

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.3
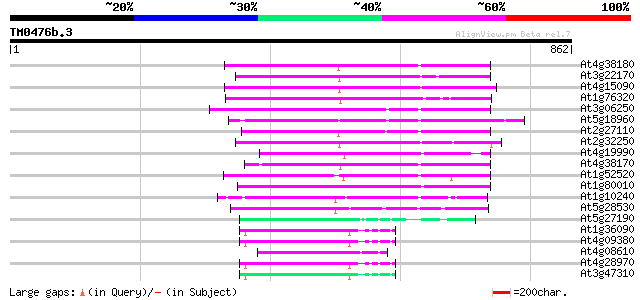
(862 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 238 1e-62
At3g22170 far-red impaired response protein, putative 214 2e-55
At4g15090 unknown protein 212 8e-55
At1g76320 putative phytochrome A signaling protein 210 3e-54
At3g06250 unknown protein 203 4e-52
At5g18960 FAR1 - like protein 197 3e-50
At2g27110 Mutator-like transposase 196 6e-50
At2g32250 Mutator-like transposase 176 4e-44
At4g19990 putative protein 172 6e-43
At4g38170 hypothetical protein 171 2e-42
At1g52520 F6D8.26 171 2e-42
At1g80010 hypothetical protein 169 8e-42
At1g10240 unknown protein 140 2e-33
At5g28530 far-red impaired response protein (FAR1) - like 117 4e-26
At5g27190 putative protein 83 6e-16
At1g36090 hypothetical protein 76 9e-14
At4g09380 putative protein 74 3e-13
At4g08610 predicted transposon protein 73 8e-13
At4g28970 putative protein 72 1e-12
At3g47310 putative protein 71 2e-12
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 238 bits (607), Expect = 1e-62
Identities = 127/415 (30%), Positives = 205/415 (48%), Gaps = 9/415 (2%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVS-DGRSALDYLASLGTKDPSMFVRHTEDADKNLEHL 389
G + + D +N + R+ + + + LDYL + +P+ F D+++ ++
Sbjct: 223 GISKVGFTEVDCRNYMRNNRQKSIEGEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNV 282
Query: 390 FWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEET 449
FW D M+++ FGD + FD TY+ N+YR P F+ VNH Q + G A I NE E +
Sbjct: 283 FWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEAS 342
Query: 450 YVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDN----- 504
+VWL L AM P ++ TD D +R AI VFP A HR C WH++ +
Sbjct: 343 FVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHVF 402
Query: 505 IKIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGK 564
+K P F F C+ + DF+R W SL++++ L ++ W+ +Y+ R W Y R
Sbjct: 403 LKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRDT 462
Query: 565 FFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGL 624
FFA T R + N+ ++ NL +F + + LE +EV+AD+ + P L
Sbjct: 463 FFADMSLTHRSDSINSYFDGYINASTNLSQFFKLYEKALESRLEKEVKADYDTMNSPPVL 522
Query: 625 KAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSRE 684
K P +E AS + TR+LF RF+ +V + + +D + + V + E+ +
Sbjct: 523 KTP---SPMEKQASELYTRKLFMRFQEELVGTLTFMASKADDDGDLVTYQVAKYGEAHKA 579
Query: 685 WLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
V + CSC E G+ C HI+AV N+ +P +L+RW++ AK
Sbjct: 580 HFVKFNVLEMRANCSCQMFEFSGIICRHILAVFRVTNLLTLPPYYILKRWTRNAK 634
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 214 bits (545), Expect = 2e-55
Identities = 117/399 (29%), Positives = 207/399 (51%), Gaps = 11/399 (2%)
Query: 347 GKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDA 406
G+ D + LD+L+ + + + + F D+ ++++FW D R NY F D
Sbjct: 225 GRTLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDV 284
Query: 407 LAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSP 466
++ D TY +NKY+ PL IF VN Q V+G A+I +E TY WL+E L A+ ++P
Sbjct: 285 VSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAP 344
Query: 467 TTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-----KIPDFLPKFKTCMFGD 521
+IT+ D M + + +FPN H L WH++ ++N+ + +F+PKF+ C++
Sbjct: 345 KVLITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKS 404
Query: 522 FQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQ 581
+ DF RKW + F L ++ W+I +Y R WA Y AG T+ R + NA
Sbjct: 405 GKDEDFARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAF 464
Query: 582 LGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRIL 641
K++ ++ +++EF++ + L+ E +AD P +K+P F E S S +
Sbjct: 465 FDKYMHKKTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPF---EKSVSEVY 521
