRTVVDYGATYLGLP 86
Query: 94 TFIPPFANLN-GSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHKL 152
P + L+ G + L+GVNYAS +AGI E+G G Q+ + +
Sbjct: 87 LVPPYLSPLSIGQNALRGVNYASAAAGILDETGRHYGARTTFNGQISQFEITIELRLRRF 146
Query: 153 -GGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYLQT 211
+ L++ + +NI +ND+ NY +P ++TS+ Y+ + YA +LI LS +
Sbjct: 147 FQNPADLRKYLAKSIIGINIGSNDYINNYLMPERYSTSQTYSGEDYADLLIKTLSAQISR 206
Query: 212 LYHFGARKSVLVGLDRLGCV 231
LY+ GARK VL G LGC+
Sbjct: 207 LYNLGARKMVLAGSGPLGCI 226
>At5g03820 putative protein
Length = 354
Score = 129 bits (324), Expect = 2e-30
Identities = 77/225 (34%), Positives = 131/225 (58%), Gaps = 8/225 (3%)
Query: 10 ILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPA 69
I+ L+T V + GE VP + + GDS+ D+GN+N L T KANF YG DF A
Sbjct: 5 IIMLMTFSVIACFYAGVGTGEPLVPALIIMGDSVVDAGNNNRLNTLIKANFPPYGRDFLA 64
Query: 70 TFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQ 127
T R+SNG+ D A+ LGF ++ P+ + NG+++L G N+ASG++G + G+
Sbjct: 65 HNATGRFSNGKLATDFTAESLGFTSYPVPYLSQEANGTNLLTGANFASGASGY--DDGTA 122
Query: 128 LGHN-VNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVF 186
+ +N + L QL +++ + + + +G ++A + S ++ ++ ++DF Q+Y++ +
Sbjct: 123 IFYNAITLNQQLKNYKEYQNKVTNIVGS-ERANKIFSGAIHLLSTGSSDFLQSYYINPIL 181
Query: 187 NTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
N R++TP QY+ L+ S ++Q LY GARK + L LGC+
Sbjct: 182 N--RIFTPDQYSDRLMKPYSTFVQNLYDLGARKIGVTTLPPLGCL 224
>At4g28780 Proline-rich APG - like protein
Length = 367
Score = 127 bits (318), Expect = 1e-29
Identities = 76/226 (33%), Positives = 122/226 (53%), Gaps = 7/226 (3%)
Query: 7 TWLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGID 66
TW+I+++ + +M T + FVFGDSL DSGN+N L T+A+A+ YGID
Sbjct: 7 TWIIMTVALSVTLFLMPQQT----NAARAFFVFGDSLVDSGNNNYLVTTARADSPPYGID 62
Query: 67 FPATFPTERYSNGRNPIDKIAQLLGFQTFIPPFA-NLNGSDILKGVNYASGSAGIRKESG 125
+P PT R+SNG N D I++ +G + +P + L G +L G N+AS GI ++G
Sbjct: 63 YPTGRPTGRFSNGLNLPDIISEQIGSEPTLPILSPELTGEKLLIGANFASAGIGILNDTG 122
Query: 126 SQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNV 185
Q + + +G Q + ++ +G DK Q ++ L + + NDF NYF P +
Sbjct: 123 VQFLNILRIGRQFELFQEYQERVSEIIGS-DKTQQLVNGALVLMTLGGNDFVNNYFFP-I 180
Query: 186 FNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
R + +++++LI + L +LY GAR+ ++ G LGCV
Sbjct: 181 STRRRQSSLGEFSQLLISEYKKILTSLYELGARRVMVTGTGPLGCV 226
>At4g10950 unknown protein
Length = 400
Score = 125 bits (314), Expect = 3e-29
Identities = 74/201 (36%), Positives = 115/201 (56%), Gaps = 11/201 (5%)
Query: 33 VPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGF 92
VP +FVFGDS DSG +N L T A+A+ L YG DF PT R+ NGR P+D +
Sbjct: 69 VPALFVFGDSSVDSGTNNFLGTLARADRLPYGRDFDTHQPTGRFCNGRIPVDYLG----- 123
Query: 93 QTFIPPFANLNGS--DILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIAH 150
F+P + G+ D+ +GVNYAS AGI SGS+LG V+ +Q+ +
Sbjct: 124 LPFVPSYLGQTGTVEDMFQGVNYASAGAGIILSSGSELGQRVSFAMQVEQFVDTFQQMIL 183
Query: 151 KLGGLDKATQCL-SQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYL 209
+G +KA++ L S ++Y++I ND+ ++++ N+ N +YTP + + L + L
Sbjct: 184 SIG--EKASERLVSNSVFYISIGVNDY-IHFYIRNISNVQNLYTPWNFNQFLASNMRQEL 240
Query: 210 QTLYHFGARKSVLVGLDRLGC 230
+TLY+ R+ V++GL +GC
Sbjct: 241 KTLYNVKVRRMVVMGLPPIGC 261
>At3g04290 GDSL-motif lipase/acylhydrolase like protein
Length = 366
Score = 119 bits (298), Expect = 2e-27
Identities = 68/195 (34%), Positives = 107/195 (54%), Gaps = 2/195 (1%)
Query: 37 FVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGFQTFI 96
FVFGDSL D+GN++ L T+A+A+ YGID+P PT R+SNG N D I++ +G + +
Sbjct: 31 FVFGDSLVDNGNNDYLVTTARADNYPYGIDYPTRRPTGRFSNGLNIPDIISEAIGMPSTL 90
Query: 97 PPFA-NLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCIAHKLGGL 155
P + +L G ++L G N+AS GI ++G Q + + + Q+ + ++ L G