Query: 642 TRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVK-RSQESSREWLVSYYTTTYEFRCSC 700
T +F +F+ ++ A + + E+ + T + + E++++++V++ T E C C
Sbjct: 522 TPAVFKKFQIEVLGA--IACSPREENRDATCSTFRVQDFENNQDFMVTWNQTKAEVSCIC 579
Query: 701 MRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
E G C H + VL ++ +IP +L+RW++ AK
Sbjct: 580 RLFEYKGYLCRHTLNVLQCCHLSSIPSQYILKRWTKDAK 618
>At4g15090 unknown protein
Length = 768
Score = 212 bits (539), Expect = 8e-55
Identities = 121/426 (28%), Positives = 209/426 (48%), Gaps = 12/426 (2%)
Query: 331 GYNNTQYILK-DLQNQVGKQRRAHVSDGRSA--LDYLASLGTKDPSMFVRHTEDADKNLE 387
GY N +L+ D+ +QV K R + +G S L+Y + ++P F + D+ L
Sbjct: 138 GYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLR 197
Query: 388 HLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKE 447
+LFW D R +Y F D ++FD TY K + PL +F VNH +Q ++G A++ +E
Sbjct: 198 NLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESM 257
Query: 448 ETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-- 505
ET+VWL++ L AM ++P ++TD DK + +A+ + PN H WH++ +
Sbjct: 258 ETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSH 317
Query: 506 ---KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFR 562
+ +FL KF C+F + +F +W +V +F L + W++ ++ R W +
Sbjct: 318 VMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMS 377
Query: 563 GKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDP 622
F AG T+ R E N+ K++ ++ LKEFL+ + L+ E ADF + + P
Sbjct: 378 DVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVADFDTCHKQP 437
Query: 623 GLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESS 682
LK+P + E + T +F +F+ ++ ++ M+ V + E
Sbjct: 438 ALKSPSPW---EKQMATTYTHTIFKKFQVEVLGVVACHPRKEKEDENMATFRV-QDCEKD 493
Query: 683 REWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHL 742
++LV++ T E C C E G C H + +L +IP +L+RW++ AK +
Sbjct: 494 DDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKDAKSGV 553
Query: 743 CETPGS 748
G+
Sbjct: 554 LAGEGA 559
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 210 bits (534), Expect = 3e-54
Identities = 123/416 (29%), Positives = 211/416 (50%), Gaps = 13/416 (3%)
Query: 332 YNNTQYILKDLQNQVGKQRRA--HVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHL 389
Y++ +I ++NQ K RR D L++L + ++P F D L ++
Sbjct: 136 YHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFLMRMQEENPKFFFAVDFSEDHLLRNV 195
Query: 390 FWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEET 449
FW D G +Y F D ++F+ +Y +KY+ PLV+F VNH Q ++G ++ ++ T
Sbjct: 196 FWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGVNHHVQPVLLGCGLLADDTVYT 255
Query: 450 YVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKI-- 507
YVWL++ L AM + P ++TD + A++ AI V P H C WH++ N+
Sbjct: 256 YVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPETRHCYCLWHVLDQLPRNLDYWS 315
Query: 508 ---PDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGK 564
F+ K C++ + +F R+W L+++F+L + PW+ +Y R WA + RG
Sbjct: 316 MWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDVPWMRSLYEERKFWAPTFMRGI 375
Query: 565 FFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGL 624
FAG R E N+ ++V +LKEFL+ + LE E +ADF + + P L
Sbjct: 376 TFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGLMLEDRYEEEAKADFDAWHEAPEL 435
Query: 625 KAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSRE 684
K+P F+ L + + ++F RF+ ++ AA +T +++ + ++VK + ++
Sbjct: 436 KSPSPFEKQML---LVYSHEIFRRFQLEVLGAAACHLT--KESEEGTTYSVK-DFDDEQK 489
Query: 685 WLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKD 740
+LV + + CSC E G C H I VL + IP + VL+RW+ A++
Sbjct: 490 YLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQMSGVFTIPINYVLQRWTNAARN 545
>At3g06250 unknown protein
Length = 764
Score = 203 bits (516), Expect = 4e-52
Identities = 117/438 (26%), Positives = 201/438 (45%), Gaps = 11/438 (2%)
Query: 307 EFDNSFEFGSNNCRRLQFTCCIGCGYNNTQYI-LKDLQNQVGKQRRAHVSDG--RSALDY 363
+ ++ E G N + T + G ++ I L DL N + R + LDY
Sbjct: 273 DHNHDLEPGKKNAGMKKITDDVTGGLDSVDLIELNDLSNHISSTRENTIGKEWYPVLLDY 332
Query: 364 LASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLV 423
S +D F D++ + +FW D R S FGDA+ FD +Y+K Y P
Sbjct: 333 FQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYRKGDYSVPFA 392
Query: 424 IFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRR 483
F NH Q ++GGA++ +E +E + WL + L AM + P +++ D D ++ A+ +
Sbjct: 393 TFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSMVADQDLPIQQAVAQ 452
Query: 484 VFPNASHRLCAWHLMCNATDNIKI--PDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLH 541
VFP HR AW + +N++ +F +++ C++ +F W SLV ++ L
Sbjct: 453 VFPGTHHRFSAWQIRSKERENLRSFPNEFKYEYEKCLYQSQTTVEFDTMWSSLVNKYGLR 512
Query: 542 ENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHH 601
+N W+ ++Y +R W AY R FF G + F G + +L+EF+ +
Sbjct: 513 DNMWLREIYEKREKWVPAYLRASFFGGIHVDGTFDPF---YGTSLNSLTSLREFISRYEQ 569
Query: 602 CLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMII 661
LE R E + DF S P L+ + +E R+ T +F F+ + + + +
Sbjct: 570 GLEQRREEERKEDFNSYNLQPFLQTK---EPVEEQCRRLYTLTIFRIFQSELAQSYNYLG 626
Query: 662 TNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLN 721
+ +S V++ + + V++ + CSC E GL C HI+ V L+
Sbjct: 627 LKTYEEGAISRFLVRKCGNENEKHAVTFSASNLNASCSCQMFEYEGLLCRHILKVFNLLD 686
Query: 722 IHNIPKSLVLERWSQGAK 739
I +P +L RW++ A+
Sbjct: 687 IRELPSRYILHRWTKNAE 704
>At5g18960 FAR1 - like protein
Length = 788
Score = 197 bits (500), Expect = 3e-50
Identities = 123/458 (26%), Positives = 213/458 (45%), Gaps = 19/458 (4%)
Query: 337 YILKDLQNQVGKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADK-NLEHLFWCDGI 395
+I K +N++GK+ + LDY S T+D F D + + +FW D
Sbjct: 335 HIKKTRENRIGKEWYPLL------LDYFQSRQTEDMGFFYAVELDVNNGSCMSIFWADSR 388
Query: 396 GRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLE 455
R S FGD++ FD +Y+K Y P NH Q ++G A++ +E +E ++WL +
Sbjct: 389 ARFACSQFGDSVVFDTSYRKGSYSVPFATIIGFNHHRQPVLLGCAMVADESKEAFLWLFQ 448
Query: 456 QLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP---DFLP 512
L AM + P +++ D D ++ A+ +VFP A HR AW + +N+ IP +F
Sbjct: 449 TWLRAMSGRRPRSIVADQDLPIQQALVQVFPGAHHRYSAWQIREKERENL-IPFPSEFKY 507
Query: 513 KFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTT 572
+++ C++ I +F W +L+ ++ L ++ W+ ++Y +R W AY R FFAG
Sbjct: 508 EYEKCIYQTQTIVEFDSVWSALINKYGLRDDVWLREIYEQRENWVPAYLRASFFAGIPIN 567
Query: 573 SRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQS 632
E F G + L+EF+ + LE R E + DF S P L+ +
Sbjct: 568 GTIEPF---FGASLDALTPLREFISRYEQALEQRREEERKEDFNSYNLQPFLQTK---EP 621
Query: 633 IELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTT 692
+E R+ T +F F+ +V + + + + +S V++ S + V++ +
Sbjct: 622 VEEQCRRLYTLTVFRIFQNELVQSYNYLCLKTYEEGAISRFLVRKCGNESEKHAVTFSAS 681
Query: 693 TYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCETPGSSPSL 752
CSC E GL C HI+ V L+I +P +L RW++ A+ S S
Sbjct: 682 NLNSSCSCQMFEHEGLLCRHILKVFNLLDIRELPSRYILHRWTKNAEFGFVRDMESGVSA 741
Query: 753 FRDSAVLVRYVCMLEACRKMCTLACPLIDDFRQTWEIV 790
A++V + EA K ++ ++ +EI+
Sbjct: 742 QDLKALMV--WSLREAASKYIEFGTSSLEKYKLAYEIM 777
>At2g27110 Mutator-like transposase
Length = 851
Score = 196 bits (497), Expect = 6e-50
Identities = 105/389 (26%), Positives = 186/389 (46%), Gaps = 10/389 (2%)
Query: 356 DGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKK 415
D + L+Y + ++P F D D + ++FW D R+ Y+ FGD + D Y+
Sbjct: 196 DAHNLLEYFKRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRC 255
Query: 416 NKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDK 475
N++R P F+ VNH Q + G A+I +E + +++WL + L AM + P +++TD D+
Sbjct: 256 NQFRVPFAPFTGVNHHGQAILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSLVTDQDR 315