Sbjct: 91 PYLSPHLTGENLLVGANFASAGIGILNDTGIQFVNIIRISKQMEYFEQYQLRVS-ALIGP 149
Query: 156 DKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQLSHYLQTLYHF 215
+ Q ++Q L + + NDF NY+L SR Y Y LI + L+ LY
Sbjct: 150 EATQQLVNQALVLITLGGNDFVNNYYLIPFSARSRQYALPDYVVYLISEYGKILRKLYEL 209
Query: 216 GARKSVLVGLDRLGC 230
GAR+ ++ G +GC
Sbjct: 210 GARRVLVTGTGAMGC 224
>At3g43570 putative protein
Length = 320
Score = 118 bits (295), Expect = 5e-27
Identities = 74/226 (32%), Positives = 114/226 (49%), Gaps = 13/226 (5%)
Query: 8 WLILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDF 67
WL L L+ V ++ Q + +P + VFGDS+ D+GN+NNLPT K NF YG D+
Sbjct: 7 WLTLVLIVVEANAVKQGKN----ATIPALIVFGDSIMDTGNNNNLPTLLKCNFPPYGKDY 62
Query: 68 PATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESG 125
P F T R+S+GR P D IA+ +G +P + N L D+LKGV +ASG G +
Sbjct: 63 PGGFATGRFSDGRVPSDLIAEKIGLAKTLPAYMNPYLKPEDLLKGVTFASGGTGYDPLT- 121
Query: 126 SQLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNV 185
+++ +++ QL++ + +S I G +KA L + V +ND Y
Sbjct: 122 AKIMSVISVWDQLIYFKEYISKIKRHFGE-EKAKDILEHSFFLVVSSSNDLAHTYLA--- 177
Query: 186 FNTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ Y YA L H+++ L+ GA+K + +GCV
Sbjct: 178 --QAHRYDRTSYANFLADSAVHFVRELHKLGAQKIGVFSAVPVGCV 221
>At1g59406 proline-rich protein, putative
Length = 349
Score = 116 bits (291), Expect = 2e-26
Identities = 75/225 (33%), Positives = 115/225 (50%), Gaps = 11/225 (4%)
Query: 9 LILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFP 68
L+ +LV + V + +T + +P + VFGDS+ D+GN+NNLPT K NF YG D+P
Sbjct: 6 LLFALVLIFVEA--NAATQGKNTTIPALIVFGDSIMDTGNNNNLPTLLKCNFPPYGKDYP 63
Query: 69 ATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGS 126
F T R+S+GR P D IA+ LG +P + N L D+LKGV +ASG G + +
Sbjct: 64 GGFATGRFSDGRVPSDLIAEKLGLAKTLPAYMNPYLKPEDLLKGVTFASGGTGYDPLT-A 122
Query: 127 QLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVF 186
++ +++ QL++ + +S I G +KA L + V +ND Y
Sbjct: 123 KIMSVISVWDQLINFKEYISKIKRHFGE-EKAKDILEHSFFLVVSSSNDLAHTYLA---- 177
Query: 187 NTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ Y YA L H+++ L+ GARK + +GCV
Sbjct: 178 -QTHRYDRTSYANFLADSAVHFVRELHKLGARKIGVFSAVPVGCV 221
>At1g59030 proline-rich protein, putative
Length = 349
Score = 116 bits (291), Expect = 2e-26
Identities = 75/225 (33%), Positives = 115/225 (50%), Gaps = 11/225 (4%)
Query: 9 LILSLVTVIVASIMQHSTVLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFP 68
L+ +LV + V + +T + +P + VFGDS+ D+GN+NNLPT K NF YG D+P
Sbjct: 6 LLFALVLIFVEA--NAATQGKNTTIPALIVFGDSIMDTGNNNNLPTLLKCNFPPYGKDYP 63
Query: 69 ATFPTERYSNGRNPIDKIAQLLGFQTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGS 126
F T R+S+GR P D IA+ LG +P + N L D+LKGV +ASG G + +
Sbjct: 64 GGFATGRFSDGRVPSDLIAEKLGLAKTLPAYMNPYLKPEDLLKGVTFASGGTGYDPLT-A 122
Query: 127 QLGHNVNLGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVF 186
++ +++ QL++ + +S I G +KA L + V +ND Y
Sbjct: 123 KIMSVISVWDQLINFKEYISKIKRHFGE-EKAKDILEHSFFLVVSSSNDLAHTYLA---- 177
Query: 187 NTSRMYTPKQYAKVLIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ Y YA L H+++ L+ GARK + +GCV
Sbjct: 178 -QTHRYDRTSYANFLADSAVHFVRELHKLGARKIGVFSAVPVGCV 221
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.336 0.145 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,981,448
Number of Sequences: 26719
Number of extensions: 280412
Number of successful extensions: 1180
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 882
Number of HSP's gapped (non-prelim): 114
length of query: 341
length of database: 11,318,596
effective HSP length: 100
effective length of query: 241
effective length of database: 8,646,696
effective search space: 2083853736
effective search space used: 2083853736
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0476b.4