Query: 476 AMRNAIRRVFPNASHRLCAWHLMCNATDN-----IKIPDFLPKFKTCMFGDFQIGDFKRK 530
A++ A +VFP A H + W ++ + + P F + C+ I +F+
Sbjct: 316 AIQIAAGQVFPGARHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESS 375
Query: 531 WESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRY 590
W S++++++L + W+ +Y R W YFR FFA + G + +V ++
Sbjct: 376 WSSVIDKYDLGRHEWLNSLYNARAQWVPVYFRDSFFAAVFPSQGYSG--SFFDGYVNQQT 433
Query: 591 NLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFR 650
L F + + +E E+EAD + P LK P +E A+ + TR++F +F+
Sbjct: 434 TLPMFFRLYERAMESWFEMEIEADLDTVNTPPVLKTP---SPMENQAANLFTRKIFGKFQ 490
Query: 651 PTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLAC 710
+V ED S V + ++ ++V++ CSC E G+ C
Sbjct: 491 EELVETFAHTANRIEDDGTTSTFRVANFENDNKAYIVTFCYPEMRANCSCQMFEHSGILC 550
Query: 711 AHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
H++ V NI +P +L RW++ AK
Sbjct: 551 RHVLTVFTVTNILTLPPHYILRRWTRNAK 579
>At2g32250 Mutator-like transposase
Length = 684
Score = 176 bits (447), Expect = 4e-44
Identities = 107/419 (25%), Positives = 184/419 (43%), Gaps = 14/419 (3%)
Query: 347 GKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDA 406
G Q D + L++ + K P F D+DK + ++FW D + +Y F D
Sbjct: 160 GLQLALEEEDLKLLLEHFMEMQDKQPGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDV 219
Query: 407 LAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSP 466
+ FD Y +N YR P F V+H Q ++G A+IG E TY WL L+A+ ++P
Sbjct: 220 VLFDTFYVRNGYRIPFAPFIGVSHHRQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAP 279
Query: 467 TTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-----KIPDFLPKFKTCMFGD 521
+ITD DK + + + VFP+ H C W ++ ++ + + F+ F C+
Sbjct: 280 GVMITDQDKLLSDIVVEVFPDVRHIFCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASS 339
Query: 522 FQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQ 581
+ F+R+W +++ +F L+EN W+ ++ R W YF G AG R +
Sbjct: 340 WTDEHFERRWSNMIGKFELNENEWVQLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASH 399
Query: 582 LGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRIL 641
K++ K+F + + L+Y E + D P L++ F E S I
Sbjct: 400 FDKYMNSEATFKDFFELYMKFLQYRCDVEAKDDLEYQSKQPTLRSSLAF---EKQLSLIY 456
Query: 642 TRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCM 701
T F +F+ + + + ++ ++ +E + + V+ + CSC
Sbjct: 457 TDAAFKKFQAEVPGVVSCQLQKEREDGTTAIFRIEDFEE-RQNFFVALNNELLDACCSCH 515
Query: 702 RMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGA-----KDHLCETPGSSPSLFRD 755
E G C H I VL ++ +P +L+RWS+ K+ C T + + F D
Sbjct: 516 LFEYQGFLCKHAILVLQSADVSRVPSQYILKRWSKKGNNKEDKNDKCATIDNRMARFDD 574
>At4g19990 putative protein
Length = 672
Score = 172 bits (437), Expect = 6e-43
Identities = 97/363 (26%), Positives = 178/363 (48%), Gaps = 24/363 (6%)
Query: 384 KNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVG-GAVI 442
++L ++FW D GR +Y+ F D ++ D T+ KN+Y+ PLV F+ VNH Q ++G G ++
Sbjct: 184 QSLRNIFWVDAKGRFDYTCFSDVVSIDTTFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLL 243
Query: 443 GNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNAT 502
+E + +VWL L+AM P ++T D+ ++ A+ VFP++ H W +
Sbjct: 244 TDESKSGFVWLFRAWLKAMHGCRPRVILTKHDQMLKEAVLEVFPSSRHCFYMWDTLGQMP 303
Query: 503 DNIKIPDFLPK-----FKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWA 557
+ + L K ++G Q DF++ W +V+ F++ +N W+ +Y R W
Sbjct: 304 EKLGHVIRLEKKLVDEINDAIYGSCQSEDFEKNWWEVVDRFHMRDNVWLQSLYEDREYWV 363
Query: 558 AAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRS 617
Y + AG T R + N+ L K+++R+ K FL+ + ++ E +++ +
Sbjct: 364 PVYMKDVSLAGMCTAQRSDSVNSGLDKYIQRKTTFKAFLEQYKKMIQERYEEEEKSEIET 423
Query: 618 GYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVK- 676
Y PGLK+P F + + TR++F +F+ ++ + G++ T +
Sbjct: 424 LYKQPGLKSPSPFGK---QMAEVYTREMFKKFQVEVLGGVACHPKKESEEDGVNKRTFRV 480
Query: 677 RSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQ 736
+ E +R ++V + + + E CSC E G +IP VL+RW++
Sbjct: 481 QDYEQNRSFVVVWNSESSEVVCSCRLFELKGEL--------------SIPSQYVLKRWTK 526
Query: 737 GAK 739
AK
Sbjct: 527 DAK 529
>At4g38170 hypothetical protein
Length = 531
Score = 171 bits (433), Expect = 2e-42
Identities = 99/385 (25%), Positives = 175/385 (44%), Gaps = 13/385 (3%)
Query: 361 LDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNK-YR 419
L+YL ++P +D ++FW D R+NY+ FGD L FD TY++ K Y+
Sbjct: 8 LNYLKRRQLENPGFLYAIEDDCG----NVFWADPTCRLNYTYFGDTLVFDTTYRRGKRYQ 63
Query: 420 RPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRN 479
P F+ NH Q + G A+I NE E ++ WL + L+AM P ++ + D+ ++
Sbjct: 64 VPFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQV 123
Query: 480 AIRRVFPNASHRLCAWHLMCNATDNIKI-----PDFLPKFKTCMFGDFQIGDFKRKWESL 534
A+ RVF R + + + P F +F C+ +F+ W+S+
Sbjct: 124 AVSRVFSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFEASWDSI 183
Query: 535 VEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKE 594
V + + +N W+ +Y R W + R F+ T N+ FV ++
Sbjct: 184 VRRYYMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDASTTMQM 243
Query: 595 FLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIV 654
++ + ++ R +E++AD+ + P +K P +E A+ + TR F +F+ V
Sbjct: 244 LIKQYEKAIDSWREKELKADYEATNSTPVMKTP---SPMEKQAASLYTRAAFIKFQEEFV 300
Query: 655 SAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHII 714
+ D+ + + V + E + VS+ + + CSC E G+ C HI+
Sbjct: 301 ETLAIPANIISDSGTHTTYRVAKFGEVHKGHTVSFDSLEVKANCSCQMFEYSGIICRHIL 360
Query: 715 AVLVDLNIHNIPKSLVLERWSQGAK 739
AV N+ +P +L RW++ AK
Sbjct: 361 AVFSAKNVLALPSRYLLRRWTKEAK 385
>At1g52520 F6D8.26
Length = 703
Score = 171 bits (432), Expect = 2e-42
Identities = 121/431 (28%), Positives = 185/431 (42%), Gaps = 30/431 (6%)
Query: 329 GCGYNNTQYILKDLQNQVGKQRRAHVSDGRSAL--DYLASLGTKDPSMFVRHTEDADKNL 386
G N + K QN G ++ G SA +Y + +P+ F + + L
Sbjct: 218 GSNVNPNSTLNKKFQNSTGSPDLLNLKRGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQL 277
Query: 387 EHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEK 446
++FW D +++ S FGD + D++Y K+ PLV F+ VNH +TT++ + E
Sbjct: 278 RNVFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGET 337
Query: 447 EETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIK 506
E+Y WLL+ L M K+SP T++TD K + AI +VFP + R H+M K
Sbjct: 338 MESYHWLLKVWLSVM-KRSPQTIVTDRCKPLEAAISQVFPRSHQRFSLTHIM------RK 390
Query: 507 IPDFL----------PKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTW 556
IP+ L F ++ ++ +F+ W +V F + EN W+ +Y R W
Sbjct: 391 IPEKLGGLHNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHNFGVIENEWLRSLYEERAKW 450
Query: 557 AAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFR 616
A Y + FFAG E ++V ++ LKEFL + L+ E +D
Sbjct: 451 APVYLKDTFFAGIAAAHPGETLKPFFERYVHKQTPLKEFLDKYELALQKKHREETLSDIE 510
Query: 617 S-GYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLG-MSMHT 674
S LK S E SRI TR +F +F+ + T G +
Sbjct: 511 SQTLNTAELKTK---CSFETQLSRIYTRDMFKKFQIEVEEMYSCFSTTQVHVDGPFVIFL 567
Query: 675 VKR------SQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKS 728
VK S+ R++ V Y + E RC C G C H + VL + IP
Sbjct: 568 VKERVRGESSRREIRDFEVLYNRSVGEVRCICSCFNFYGYLCRHALCVLNFNGVEEIPLR 627
Query: 729 LVLERWSQGAK 739
+L RW + K
Sbjct: 628 YILPRWRKDYK 638
>At1g80010 hypothetical protein
Length = 696
Score = 169 bits (427), Expect = 8e-42
Identities = 115/401 (28%), Positives = 181/401 (44%), Gaps = 14/401 (3%)
Query: 350 RRAHVSDGRSAL-DYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALA 408
RR + G AL D+ + P+ D +L ++FW D R YS FGD L
Sbjct: 235 RRLELRGGFRALQDFFFQIQLSSPNFLYLMDLADDGSLRNVFWIDARARAAYSHFGDVLL 294
Query: 409 FDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTT 468
FD T N Y PLV F +NH T ++G ++ ++ ETYVWL L M + P
Sbjct: 295 FDTTCLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCMLGRPPQI 354
Query: 469 VITDGDKAMRNAIRRVFPNASHRLCAWHLM---CNATDNIKIPDFLP-KFKTCMFGDFQI 524
IT+ KAMR A+ VFP A HRL H++ C + ++ D P ++G ++
Sbjct: 355 FITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQLQDSDLFPMALNRVVYGCLKV 414
Query: 525 GDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCE-GFNAQLG 583
+F+ WE ++ F + N I DM+ R WA Y + F AG T
Sbjct: 415 EEFETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFS 474
Query: 584 KFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTR 643
+V +L+EFL+ + L+ RE D S P LK ++S +++ T
Sbjct: 475 GYVHENTSLREFLEGYESFLDKKYTREALCDSESLKLIPKLKTTHPYES---QMAKVFTM 531
Query: 644 QLFFRFRPTIVSAADMI-ITNCEDTLGMSMHTVK-RSQESSREWLVSYYTT-TYEFRCSC 700
++F RF+ + + + +T S + VK R + R++ V Y T+ + RC C
Sbjct: 532 EIFRRFQDEVSAMSSCFGVTQVHSNGSASSYVVKEREGDKVRDFEVIYETSAAAQVRCFC 591
Query: 701 M--RMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
+ G C H++ +L + +P +L+RW + K
Sbjct: 592 VCGGFSFNGYQCRHVLLLLSHNGLQEVPPQYILQRWRKDVK 632
>At1g10240 unknown protein
Length = 680
Score = 140 bits (354), Expect = 2e-33
Identities = 105/424 (24%), Positives = 184/424 (42%), Gaps = 22/424 (5%)
Query: 320 RRLQFTCCIGCGYNNTQYILKDLQNQVGKQRRAHVSDGRSALDYLA---SLGTKDPSMFV 376
R L+ C+ G+ + KD++N + ++ D +D+L S+ KDP+
Sbjct: 192 RLLELEKCVEPGF--LPFTEKDVRNLLQSFKKLDPEDEN--IDFLRMCQSIKEKDPNFKF 247
Query: 377 RHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTV 436
T DA+ LE++ W +Y +FGDA+ FD T++ + PL I+ VN+
Sbjct: 248 EFTLDANDKLENIAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCF 307
Query: 437 VGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWH 496
G ++ +E ++ W L+ M+ K+P T++TD + ++ AI P H LC W
Sbjct: 308 FGCVLLRDENLRSWSWALQAFTGFMNGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWM 367
Query: 497 LM------CNATDNIKIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMY 550
++ NA + D+ +F ++ + +F+ W +V F LH N I ++Y
Sbjct: 368 VVGKFPSWFNAGLGERYNDWKAEFYR-LYHLESVEEFELGWRDMVNSFGLHTNRHINNLY 426
Query: 551 ARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHRE 610
A R W+ Y R F AG T R + NA + +F+ + L F++ +++
Sbjct: 427 ASRSLWSLPYLRSHFLAGMTLTGRSKAINAFIQRFLSAQTRLAHFVEQVAVVVDFKDQAT 486
Query: 611 VEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGM 670
+ + + LK +E A+ +LT F + + +V AA ++ +
Sbjct: 487 EQQTMQQNLQNISLKTG---APMESHAASVLTPFAFSKLQEQLVLAAHYASFQMDEGYLV 543
Query: 671 SMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLV 730
HT + R+ V + CSC E G C H + VL N +P +
Sbjct: 544 RHHT---KLDGGRK--VYWVPQEGIISCSCQLFEFSGFLCRHALRVLSTGNCFQVPDRYL 598
Query: 731 LERW 734
RW
Sbjct: 599 PLRW 602
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 117 bits (292), Expect = 4e-26
Identities = 95/406 (23%), Positives = 166/406 (40%), Gaps = 22/406 (5%)
Query: 340 KDLQNQVGKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMN 399
K +Q SD L+ L +D T D ++ +E++ W G
Sbjct: 241 KSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRG 300
Query: 400 YSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLE 459
YS+FGD + FD +Y+ Y L +F +++ + ++G ++ +E ++ W L+ +
Sbjct: 301 YSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFVR 360
Query: 460 AMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMC------NATDNIKIPDFLPK 513
M + P T++TD D +++AI R PN +H + H++ + T +F
Sbjct: 361 FMRGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKLASWFSQTLGSHYEEFRAG 420
Query: 514 F-KTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTT 572
F C G+ + +F+++W+ LV F L + +Y+ R +W R F A T+
Sbjct: 421 FDMLCRAGN--VDEFEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVAQTMTS 478
Query: 573 SRCEGFNAQLGKFVKRRYNLKEFLQHF--HHCLEYMRHREVEADFRSGYGDPGLKAPPIF 630
FN + F+KR + +Q L+ + + P LK
Sbjct: 479 E----FNLSIDSFLKRVVDGATCMQLLLEESALQVSAAASLAKQILPRFTYPSLKT---C 531
Query: 631 QSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYY 690
+E A ILT F + +V + + + V ++ E V +
Sbjct: 532 MPMEDHARGILTPYAFSVLQNEMVLSVQYAVAE----MANGPFIVHHYKKMEGECCVIWN 587
Query: 691 TTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQ 736
E +CSC E G+ C H + VL N +IP+ L RW Q
Sbjct: 588 PENEEIQCSCKEFEHSGILCRHTLRVLTVKNCFHIPEQYFLLRWRQ 633
>At5g27190 putative protein
Length = 779
Score = 83.2 bits (204), Expect = 6e-16
Identities = 85/370 (22%), Positives = 141/370 (37%), Gaps = 41/370 (11%)
Query: 353 HVSDGRSALD---YLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAF 409
H S +S D YL L +P DA + ++LF G + V +
Sbjct: 337 HCSPEKSYTDLYSYLHVLKEVNPGTISYVEVDAQQKFKYLFVALGACIEGFKVMRKVVIV 396
Query: 410 DATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTV 469
DAT+ K Y LV + + + ++ AVI E + ++ W +L +
Sbjct: 397 DATFLKTVYGGMLVFATAQDPNHHNYIIASAVIDRENDASWSWFFNKLKTVIPDVPGLVF 456
Query: 470 ITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKI--PDFLPKFKTCMFGDFQIGDF 527
++D +++ +I +VFPNA H C WHL N +K + F C Q +F
Sbjct: 457 VSDRHQSIIKSIMQVFPNARHGHCVWHLSQNVKVRVKTEKEEAAANFIACAHVYTQF-EF 515
Query: 528 KRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVK 587
R++ F + P+ +D +A WA YF G + T++ CE N+ +
Sbjct: 516 TREYARFRRRFP-NVGPY-LDRWAPVENWAMCYFEGDRY-NIDTSNACESLNSTFER--A 570
Query: 588 RRYNLKEFLQHFHHCLE--YMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQL 645
R+Y L L + + +HR+ +S++ S +R L +
Sbjct: 571 RKYFLLPLLDAIIEKISEWFNKHRK--------------------ESVKYSKTRGLVPVV 610
Query: 646 FFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMES 705
A M +T +++ +R E +SYY CSC R +
Sbjct: 611 ENEIHSLCPIAKTMPVT--------ELNSFERQYSVIGEGGISYYVDLLRKTCSCRRFDV 662
Query: 706 IGLACAHIIA 715
C H IA
Sbjct: 663 DKYPCVHAIA 672
>At1g36090 hypothetical protein
Length = 645
Score = 75.9 bits (185), Expect = 9e-14
Identities = 61/256 (23%), Positives = 106/256 (40%), Gaps = 29/256 (11%)
Query: 353 HVSDGRSA--------LDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFG 404
HVSD R YL L +P + +K ++LF G +
Sbjct: 309 HVSDVRGTPERSYTMLFSYLYMLEKVNPGTVTYVELEGEKKFKYLFIALGACIEGFRAMR 368
Query: 405 DALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKK 464
+ DAT+ K Y LVI + + + + +I +EK+ +++W LE+L
Sbjct: 369 KVIVVDATHLKTVYGGMLVIATAHDPNHHHYPLAFGIIDSEKDVSWIWFLEKLKTVYSDV 428
Query: 465 SPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKI--PDFLPKFKTCMFG-- 520
I+D ++++ A++ V+PNA H C WHL N D +KI KF+ C
Sbjct: 429 PRLVFISDRHQSIKKAVKTVYPNALHAACIWHLCQNMRDRVKIDKDGAAVKFRDCAHAYT 488
Query: 521 ----DFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCE 576
+ + G F W V+ +++ + + W+ +F+G + T++ E
Sbjct: 489 ESEFEKEFGHFTSLWPKAVD--------FLVKVGFEK--WSRCHFKGDKY-NIDTSNSAE 537
Query: 577 GFNAQLGKFVKRRYNL 592
N K R+Y+L
Sbjct: 538 SINGVFKK--ARKYHL 551
>At4g09380 putative protein
Length = 960
Score = 74.3 bits (181), Expect = 3e-13
Identities = 60/256 (23%), Positives = 104/256 (40%), Gaps = 29/256 (11%)
Query: 353 HVSDGRSA--------LDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFG 404
HVSD R YL L +P + +K ++LF G +
Sbjct: 349 HVSDVRGTPERSYTMLFSYLYMLEKVNPGTVTYVELEGEKKFKYLFIALGACIEGFRAMR 408
Query: 405 DALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKK 464
+ DAT+ K Y LVI + + + + +I +EK+ +++W LE L
Sbjct: 409 KVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLENLKTVYSDV 468
Query: 465 SPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKI--PDFLPKFKTCMFG-- 520
I+D ++++ A++ V+PNA H C WHL N D +KI KF+ C
Sbjct: 469 PGLVFISDRHQSIKKAVKTVYPNALHAACIWHLCQNMRDRVKIDKDGAAVKFRDCAHAYT 528
Query: 521 ----DFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCE 576
+ + G F W + +++ + + W+ +F+G + T++ E
Sbjct: 529 ESEFEKEFGHFTSLWPKAAD--------FLVKVGFEK--WSRCHFKGDKY-NIDTSNSAE 577
Query: 577 GFNAQLGKFVKRRYNL 592
N K R+Y+L
Sbjct: 578 SINGVFKK--ARKYHL 591
>At4g08610 predicted transposon protein
Length = 907
Score = 72.8 bits (177), Expect = 8e-13
Identities = 50/202 (24%), Positives = 86/202 (41%), Gaps = 6/202 (2%)
Query: 381 DADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGA 440
DA + ++LF G+ + V + DAT+ K Y LV + + + ++ A
Sbjct: 387 DAQQKFKYLFVSLGVCIEGFKVMRKVVIVDATFLKTVYGDMLVFATAQDPNHHNYIIASA 446
Query: 441 VIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCN 500
VI E + ++ W +L + + ++D +++ +I VFPNA H C WHL N
Sbjct: 447 VIDRENDASWSWFFNKLKTVIPDELGLVFVSDRHQSIIKSIMHVFPNARHGHCVWHLSQN 506
Query: 501 ATDNIKI--PDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAA 558
+K + F C Q +F R++ F +D++A WA
Sbjct: 507 VKVRVKTEKEEAAANFIACAHVYTQF-EFTREYARFRRRF--PNAGTYLDIWAPVENWAM 563
Query: 559 AYFRGKFFAGFRTTSRCEGFNA 580
YF G + T++ CE N+
Sbjct: 564 CYFEGDRY-NIDTSNVCESLNS 584
>At4g28970 putative protein
Length = 914
Score = 72.4 bits (176), Expect = 1e-12
Identities = 59/256 (23%), Positives = 104/256 (40%), Gaps = 29/256 (11%)
Query: 353 HVSDGRSA--------LDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFG 404
HVSD R YL L +P + +K ++LF G +
Sbjct: 349 HVSDVRGTPERSYTMLFSYLYMLEKVNPGTVTYVELEGEKKFKYLFIALGACIEGFRAMR 408
Query: 405 DALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKK 464
+ DAT+ K Y LVI + + + + +I +EK+ +++W LE+L
Sbjct: 409 KVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLEKLKTVYSDV 468
Query: 465 SPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKI--PDFLPKFKTCMFG-- 520
I+D ++++ A++ V+PNA H C WHL N D + I KF+ C
Sbjct: 469 PGLVFISDRHQSIKKAVKTVYPNALHAACIWHLCQNMRDRVTIDKDGAAVKFRDCAHAYT 528
Query: 521 ----DFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCE 576
+ + G F W + +++ + + W+ +F+G + T++ E
Sbjct: 529 ESEFEKEFGHFTSLWPKAAD--------FLVKVGFEK--WSRCHFKGDKY-NIDTSNSAE 577
Query: 577 GFNAQLGKFVKRRYNL 592
N K R+Y+L
Sbjct: 578 SINGVFKK--ARKYHL 591
>At3g47310 putative protein
Length = 735
Score = 71.2 bits (173), Expect = 2e-12
Identities = 58/256 (22%), Positives = 103/256 (39%), Gaps = 29/256 (11%)
Query: 353 HVSDGRSA--------LDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFG 404
HVSD R YL L +P + +K ++LF G +
Sbjct: 298 HVSDVRGTPERSYTMLFSYLYMLEKVNPGTVTYVELEGEKKFKYLFIALGACIEGFRTMR 357
Query: 405 DALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKK 464
+ DAT+ K Y LVI + + + + +I +E + +++W LE+L
Sbjct: 358 KVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAFGIIDSENDVSWIWFLEKLKTVYSDV 417
Query: 465 SPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKI--PDFLPKFKTCMFG-- 520
I+D ++++ ++ V+PNA H C WHL N D +KI KF+ C
Sbjct: 418 PGLVFISDRHQSIKKVVKTVYPNALHAACIWHLCQNMRDRVKIDKDGAAVKFRDCAHAYT 477
Query: 521 ----DFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCE 576
+ + G F W + +++ + + W+ +F+G + T++ E
Sbjct: 478 ESEFEKEFGHFTSLWPKAAD--------FLVKVGFEK--WSRCHFKGDKY-NIDTSNSAE 526
Query: 577 GFNAQLGKFVKRRYNL 592
N K R+Y+L
Sbjct: 527 SINGVFKK--ARKYHL 540
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.351 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,300,126
Number of Sequences: 26719
Number of extensions: 768611
Number of successful extensions: 3379
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 76
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 3159
Number of HSP's gapped (non-prelim): 189
length of query: 862
length of database: 11,318,596
effective HSP length: 108
effective length of query: 754
effective length of database: 8,432,944
effective search space: 6358439776
effective search space used: 6358439776
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0476b.3


